Abstract
We have recently shown that an MCF-7 tumor can be imaged in a mouse by PET with 64Cu-labeled Peptide nucleic acids (PNAs) tethered to the permeation peptide Lys4 that recognize the uniquely overexpressed and very abundant upstream of N-ras or N-ras related gene (unr mRNA) expressed in these cells. Herein we describe how the high affinity antisense PNAs to the unr mRNA were identified and characterized. First, antisense binding sites on the unr mRNA were mapped by an reverse transcriptase random oligonucleotide library (RT-ROL) method that we have improved, and by a serial analysis of antisense binding sites (SAABS) method that we have developed which is similar to another recently described method. The relative binding affinities of oligodeoxynucleotides (ODNs) complementary to the antisense binding sites were then qualitatively ranked by a new Dynabead-based dot blot assay. Dissociation constants for a subset of the ODNs were determined by a new Dynabead-based solution assay and were found to be 300 pM for the best binders in 1 M salt. PNAs corresponding to the ODNs with the highest affinities were synthesized with an N-terminal CysTyr and C-terminal Lys4 sequence. Dissociation constants of these hybrid PNAs were determined by the Dynabead-based solution assay to be about 10 pM for the highest affinity binders.
INTRODUCTION
One general approach to targeting cancer cells with drugs and probes make use of a cancer cell-specific or overexpressed surface receptor that can bind to a ligand and internalize the appended drug or probe (1–6). Another approach makes use of antibody drug or probe conjugates to recognize a cancer cell-specific or overexpressed surface protein (7,8). Other approaches involve the use of enzymes overexpressed by a cancer cell to selectively activate a prodrug or probe (9). In all these approaches a cancer cell-specific or overexpressed protein or enzyme has to be identified and an appropriate ligand, antibody or substrate developed to recognize it, necessitating a great deal of research time and expense. Furthermore, patient-specific variability and spontaneous mutations of the target proteins during treatment could compromise the effectiveness of a given probe or drug conjugate.
Cancer cells that have unique or overexpressed proteins, also have unique or overexpressed mRNA precursors. Unlike designing a ligand for a protein, designing ligands that can recognize an overexpressed mRNA is straightforward in principle, requiring nothing more than a Watson–Crick complementary antisense oligodeoxynucleotide (ODN) or analog. Such antisense-based agents could be readily be made patient-specific and could easily be adapted to deal with spontaneous mutations during the course of treatment. There are, however, a number of considerations that must still be taken into account in order to make use of an antisense strategy for targeting probes and drugs to a cell.
The first consideration is that messenger RNA molecules are inside a cell, and before an antisense ODN or analog can recognize and bind to the mRNA it must be able to enter the cell. Recently, a class of peptide sequences, known as protein transduction domains, or permeation peptides have been identified that enable a wide variety of attached molecules to enter cells, including ODNs and analogs (10–12). As a result, there have been a number of reports of attempts to image cells by way of antisense ODNs or analogs attached to permeation peptides, but often the selectivity is poor and/or the signal is weak (13–18). The limited success of this approach to date may be due to a number of factors, among which are: targeting an mRNA of low abundance, low binding affinity of the antisense agent to the target sequence due to its inaccessibility and/or conformation (19), and the use of cleavable linkers between the antisense agent and the permeation peptide which would prevent the antisense agent from exiting cells deficient in the targeting mRNA.
In principle, much better cell discrimination with antisense Peptide nucleic acids (PNAs) could be achieved by targeting high affinity binding sites on a uniquely overexpressed mRNA that is also very abundant. In addition, the antisense agent should be irreversibly linked to the permeation peptide to allow it to equilibrate with the extracellular media and selectively concentrate in cells having high concentrations of the mRNA. Recently, we showed that we could image an MCF-7 tumor in a mouse by PET with 64Cu-labeled PNAs bearing a Lys4 permeation peptide, and that the best image was obtained with an antisense PNA for the upstream of N-ras or N-ras related gene (unr mRNA) that is highly abundant and uniquely overexpressed in that tumor type (20). Antisense PNAs for the unr mRNA could also be used as part of a nucleic acid triggered prodrug and probe activation (NATPA) system that we have been developing (21–25). In this paper, we describe the general methodologies that we used to map and characterize high affinity antisense ODN and PNA binding sites on the unr mRNA that were used in the design of the PET MCF-7 imaging agents.
MATERIALS AND METHODS
General
Image clone 5285557 containing the unr mRNA sequence (D1S155E, NM_007158.2, GI: 20070240 and ATTC clone #7020864) was obtained from ATTC. pZErO-1 vector used for serial analysis of antisense binding sites (SAABS) analysis was from Invitrogen (Carlsbad, CA). Streptavidin coated Dynabeads were Dynabead M-280 Streptavidin from Dynal Biotech (Brown Deer, WI). Diisopropylethylamine (DIPEA), trifluoroacetic acid (TFA), m-cresol and diethyl ether (anhydride) were purchased from Aldrich. α-N-9-Fluorenylmethoxycarbonyl (Fmoc) protected amino acids (Cys(Trt)-OH, Tyr(tBu)-OH and Lys(Boc)-OH) were purchased from NovaBiochem. O-(7-Azabenzo-triazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU), Fmoc-XAL PEG-PS resin, PNA building blocks (Fmoc-A-(Bhoc)-OH, Fmoc-C-(Bhoc)-OH, Fmoc-G-(Bhoc)-OH and Fmoc-T-OH) and other reagents and solvents for PNA and peptide synthesis were purchased from PerSeptive Biosystems. UV spectral data were acquired on a Bausch and Lomb Spectronic 1001 spectrophotometer or Varian Cary 100 Bio UV-Visible Spectrophotometer. Matrix-assisted laser desorption ionization (MALDI) mass spectra of PNA-peptide conjugates were measured on a PerSeptive Voyager RP MALDI-time of flight (TOF) mass spectrometer using sinapinic acid as a matrix and calibrated versus insulin (average [M + H+] = 5734.5) that was present as an internal standard. High-pressure liquid chromatography (HPLC) of PNAs was carried out on Beckman Coulter System Gold 126 HPLC system. ODN and PNA synthesis was carried out on an automated ABI 8909 synthesizer (Applied Biosystems) with a PNA option.
Substrate preparation
Human unr mRNA
Transcription reactions were carried out using the Promega RiboMAX Large Scale RNA production system utilizing the T7 promoter at the 5′ end of the unr gene in a pBluescriptR plasmid (IMAGE clone 5285557) and linearizing the plasmid at the unique NdeI site at the 3′ end of the gene before transcription. Transcription was allowed to proceed with T7 RNA polymerase and NTPs for 3 h at 37°C, and was followed by DNase I treatment, phenol:chloroform:isoamylalcohol (25:24:1) extraction and ethanol precipitation. NTPs were then removed with a NucAway spin column (Ambion, Austin TX). The final purified unr RNA was quite homogeneous and corresponded in size to the expected 3.4 Kb transcript. For the SAABS and Dynabead-based-assays, the unr mRNA was transcribed in the presence of Bio-UTP.
ODNs
ODNs were either obtained from Integrated DNA Technologies (IDT, Coralville, IA) or were synthesized by standard solid phase phosphoramidite synthesis. Random sequences were synthesized using a mixture of phosphoramidites in a ratio of 1.5:1.25:1.15:1.0 (A:C:G:T). All ODNs were purified by 15 or 20% denaturing polyacrylamide gels before and after any 32P-radiolabeling. The reverse transcriptase primer used in the reverse transcriptase random oligodeoxynucleotide library (RT-ROL) assay (XN9-tag) was: d(GGATTTGCTGGTGCAGTACANNNNNNNNNX). The primers used in the PCR amplification step of the RT-ROL assay were: unr1: d(GCTGAGCTGTTGGGTATGAAG), unr2: d(TCATCCTTTGAAACGTGTGC), unr3: d(ACGAACGTAATGGGGAAGTG); unr4: d(AAATCCAAGGTGACCCTGCT); unr5: d(TGACTGTGGGGTGAAACTGA); unr6: d(GAGGGCGATATGAAAGGTGA); unr7: d(AACCACATCCACAAAGCACA). The ODNs used in the SAABS assay were: S1: d(GGATTTGCTGGTGCAACATG); CS1: d(CATGTTGCACCAGCAAATCC); CS2: d(CCTCGAATTCAAGCTTCATG); S1-ROL-S2: d(GGATTTGCTGGTGCAACATGN8CATGAAGCTTGAATTCGAGG).
PNA-peptide conjugates
The hybrid PNA-peptides were synthesized with a capping step in 2 µmol scale using Fmoc chemistry and the manufacturer's protocols, reagents and PNA monomers. After completion of automated synthesis, PNAs were cleaved from the solid support and the bases were deprotected using trifluoroacetic acid:m-cresol (4:1) for 2 h and then precipitated with diethyl ether. The hybrid PNA-peptides were purified by reverse phase chromatography on a Microsorb-MV 300-5 C-18 column, (250 × 4.6 mm column, 300 Å pore size, Varian Inc.) with a 0–5% B/5 min, 5–60% B/30 min, 60–95% B/5 min, 95% B for 5 min, 95–0%/10 min, where A = 0.1% TFA in water, B = 0.08% TFA in CH3CN. The fractions containing the products were evaporated to dryness in a Savant Speedvac, and redissolved in pure water. Peptide-PNA conjugates were quantified by spectrophotometric A260 values and characterized by MALDI-TOF (Table 2).
Table 2.
Binding affinity of Cys-Tyr-PNA-Lys4 by the Dynabead-based solution binding assay
| Entry | Site | Code | Calcd M + 1a | Obsvd M + 1a | PNA sequence | Kd (pM) |
|---|---|---|---|---|---|---|
| 1 | 899 | S3-2 | 5571 | 5576 | TATCTCCAGTTTCCAGCT | 6 ± 3 |
| 2 | 673 | 7-2 | 5588 | 5594 | TTTCCCAGTCCGTCGGTC | 12 ± 4 |
| 3 | 476 | 5-2 | 5641 | 5646 | CATTATGTCCATTGTTGT | 17 ± 6 |
| 4 | 2848 | 50-2 | 5779 | 5784 | TGGTGTGCTTTGTGGATG | 20 ± 7 |
| 5 | 1414 | S5-2 | 5651 | 5659 | TAATTTGTCACGTCGGTC | 49 ± 6 |
| 6 | 1927 | 29 | 5592 | 5594 | CCTCTGTTTGTCACTAAT | 300 ± 30 |
| 7 | 2020 | 32 | 5603 | 5604 | CCAAAATTATCCTTCAGA | 380 ± 60 |
| 8 | 2114 | 35 | 5587 | 5591 | TGTCCCCCAGTTCCAGGC | 680 ± 100 |
| 9 | 1389 | 20 | 5658 | 5660 | TATTAAACCTAACATGGT | 820 ± 190 |
| 10 | 1263 | 1263 | 5653 | 5655 | TTTTTACTGGGTACTTTT | 340 ± 60 |
| 11 | 1012 | 1012 | 5604 | 5608 | CTTCATGGCACAAACTAC | 650 ± 170 |
| 12 | 1526 | 1526 | 5637 | 5640 | CACACTTGATGAAACCAA | 1000 ± 310 |
| 13 | 1653 | 1653 | 5680 | 5683 | ATAGCATGATTTCTTTGA | 1000 ± 190 |
| 14 | nab | 50-2S | 5562 | 5568 | CATCCACAAAGCACACCA | >10 000 |
The site refers to the position in the mRNA that is complementary to the 3′ end of the PNA sequence shown (the start codon is at 448). The code number X-Y is the same as that for the corresponding ODN listed in Table 1.
aCalculated MW is for the average M + 1 ion, observed MW is for the M + 1 ion detected by MALDI-TOF.
bnot applicable.
Radioiodination of PNA-peptide conjugates
The PNA-peptides were radioiodinated by a procedure described for iodination of PNAs with 125I (13). Thus Na131I (12 µl, 5 mCi/120 µl solution from Amersham) was added to 2 µl of water and 2 µl of 1 M sodium phosphate (pH 7), then 2 µl of 100 µM NaI was added, followed by 4 µl of 1 mM chloramine-T and 4 µl of 100 µM PNA. The reaction was mixed by micropipetting, and allowed to stand for 5 min at room temperature and quenched with 4 µl of 10 mM sodium metabisulfite. The reaction mixture was then diluted with 80 µl of water and placed on to a Waters Sep-Pak cartridge (Vac C18, 6 cc, Part #WAT036905) that had been prewashed with 5 ml of 0.1% TFA in water. The unreacted Na131I (average of 27% of the total radioactivity) was washed out with 10 ml of 0.1% TFA in water. This was followed by 5–10 ml of 5% acetonitrile in 0.1% TFA/water, which contained little radioactivity. The radiolabeled PNA-peptide (average of 42% of the radioactivity) was eluted with 5 ml of 40% CH3CN in 0.1% TFA/water, leaving an average of 31% of the total radioactivity on the column. The average recovery of the labeled PNA-peptide was 57%. An aliquot of the fraction containing the labeled PNA-peptide was then diluted down to make a 100 µM stock solution. The actual concentration was then determined based on the activity of the solution in comparison to the 5 ml stock solution.
Mapping assays
Modified RT-ROL mapping assay
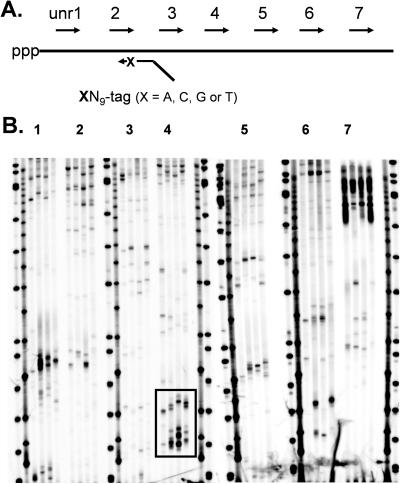
The ODNs used for mapping the antisense binding sites on the unr mRNA by the modified RT-ROL assay are shown in Figure 2. The mapping was carried out by heating the RNA at 65°C for 5 min with dNTPs followed by a quickly chilling on ice for 1 min and then adding the random ODN library primer XN9-tag, RNasIn (Promega, Madison WI) RNase inhibitor and SuperScript II reverse transcriptase (Invitrogen Life Technologies) for another 5 min. The reaction was incubated at 42°C for 50 min, and the enzyme inactivated by heating at 70°C for 15 min. Then 5′-endlabeled PCR primer, dNTPs and Taq polymerase were added and subjected to an initial incubation at 94°C for 3 min, followed by 30 cycles of PCR (1 min 94°C, 1 min 55°C and 1 min 72°C). The products were then separated on 6% denaturing polyacrylamide gels.
Figure 2.
(A) Primers used to map ODN accessible sites on unr transcript. Sequences of unrX is given in the experimental section. (B) Phosphorimage of polyacrylamide gels of the PCR products for each section (X = 1–7) of the unr transcript. Boxed site is analyzed in Figure 3.
SAABS assay
Streptavidin coated Dynabeads were first washed with solution A (DEPC-treated 0.1 M NaOH and DEPC-treated 0.05 M NaCl), followed by three washes with solution B (DEPC-treated 0.1 M NaCl), and then washed two times with 100 µl of hybridization solution [1M NaCl and Tris–HCl (pH 7.4)]. The beads were then resuspended in 50 µl of hybridization solution. The biotinylated RNA (3 µl of 3 µg/µl) was incubated with 45 µl of hybridization solution at 65°C for 5 min, and then cooled at room temperature for 10 min. The ODN containing the random 8mer (S1-ROL-S2) was then annealed with the complementary ODNs CS1 and CS2 in hybridization buffer (50 µl) by heating at 65°C for 5 min, and then cooled to room temperature for 10 min. The RNA solution was then mixed with S1-ROL-S2•CS1•CS2 together with 2 µl RNasIN, and incubated at 27°C for 1 h. The solution of beads was then added and resuspended every 5 min by pipetting for a total of 15 min. Following the incubation, the beads were separated with a magnet and washed 6 times with the hybridization solution and then resuspended in 100 µl of H2O. Thirty cycles of PCR were then carried out on the beads with Taq DNA polymerase, and S1 and CS2 primers. After amplification, the reaction mix was directly loaded on to a 12% denaturing PAGE gel and electrophoresed to separate the 48 bp PCR product. The 48 bp PCR band was excised and the eluted product digested with NlaIII at 37°C overnight to release the 12mer product from the tags which was purified by 20% denaturing PAGE gel. The 12mer products were ligated with T4 DNA ligase and ATP at 16°C for 2 h. Concatemers were isolated using the Qiaquick Gel Extraction Kit (Qiagen, Valencia, CA), following the manufacturer's manual. The mixture was then electrophoresed on a 1% agarose gel (TAE) and fractions with 300–500 bp length were excised. Concatemers were ligated into the SphI site of pZErO-1 (Invitrogen) with T4 DNA ligase and ATP for 4 h at 16°C and then transfected into Escherichia coli, following the manufacturer's manual (Oneshot® Top10, Invitrogen). The transfectants were plated on low salt Luria–Bertani (LB) plates containing 50 µg/ml Zeocin™ (Invivogen, San Diego, CA) and incubated for about 18 h at 37°C. Zeocin™ resistant transformants were picked using pipette tips, incubated in 5 ml SOB containing 50 µl/ml Zeocin™ and grown overnight at 37°C. Plasmid DNA was prepared by the Plasmid mini prep kit (Qiagen). Plasmids containing sizable inserts were then forwarded for sequence analysis. The sequences were analyzed by a computer program written expressly for this purpose. The program first extracts the 8mer sequences from between Nla III sites, and then scans for a perfect match between either the sense or antisense 8mer sequences and the mRNA sequence. If either the sense or antisense 8mer sequence matches uniquely, each matching base in the mRNA is assigned one point. If the sequence matches n different sites, each matching base at each site is assigned 1/n points. This sequence of steps is repeated for sequences only matching 6–7 bases.
Binding assays
Dynabead-based dot blot assay
Strepavidin-coated Dynabeads were washed following the manufacturer's protocol, and then washed two times with hybridization buffer [1 M NaCl, 0.5 mM EDTA and 20 mM sodium cacodylate (pH 7.0)]. After the last wash, the beads were resuspended in hybridization buffer in the presence of biotinylated unr mRNA, incubated for 30 min at 37°C, and mixed occasionally by flicking the tube with a finger. The tube containing the beads was then placed on a magnetic stand for 1 min, following which the supernatant was removed and discarded. The beads were then washed three times with hybridization buffer and then resuspended in hybridization buffer in the presence of 1 pmol of 32P-labeled ODN, incubated for 30 min at 37°C with periodic agitation by flicking. After washing three times as described above, the beads were resuspended in 20 µl hybridization buffer, and 2 µl was spotted on a Nylon membrane.
Dynabead-based ODN binding assay
The radiolabeled ODN (100 pM) was incubated with the 0.003–10 nM of biotinylated mRNA and 1 µl of RNasIN inhibitor for 4 h at 37°C in a total volume of 100 µl. Then streptavidin coated Dynabeads were added and mixed for 30 min. The beads were then separated by a magnet and the solution removed. The beads were resuspended in 100 µl of water and both solutions counted by liquid scintillation to give free and bound fractions directly.
Dynabead-based PNA binding assay
The binding assay was carried out as described above for the ODNs in Bio-Rad EZ Micro test tubes in triplicate. Specifically, the radiolabeled PNA (13–26 pM) was incubated with 0.1–1000 pM of biotinylated mRNA and 1 µl of RNasIN inhibitor for 4 h at 37°C in a total volume of 100 µl of 0.1 M NaCl, 50 mM EDTA and 2 mM cacodylic acid. Then 5 µl of streptavidin coated Dynabeads were added and mixed for 2 h. The beads were separated by a magnet and the supernatant transferred to a liquid scintillation vial. The beads were washed with 100 µl buffer three times and the washes transferred to the first scintillation vial. The beads were then transferred to a liquid scintillation vial by suspending three times in 100 µl of buffer. Liquid scintillation fluid (5 ml of CytoScint plus) was then added and both scintillation vials counted by liquid scintillation to give free and bound fractions directly.
Binding data analysis
The dissociation constants were determined by non-linear least squares fitting of the fraction bound versus RNA concentration data to the analytical expression shown below for a two state binding equilibrium using the Kalaidagraph program.
FB is the experimentally determined fraction of bound ODN or PNA ligand L, NSB = fraction of ligand L that is non-specifically bound (fitted parameter), SB is the fraction of ligand that can be specifically bound and was set equal to (1−NSB) when the ligand was an ODN, [L] = total ligand concentration, Kd = dissociation constant of the ligand (fitted parameter) and [RNA] = total RNA concentration. A lower limit of the binding constant could only be estimated for incomplete binding curves. For the PNA binding analysis, three sets of data were averaged and weighted according to their standard deviations and SB was also fitted to correct for the presence radioactive material that appeared to show no binding affinity for the RNA (the curves plateaued at values below 100%).
RESULTS AND DISCUSSION
Selection of a cancer-specific overexpressed mRNA
To validate our antisense cell targeting approach, we searched the NCBI serial analysis of gene expression (SAGE) database (http://www.ncbi.nlm.nih.gov/SAGE/) for abundant mRNAs that are >10-fold overexpressed in standard cancer cell lines compared to any normal cell lines. In doing so we discovered that the well studied MCF-7 breast cancer cell line contains an almost 10-fold higher amount of the unr (upstream of N-RAS) mRNA (GI: 41282241) (26,27), than normal cell lines, and is present at a level of about 5000–6000 tags/million, or roughly the same number of copies per cell. We found a full length cDNA clone from the I.M.A.G.E. consortium (5285557 from the NIH_MGC_96 library, UniGene Library 6001), and obtained the pBluescriptR vector containing the cDNA through the American Tissue Culture Collection (ATCC), #7020864). We were able to produce the 3.4 kB unr mRNA in high yield and homogeneity by in vitro transcription with T7 RNA polymerase.
Modified RT-ROL assay for identifying antisense binding sites
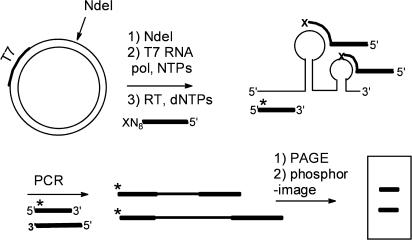
Because mRNA is folded in vivo, much of its sequence is inaccessible to an antisense ODN either because of intramolecular base pairing to a complementary RNA sequence within the mRNA, or because it is buried within the three dimensional structure and physically inaccessible (28). It is also likely that many otherwise accessible sites are also blocked by binding to proteins in vivo. A number of chemical, physical and enzymatic techniques have been developed to map the accessible sites on short folded mRNAs, but most are unsuitable for mapping sites on long mRNA sequences, either due to electrophoretic resolution problems or the expense associated with making the required custom DNA arrays (29). To identify the sites on the unr mRNA that could potentially bind PNAs in vivo, we adopted and improved upon a recently reported length-independent RT-ROL method. This method uses a random oligonucleotide library attached to a PCR tag to prime RT followed by PCR amplification with primers corresponding to the PCR tag and different sections of the target mRNA (30).
In the original method, the priming end was a completely random sequence, making it difficult to assign the PCR products to a specific-site on the mRNA because of the difficulty in determining the precise length of the PCR product. To improve the precision of the method we synthesized a set of four separate random 9mer primers terminating in either A, C, G or T (Figure 1) thereby making it much easier to assign the PCR product bands to specific sequences by the pattern of bands produced. The entire unr mRNA was mapped in this way using 7 forward primers (unrX) for each section of the transcript and the PCR tag on the random ODN library (Figure 2A). The pattern of PCR bands formed (Figure 2B) were analyzed in comparison to the mRNA sequence to reveal the ODN binding sites (Figure 3). Approximately 50 antisense sites on the unr mRNA were identified by this RT-ROL assay.
Figure 1.
Modified RT-ROL assay. In this assay antisense binding sites are identified by the ability of a random 9mer oligodeoxynucleotide library (ROL) terminating a specific base and a PCR tag to prime complementary DNA (cDNA) synthesis by reverse transcriptase (RT). The cDNA is then amplified by PCR with a radiolabeled primer having the same sense as the RNA, and an unlabelled primer having the same sequence as the PCR tag.
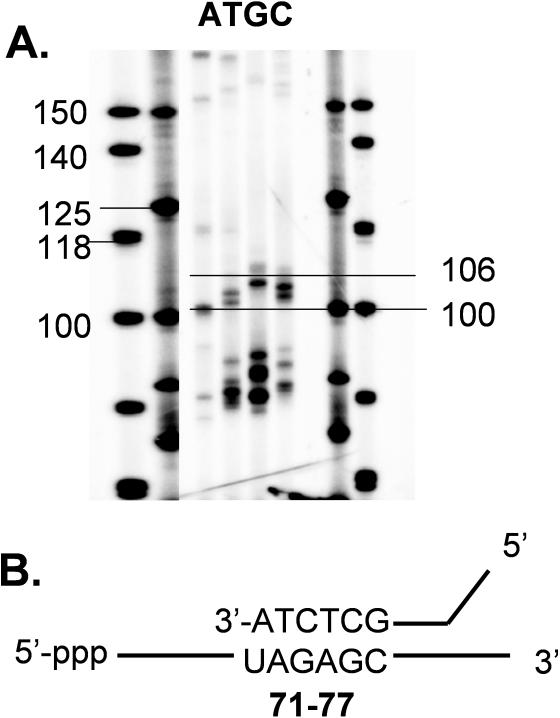
Figure 3.
Interpretation of the RT-ROL gel. (A) Blowup of boxed section of gel in Figure 2 showing the approximate alignment of the bands with standard length markers. (B) Alignment of the observed bands with the mRNA sequence, taking into account the sequence of the mRNA and the terminal nucleotide on the primer, along with the fact that the PCR tag adds an additional 20 nt to the extra 9 random nucleotides in the PCR amplified product band.
Serial analysis of antisense binding sites
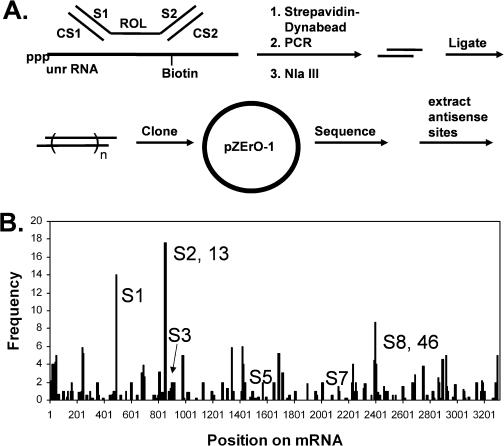
A potential problem with the RT-ROL assay is that the priming of RT might be sensitive to secondary and tertiary structure, and as a result, the intensity of the PCR bands produced in the RT-ROL assay might not correspond directly to the actual binding affinity of the ODN. To circumvent this problem we developed an enzyme independent method for mapping antisense binding sites that adapts methodology used in SAGE to determine which members of a random library of ODNs bind to an mRNA molecule and with what frequency. The method, which we call SAABS for serial analysis of antisense binding sites is shown in Figure 4A. In the first step, members of a random oligonucleotide library that bind to the mRNA are separated from those that do not. In the second step, the selected sequences are PCR amplified, restricted, concatenated by ligation and then cloned. In the third step, the clones are sequenced and the sequences retrieved from between the restriction sites and then aligned with the RNA sequence by a computer program.
Figure 4.
SAABS assay. (A) SAABS procedure. A random 8mer oligodeoxynucleotide library (ROL) flanked by two PCR tags is first incubated with biotinylated mRNA. The mRNA bound ODN is then separated from free ODN by binding the biotinylated mRNA to a streptavidin coated Dynabead, which is then separated from the unbound sequence by a magnetic field. The bound sequence is then PCR amplified with S1 and CS2, restricted with NlaIII, concatenated by ligation, cloned in pZErO-1 and sequenced. (B) Frequency distribution of the antisense binding sites on the unr mRNA obtained from the SAABS assay. The 8mer sequences were retrieved from the sequenced clones and aligned with the mRNA sequence. Some of the sites identified correspond to sites found by the RT-ROL assay (13 and 46), whereas others were uniquely detected by the SAABS assay and denoted with an S prefix (S1, S3, S5 and S7).
Recently, another group has reported a similar strategy for identifying antisense binding sites, which they called MAST for mRNA accessible site tagging (31). Our method is very similar to theirs, which also uses streptavidin coated Dynabeads, except that we use an 8mer random nucleotide library instead of an 18mer library, add the Dynabeads after binding of the ODNs to the biotinylated mRNA, and concatenate the antisense sequence tags before sequencing, which they do not.
In our procedure (Figure 4A) a random 8mer ODN library linked to two PCR tags is incubated with a biotinylated mRNA molecule that is bound to a strepavidin-coated Dynabead. The PCR tags are prevented from hybridizing to the mRNA by binding to the complementary ODNs CS1 and CS2. Following incubation with the mRNA, the Dynabeads are separated by a magnet and washed to remove unbound ODNs. The bound ODNs are then PCR amplified with primers S1 and CS2, restricted with NlaIII to give 12mers that are isolated by denaturing 20% PAGE, concatenated by ligation, cloned into the pZErO-1 vector and transfected into E.coli. Plasmids containing inserts are then sequenced, and the 8mer sequence between the NlaIII restriction sites were extracted by a computer program and both strands matched to complementary sites on the RNA sequence.
The program scored each matching nucleotide in the mRNA by the number of times that it was matched to an 8mer sequence divided by the number of other sites that also matched that same antisense sequence. If a complete match was not found, the process was repeated for all possible 7mer and then 6mer sequences. The relative frequency of the sequences retrieved by the SAABS method are plotted against their position as shown in Figure 4B. Some of these sites were not detected by the RT-ROL assay and are labeled with a S. Presumably, these sites correspond to binding sites that reverse transcriptase may not have been able to extend. Fifteen of the highest frequency sites uniquely determined by this method were chosen for further analysis.
Affinity screening of potential antisense ODNs by a Dynabead-based dot blot assay
To determine the relative affinity of ODNs for the antisense binding sites determined by the RT-ROL and SAABS method we developed a sensitive and semi-quantitative dot blot assay. In our first attempts, we tried to use a standard dot blotting method that involves photocrosslinking of antisense ODNs to a nylon membrane and then quantifying the amount of radiolabeled mRNA that becomes bound. Unfortunately, this technique was not very reproducible, probably because of the non-uniformity of UV crosslinking of ODNs of varying sequence to the nylon membrane.
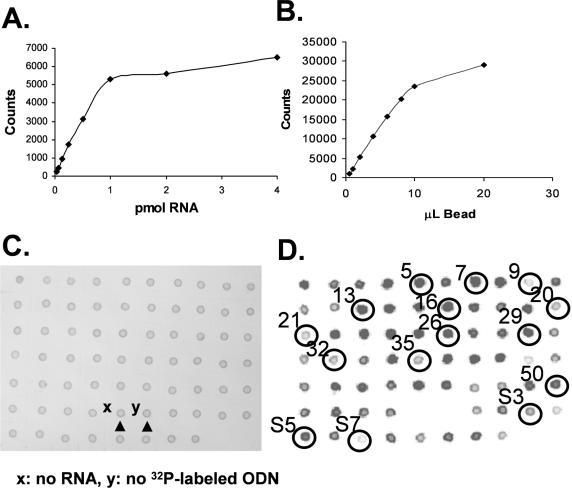
In developing a better dot blotting method for screening ODN binding sites, we investigated the use of Dynabeads to anchor biotinylated unr mRNA to the blot. We found that the binding capacity of the streptavidin coated beads for biotinylated unr mRNA produced by transcription in the presence of Bio-dUTP, to be about 1 pmol of RNA/20 µl of Dynabeads (Figure 5A) and that the Dynabead bound mRNA could bind about one half an equivalent of a radiolabeled antisense ODN under saturating conditions (Figure 5B).
Figure 5.
Dynabead-based dot blot assay to determine relative binding affinity of ODNs. (A) Determining the loading capacity of the streptavidin coated Dynabead by titrating 20 µl of bead solution in 40 µl total volume of 0.5 M NaCl with biotinylated radiolabeled unr mRNA. (B) Determining the µl of Dynabead bound RNA needed to completely bind 1 pmol of ODN5 in a total volume of 40 µl. (C and D) Solutions of RNA were incubated with 1 pmol of ODN [1–54 from RT-ROL assay and 57–68 (S1-S12) from the SAABS assay] and then incubated with 10 µl of Dynabeads and spotted on Nylon membrane. (C) is a photograph of blot showing equal loading of beads. (D) is a radiogram showing relative amounts of retained ODN. ODNs corresponding to circled spots were further studied by quantitative methods.
The Dynabead-based dot blot assay was then used to qualitatively assess the relative binding affinities of 20mer ODNs for the accessible antisense sites identified by the RT-ROL and SAABS method (Figure 5C and D). High and low affinity ODNs were selected for the quantitative binding assays to provide a range of affinities for comparison of the ODN and PNA binding affinities. The sequences were further optimized by shifting the sequence to the 5′ or 3′ end to minimize self-complementarity and shortening to 15mers to minimize unfavorable electrostatic interactions. These second generation ODNs are labeled X-2, where X = the parental ODN. On the basis of this assay, about 15 ODNs were selected for quantitative assays. We also selected an additional four antisense ODNs corresponding to sites that were not detected by either the RT-ROL or SAABS assay, and a sense sequence (PNA 50S), and two unrelated sequences L-15 and V-12.
Quantitative Dynabead-based assay of antisense ODN binding affinity
Initially we attempted to obtain ODN binding constants for the unr mRNA by a Centricon centrifugal filtration assay (32). In this assay 5′ 32P-radiolabeled ODN is incubated with increasing concentrations of mRNA, and bound ODN is separated from free ODN by centrifugation through a filter that does not allow the mRNA or any ODN-bound mRNA to pass through the filter. Unfortunately, this method did not work very well in our hands due to problems with non-specific binding to the filter.
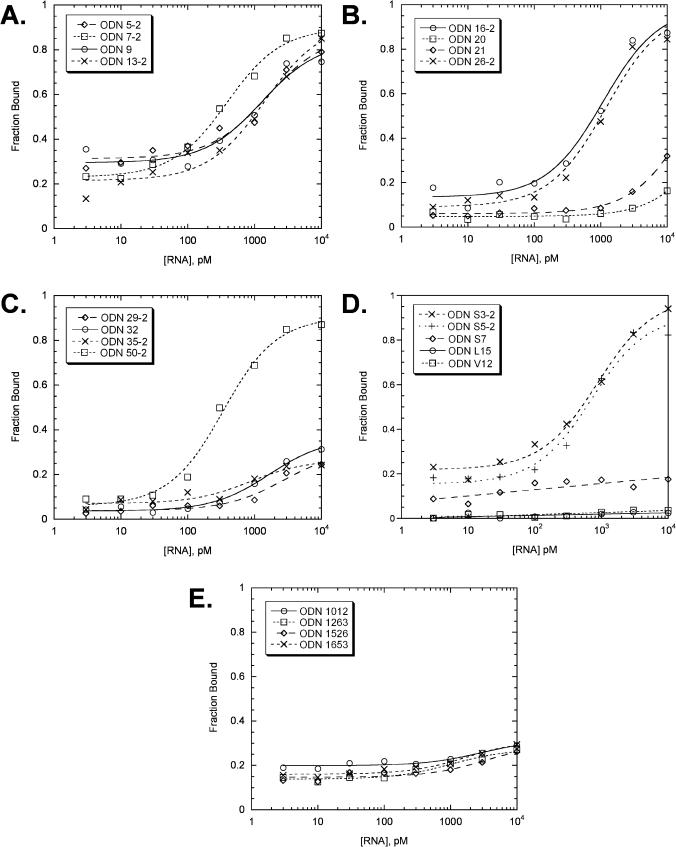
To get around the non-specific binding problem, we developed an assay method which makes use of a magnetic field and Dynabeads to separate ODNs bound to mRNA from free ODNs in solution without the use of a filter. Following equilibration of the ODN with biotinylated unr mRNA, the mRNA and any bound ODN was bound to streptavidin coated Dynabeads and then separated from free ODN using a magnetic field. The Dynabead method gave much better results than the Centricon-based method, though it appeared that there was a variable amount of non-specific binding which resulted in measurable radioactivity in the Dynabead fraction at low concentrations of mRNA where no ODN should have been bound (Figure 6). This non-specific binding could be easily subtracted during the non-linear least squares fitting procedure used to determine the dissociation constants (see experimental section).
Figure 6.
Curve fits of the data from the Dynabead ODN binding assay. (A–C) antisense ODNs for sites identified by the RT-ROL assay, (D) antisense ODNs for sites identified by the SAABS assay, and two unrelated ODNs, L-15 and V-12, (E) antisense ODNs for sites not identified by either the RT-ROL or SAABS assay.
As can be seen in Table 1, five ODNs had Kds of 1 nM or less (entries 1–5), and 9 had Kds of <3 nM (entries 1–9). When the relative binding affinities for the ODNs are compared to the relative intensities of the spots in the dot blot assay there appears to be a reasonable correlation with a few exceptions. All those with dissociation constants >10 nM (entries 10–15) appeared weak on the dot blot assay with the exception of 35-2 which appeared strong. Likewise all the ODNs with dissociation constants of <3 nM appeared strong with the exception of S3-2 and 9 which appeared weak. For the most part then, the dot blot method appears to be a reasonable way of rapidly screening for high affinity binders, which can then be verified by the quantitative assay. Antisense ODNs corresponding to sites not detected by either the RT-ROL or SAABS assay showed little if any affinity (Figure 6E, Table 1, entries 16–19).
Table 1.
Binding affinity of the antisense ODNs by the Dynabead assay
| Entry | Site | Code | ODN sequence | Kd (nM) |
|---|---|---|---|---|
| 1 | 2851 | 50-2 | TGGTGTGCTTTGTGG | 0.3 ± 0.1 |
| 2 | 676 | 7-2 | TTTCCCAGTCCGTCG | 0.3 ± 0.1 |
| 3 | 1414 | S5-2 | TTTGTCACGTCGGTC | 0.7 ± 0.2 |
| 4 | 901 | S3-2 | ATCTCCAGTTTCCAG | 0.8 ± 0.2 |
| 5 | 1145 | 16-2 | CATTTCTGTCCTTGA | 1.0 ± 0.3 |
| 6 | 1802 | 26-2 | CATCCTCAGCCTCCT | 1.1 ± 0.3 |
| 7 | 727 | 9 | ATTCGTTCTTCAGGGAGGAT | 1.2 ± 0.6 |
| 8 | 839 | 13-2 | CACTTCCCCATTACG | 1.4 ± 0.6 |
| 9 | 476 | 5-2 | TATGTCCATTGTTGT | 1.5 ± 0.7 |
| 10 | 2020 | 32 | CCAAAATTATCCTTCAGAGT | >10 |
| 11 | 1396 | 21 | TCTGTTGAAATATTAAACCT | >10 |
| 12 | 1927 | 29-2 | CCTCTGTTTGTCACT | >10 |
| 13 | 2114 | 35-2 | TGTCCCCCAGTTCCA | >10 |
| 14 | 1389 | 20 | AATATTAAACCTAACATGGT | >10 |
| 15 | 2115 | S7 | ATGTCCCCCAGTTCC | >100 |
| 16 | 1012 | 1012 | TCCTTCATGGCACAAACTAC | >100 |
| 17 | 1263 | 1263 | GGTTTTTACTGGGTACTTTT | >100 |
| 18 | 1526 | 1526 | CACACACTTGATGAAACCAA | >100 |
| 19 | 1653 | 1653 | TAATAGCATGATTTCTTTGA | >100 |
| 20 | na | V-12 | CGATTGGAGCGC | >100 |
| 21 | na | L-15 | AGATCGCAACTCATA | >100 |
The antisense site refers to the position in the mRNA (start codon at 448) that is complementary to the 3′ end of the corresponding ODN. The code number X-Y refers to ODN X used in the dot blot experiments, and if followed by a 2 refers to an optimized second generation sequence that overlaps the original sequence. The S prefix refers to a site uniquely identified by the SAABS assay.
Design and synthesis of the antisense PNAs
Because ODNs are readily degraded in vivo and can additionally lead to cleavage of the mRNA transcript to which they are bound via the combined action of RNAse H, they are not suitable for in vivo targeting purposes. PNAs are, however, ideally suited for this purpose because they are nucleic acid analogs in which the enzymatically labile phosphodiester backbone has been replaced by an unnatural peptide backbone (33,34). As a result, PNAs are highly resistant to enzymatic degradation (35), do not activate RNAse H activity (36), have a high affinity for complementary mRNA and can invade duplex regions (37).
Therefore, we designed hybrid PNAs corresponding to the four ODNs that showed the highest binding affinity, and ODN 5-2 which showed slightly less affinity. We also synthesized PNAs corresponding to four ODNs showing low affinity and to the four ODNs corresponding to sites not detected by either method. The sense sequence, PNA50-2S, corresponding to the PNA50-2 binding site was also made as a control. The PNAs were synthesized with a recently reported four lysine permeation peptide (38,39) at the C-terminus, which was used for our PET imaging studies in mice (20), and to increase water solubility and mRNA binding affinity. A cysteine-tyrosine sequence was added to the N-terminal end to enable subsequent attachment of reporter groups to the cysteine and radioiodination of the tyrosine. The PNAs were synthesized by standard solid phase Fmoc chemistry on an ABI Expedite 8909 automated synthesizer, purified by reverse phase HPLC, and characterized by molecular weight determination by MALDI-TOF (Table 2).
Quantitative Dynabead-based assay of antisense PNA binding affinity
The binding affinity of the PNAs for the unr mRNA were determined by monitoring bound and free 131I-labeled PNA as a function of mRNA concentration using the Dynabead assay method that we developed to quantify ODN binding to the mRNA (Figure 7). We chose 131I to label the PNAs because of the high specific activity in which it can be obtained and its short half-life of 8 days. The PNAs were radiolabeled by a procedure previously used to label PNAs with 125I and Chloramine-T (13).
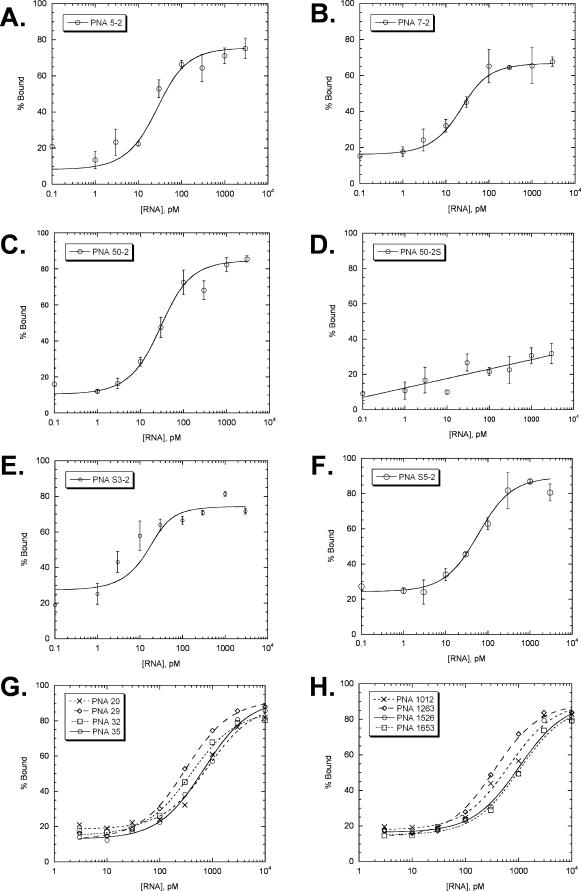
Figure 7.
Curve fits to the PNA binding data from the Dynabead assay. (A–C) antisense PNAs corresponding to high affinity antisense ODNs identified by the RT-ROL assay, (D) sense PNA corresponding to PNA50-2, (E and F) antisense PNAs corresponding to high affinity antisense ODNs identified by the SAABS assay, (G) antisense PNAs corresponding to low affinity antisense ODNs identified by the RT-ROL assay, (H) antisense PNAs corresponding to sites not identified by either assay. The error bars represent the standard deviation of the average of three experiments and are not shown for (G) and (H) for clarity.
We also checked the integrity of the PNA produced under these conditions by repeating the labeling experiment with a short PNA test sequence, DOTA-Tyr-ATGC-Lys (20), with non-radioactive iodine by the chloramine-T method and also by IODO-beads and analyzed the products by MALDI. We found that at a 5 min reaction time, both methods lead primarily to the mono-iodinated compound and some of the diiodinated compound (tyrosine can react twice), but that at longer times, other products are produced (data not shown). Thus the 5 min time period was used for radiolabeling the Cys-Try-PNA-Lys4 by 131I.
In the first set of PNA binding experiments we discovered that we were not recovering all the radioactivity and were able to trace the loss to non-specific binding of the PNA to the microfuge tubes that we were using for the incubations. After ordering and testing a number of tubes and microtiter plates advertised as minimizing binding of peptides and nucleic acids, we found that Corning NBS microtiter plates and the more convenient Bio-Rad EZ Micro 1.5 ml test tubes had the lowest non-specific binding affinity for the PNAs.
Triplicate sets of data were obtained and a plot of % bound as a function of RNA concentration was fit to a simple two state binding equilibrium as we had done for the ODNs (Figure 7). The Kds for the PNAs are tabulated in Table 2. As expected, the PNAs corresponding to the high affinity ODNs show very high binding affinity (low Kds) for the unr mRNA with Kds that range from 7 to 50 pM at 0.1 M salt (entries 1–5). The PNAs corresponding to the low affinity ODNs, showed higher Kds of 300–820 pM (entries 6–9). Surprisingly, the PNAs corresponding to sites not detected by either the RT-ROL or SAABS assays also bound with similar Kds of 340–1000 pM (entries 10–13) found for the low affinity sites. The sense PNA 50-2S, appeared to have essentially no affinity for the RNA (entry 14).
The binding affinities of the PNAs for the unr mRNA are much greater (lower Kds) than those of the corresponding ODNs. The difference in binding affinity between the PNAs and ODNs is likely to be much greater in vivo because the PNA dissociation constants were obtained at the physiological concentration of 0.1 M NaCl, whereas the ODN dissociation constants were acquired at 1 M salt. At 0.1 M salt the Kds for the ODNs are expected to be greater due to electrostatic repulsions, whereas the PNAs are expected to have lower Kds due to favorable electrostatic attraction with the Lys4 tail. PNA 50-2S, which is identical in sequence to the mRNA target, showed no significant binding in the range of RNA concentrations that bound tightly to the antisense sequences.
The significant binding affinity of the PNAs to sites not detected by either the RT-ROL or SAABS assay (Kds of 340–1000 pM) might be attributable to the ability of PNA to invade and bind to regions of folded RNA that might not bind ODNs because of electrostatic repulsion. Even though, the best of these PNAs (PNA 1927) has >50-fold less affinity than the highest affinity PNA identified by the SAABS assay (PNA S3-2), and >25-fold less than the highest affinity PNA found by the RT-ROL assay (PNA 7-2). Likewise, PNAs corresponding to low affinity antisense ODNs bind with less affinity (300–820 pM) than those PNAs corresponding to the high affinity antisense ODNs. These results suggest that accessibility of a site to an ODN may be a prerequisite for high affinity PNA binding. Thus it would appear that the RT-ROL and SAABS assay offer a more direct and efficient way to find high affinity antisense PNAs than might be found by simply screening PNAs to randomly selected sites.
CONCLUSION
We have identified high affinity antisense binding sites on unr mRNA produced in vitro through an improved RT-ROL and SAABS mapping methods, and have quantified their binding affinity by a new Dynabead-based solution assay. The two mapping methods identified many of the same antisense binding sites, but there were also a few high affinity sites identified by the SAABS method that were not detected by the RT-ROL method, presumably because reverse transcriptase was unable to bind or extend an ODN at that site. The affinity of PNAs for these sites on the unr mRNA in vivo remains to be determined, although our recent results with PET imaging MCF-7 tumors in mice with derivatives of PNA50-2, PNA5-2 and PNA7-2 and PNA50-2S are consistent with a contribution from an antisense mechanism. The methods that we improved upon and developed for mapping and quantifying the antisense binding sites might also be useful for designing siRNA agents, and for guiding RNA folding calculations.
Acknowledgments
We want to thank Demetrios Sarantites for help with the radioiodination experiments. This research was supported by an NCI/NASA Biomolecular Sensors contract (N01-CO-27103) and by the National Institutes of Health Program of Excellence in Nanotechnology (1 U01 HL080729-01). Funding to pay the Open Access publication charges for this article was provided by NIH HL080729.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dyba M., Tarasova N.I., Michejda C.J. Small molecule toxins targeting tumor receptors. Curr. Pharm. Des. 2004;10:2311–2334. doi: 10.2174/1381612043384024. [DOI] [PubMed] [Google Scholar]
- 2.Brannon-Peppas L., Blanchette J.O. Nanoparticle and targeted systems for cancer therapy. Adv. Drug Deliv. Rev. 2004;56:1649–1659. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka T., Shiramoto S., Miyashita M., Fujishima Y., Kaneo Y. Tumor targeting based on the effect of enhanced permeability and retention (EPR) and the mechanism of receptor-mediated endocytosis (RME) Int. J. Pharm. 2004;277:39–61. doi: 10.1016/j.ijpharm.2003.09.050. [DOI] [PubMed] [Google Scholar]
- 4.Bennasroune A., Gardin A., Aunis D., Cremel G., Hubert P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit. Rev. Oncol. Hematol. 2004;50:23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Zhao X.B., Lee R.J. Tumor-selective targeted delivery of genes and antisense oligodeoxyribonucleotides via the folate receptor. Adv. Drug Deliv. Rev. 2004;56:1193–1204. doi: 10.1016/j.addr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Shadidi M., Sioud M. Selective targeting of cancer cells using synthetic peptides. Drug Resist. Updat. 2003;6:363–371. doi: 10.1016/j.drup.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 7.van Dongen G.A., Visser G.W., Vrouenraets M.B. Photosensitizer-antibody conjugates for detection and therapy of cancer. Adv. Drug Deliv. Rev. 2004;56:31–52. doi: 10.1016/j.addr.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Goodwin D.A., Meares C.F. Advances in pretargeting biotechnology. Biotechnol. Adv. 2001;19:435–450. doi: 10.1016/s0734-9750(01)00065-9. [DOI] [PubMed] [Google Scholar]
- 9.Rooseboom M., Commandeur J.N., Vermeulen N.P. Enzyme-catalyzed activation of anticancer prodrugs. Pharmacol. Rev. 2004;56:53–102. doi: 10.1124/pr.56.1.3. [DOI] [PubMed] [Google Scholar]
- 10.Wadia J.S., Dowdy S.F. Protein transduction technology. Curr. Opin. Biotechnol. 2002;13:52–56. doi: 10.1016/s0958-1669(02)00284-7. [DOI] [PubMed] [Google Scholar]
- 11.Moulton H.M., Moulton J.D. Peptide-assisted delivery of steric-blocking antisense oligomers. Curr. Opin. Mol. Ther. 2003;5:123–132. [PubMed] [Google Scholar]
- 12.Kabouridis P.S. Biological applications of protein transduction technology. Trends Biotechnol. 2003;21:498–503. doi: 10.1016/j.tibtech.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H.J., Boado R.J., Braasch D.A., Corey D.R., Pardridge W.M. Imaging gene expression in the brain in vivo in a transgenic mouse model of Huntington's disease with an antisense radiopharmaceutical and drug-targeting technology. J. Nucl. Med. 2002;43:948–956. [PubMed] [Google Scholar]
- 14.Lewis M.R., Jia F., Gallazzi F., Wang Y., Zhang J., Shenoy N., Lever S.Z., Hannink M. Radiometal-labeled peptide-PNA conjugates for targeting bcl-2 expression: preparation, characterization, and in vitro mRNA binding. Bioconjug. Chem. 2002;13:1176–1180. doi: 10.1021/bc025591s. [DOI] [PubMed] [Google Scholar]
- 15.Rao P.S., Tian X., Qin W., Aruva M.R., Sauter E.R., Thakur M.L., Wickstrom E. 99mTc-peptide-peptide nucleic acid probes for imaging oncogene mRNAs in tumours. Nucl. Med. Commun. 2003;24:857–863. doi: 10.1097/01.mnm.0000084583.29433.df. [DOI] [PubMed] [Google Scholar]
- 16.Tian X., Aruva M.R., Rao P.S., Qin W., Read P., Sauter E.R., Thakur M.L., Wickstrom E. Imaging oncogene expression. Ann. N. Y. Acad. Sci. 2003;1002:165–188. doi: 10.1196/annals.1281.015. [DOI] [PubMed] [Google Scholar]
- 17.Gallazzi F., Wang Y., Jia F., Shenoy N., Landon L.A., Hannink M., Lever S.Z., Lewis M.R. Synthesis of radiometal-labeled and fluorescent cell-permeating peptide-PNA conjugates for targeting the bcl-2 proto-oncogene. Bioconjug. Chem. 2003;14:1083–1095. doi: 10.1021/bc034084n. [DOI] [PubMed] [Google Scholar]
- 18.Shi N., Boado R.J., Pardridge W.M. Antisense imaging of gene expression in the brain in vivo. Proc. Natl Acad. Sci. USA. 2000;97:14709–14714. doi: 10.1073/pnas.250332397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohail M., Southern E.M. Selecting optimal antisense reagents. Adv. Drug Deliv. Rev. 2000;44:23–34. doi: 10.1016/s0169-409x(00)00081-8. [DOI] [PubMed] [Google Scholar]
- 20.Sun X., Fang H., Li X., Rossin R., Welch M.J., Taylor J.S. MicroPET imaging of MCF-7 tumors in mice via unr mRNA-targeted peptide nucleic acids. Bioconj. Chem. 2005;16:294–305. doi: 10.1021/bc049783u. [DOI] [PubMed] [Google Scholar]
- 21.Ma Z., Taylor J.S. PNA-based RNA-triggered drug-releasing system. Bioconjug. Chem. 2003;14:679–683. doi: 10.1021/bc034013o. [DOI] [PubMed] [Google Scholar]
- 22.Ma Z., Taylor J.S. Nucleic acid triggered catalytic drug release. Proc. Natl Acad. Sci. USA. 2000;97:11159–11163. doi: 10.1073/pnas.97.21.11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Z., Taylor J.S. Nucleic acid triggered catalytic drug and probe release: a new concept for the design of chemotherapeutic and diagnostic agents. Bioorg. Med. Chem. 2001;9:2501–2510. doi: 10.1016/s0968-0896(01)00245-0. [DOI] [PubMed] [Google Scholar]
- 24.Cai J., Li X., Yue X., Taylor J.S. Nucleic acid-triggered fluorescent probe activation by the Staudinger reaction. J. Am. Chem. Soc. 2004;126:16324–16325. doi: 10.1021/ja0452626. [DOI] [PubMed] [Google Scholar]
- 25.Cai J., Li X., Taylor J.S. Improved nucleic acid triggered probe activation through the use of a 5-thiomethyluracil peptide nucleic acid building block. Org. Lett. 2005;7:751–754. doi: 10.1021/ol0478382. [DOI] [PubMed] [Google Scholar]
- 26.Jeffers M., Paciucci R., Pellicer A. Characterization of unr; a gene closely linked to N-ras. Nucleic Acids Res. 1990;18:4891–4899. [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrer N., Garcia-Espana A., Jeffers M., Pellicer A. The unr gene: evolutionary considerations and nucleic acid-binding properties of its long isoform product. DNA Cell. Biol. 1999;18:209–218. doi: 10.1089/104454999315420. [DOI] [PubMed] [Google Scholar]
- 28.Sohail M., Southern E.M. Hybridization of antisense reagents to RNA. Curr. Opin. Mol. Ther. 2000;2:264–271. [PubMed] [Google Scholar]
- 29.Smith L., Andersen K.B., Hovgaard L., Jaroszewski J.W. Rational selection of antisense oligonucleotide sequences. Eur. J. Pharm. Sci. 2000;11:191–198. doi: 10.1016/s0928-0987(00)00100-7. [DOI] [PubMed] [Google Scholar]
- 30.Allawi H.T., Dong F., Ip H.S., Neri B.P., Lyamichev V.I. Mapping of RNA accessible sites by extension of random oligonucleotide libraries with reverse transcriptase. RNA. 2001;7:314–327. doi: 10.1017/s1355838201001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H.Y., Mao J., Zhou D., Xu Y., Thonberg H., Liang Z., Wahlestedt C. mRNA accessible site tagging (MAST): a novel high throughput method for selecting effective antisense oligonucleotides. Nucleic Acids Res. 2003;31:e72. doi: 10.1093/nar/gng072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walton S.P., Stephanopoulos G.N., Yarmush M.L., Roth C.M. Thermodynamic and kinetic characterization of antisense oligodeoxynucleotide binding to a structured mRNA. Biophys. J. 2002;82:366–377. doi: 10.1016/S0006-3495(02)75401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlmann E., Peyman A., Breipohl G., Will D.W. PNA: synthetic polyamide nucleic acids with unusual binding properties. Angew. Chem. Int. Ed. 1998;37:2796–2823. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2796::AID-ANIE2796>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 34.Ray A., Norden B. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB. J. 2000;14:1041–1060. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- 35.Uhlmann E., Peyman A. Antisense oligonucleotides: a new therapeutic principle. Chem. Rev. 1990;90:543–584. [Google Scholar]
- 36.Gee J.E., Robbins I., van der Laan A.C., van Boom J.H., Colombier C., Leng M., Raible A.M., Nelson J.S., Lebleu B. Assessment of high-affinity hybridization, RNase H cleavage, and covalent linkage in translation arrest by antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 1998;8:103–111. doi: 10.1089/oli.1.1998.8.103. [DOI] [PubMed] [Google Scholar]
- 37.Peffer N.J., Hanvey J.C., Bisi J.E., Thomson S.A., Hassman C.F., Noble S.A., Babiss L.E. Strand-invasion of duplex DNA by peptide nucleic acid oligomers. Proc. Natl Acad. Sci. USA. 1993;90:10648–10652. doi: 10.1073/pnas.90.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sazani P., Kang S.H., Maier M.A., Wei C., Dillman J., Summerton J., Manoharan M., Kole R. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001;29:3965–3974. doi: 10.1093/nar/29.19.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sazani P., Gemignani F., Kang S.H., Maier M.A., Manoharan M., Persmark M., Bortner D., Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]