Abstract
Polar pili biogenesis in Caulobacter involves the asymmetric localization of the CpaE and CpaC components of the pili-specific secretion apparatus to one pole of the predivisional cell followed by the biosynthesis of the pili filaments in the daughter swarmer cell. The histidine kinase signaling protein, PleC, that controls the temporal accumulation of the PilA pilin subunit is asymmetrically localized to the pole at which pili are assembled. Here we identify a protein, PodJ, that provides the positional information for the polar localization of both PleC and CpaE. The PodJ protein was found to exist in two forms, a truncated 90-kDa and a full-length 110-kDa form, each controlling a different aspect of polar development and each localizing to the cell poles at a specific time in the cell cycle. When active PleC is delocalized in a ΔpodJ mutant, the accumulation of PilA, the downstream target of PleC signaling, is impaired, providing evidence that the polar localization of this histidine kinase stimulates the response signaled by a two-component system.
Cell division in Caulobacter crescentus yields a swarmer cell and a stalked cell, each with different morphologies and cell fates (1). Only the stalked cell initiates DNA replication. The swarmer cell acquires the ability to replicate its chromosome once it completes an obligate differentiation into a stalked cell (Fig. 1A). During this transition a flagellum, pili, and the chemosensory apparatus (2) are lost from the swarmer cell pole and replaced by a stalk with holdfast material at its tip (3). As the stalked cell grows and replicates its DNA, a new flagellum, chemotaxis apparatus, and pilus secretion apparatus are assembled at the pole opposite the stalk. An asymmetric cell division then yields a stalked cell and a new flagellated swarmer cell that assembles pili filaments (Fig. 1A).
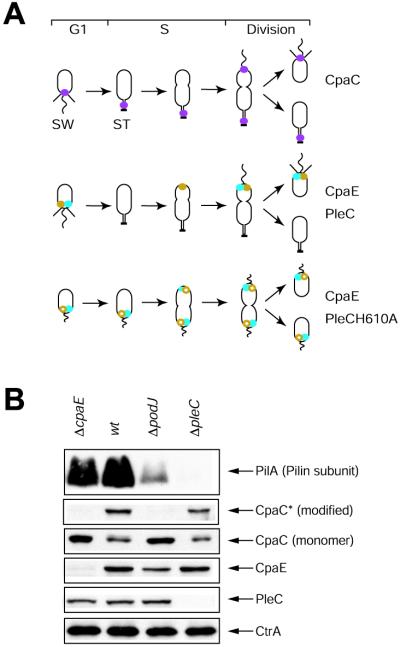
Figure 1.
(A) Schematic drawings of the wild-type (wt) C. crescentus cell cycle showing the subcellular location of the CpaC pilus secretion channel (purple dots), the CpaE pilus assembly protein (blue dots), the PleC histidine kinase (gold dots), and the cell cycle of the pleC(H610A) mutant (Bottom), showing the location of the inactive form of PleC, PleCH610A (open gold circles), and CpaE (blue dots). The swarmer cell (SW) bears polar pili and a polar flagellum, and the stalked cell (ST) shows the hold-fast at the tip of the stalk (thick bar). The pleC(H610A) mutant is pili-less and stalkless and bears a paralyzed flagellum (short wavy line). (B) Steady-state levels of proteins required for pilus assembly (PilA, CpaC*, CpaC, CpaE, PleC, and CtrA) in cell extracts from wild-type, ΔcpaE, ΔpleC and ΔpodJ mutant cells determined by immunoblotting. As observed (4), the loss of CpaC* in the ΔpodJ strain is accompanied by an increase in the steady-state level of the CpaC monomer.
To determine the molecular mechanisms that control polarity, we have initiated a study of polar pili biogenesis (4, 5). Pili serve as bacteriophage receptor sites in Caulobacter and are known to mediate adhesion, motility, biofilm formation, and DNA transfer in other bacteria (6, 7). Pili are formed by the polymerization of a pilin subunit into a filament that is anchored in the inner membrane, spans the periplasm, and exits the outer membrane through a specialized pilus secretion channel formed by a protein belonging to the secretin family (7–9). Pili biogenesis in Caulobacter requires at least nine genes (4, 5). Five genes encoding components of the pilus secretion machinery, cpaB-F, including the CpaC secretin homologue, are transcribed in the predivisional cell. A gene encoding a homologue of a prepilin peptidase, cpaA, is transcribed next, followed by the transcription of pilA, encoding the PilA pilus subunit (10). CpaC is localized to the incipient swarmer cell pole of the predivisional cell before the accumulation of PilA. The polar positioning of CpaC requires the CpaE assembly protein that is located at the incipient swarmer cell pole. CpaE is lost at the swarmer-to-stalked cell transition coincident with the loss of pili, but CpaC is retained at the tip of the newly formed stalk (4).
Localization to the cell pole is also a feature of the two-component signaling proteins that regulate polar morphogenesis including the PleC, DivJ, and CckA histidine kinases and the DivK response regulator (11–13). PleC and the essential response regulator CtrA positively control the accumulation of PilA (4, 5, 10, 14). The localization pattern of PleC parallels that of CpaE, raising the possibility that it is controlled by a common mechanism. Here we report the identification of a localization factor, PodJ, that provides the positional information for the polar location of PleC and CpaE. The PodJ protein exists in two forms, full length (PodJL) and truncated (PodJS), each localizing to the cell pole at different stages in the cell cycle where they perform different biological functions. Moreover, in the absence of PodJ, the CpaC secretin homologue fails to localize to the cell pole. Because the positioning of CpaC directly depends on CpaE (4), the observed requirement of PodJ for CpaC localization is probably indirect due to the delocalization of CpaE in the absence of PodJ. Thus, the mere presence of CpaE activity within the cell is not sufficient for proper execution of its function, but rather the function needs to be performed at the correct subcellular site.
PilA is a target of the PleC signaling pathway in that PilA fails to accumulate in a pleC deletion mutant or in a mutant strain in which the normally phosphorylated histidine has been changed to an alanine (4). We show here that PilA accumulation is also impaired in a ΔpodJ mutant, which fails to localize active PleC to the swarmer cell pole, providing evidence that the activity and localization of the PleC histidine kinase both are required for the accumulation of PilA. Thus, the localization of the PleC histidine kinase to the cell pole activates the phosphorelay that regulates a downstream target.
Experimental Procedures
Bacterial Growth Conditions and Strains.
C. crescentus CB15N and CB15 and their derivatives were grown in M2G for immunoblotting, synchronization, and fluorescence microscopy experiments (4, 15). Synchronization and bacteriophage ΦCr30-mediated generalized transductions were performed as described (15, 16). Rosette formation was assayed by light microscopy in CB15 derivatives. Chemotaxis was assayed as described (13). The presence of pili was assayed by sensitivity to bacteriophage ΦCbK as described (5). The ΔcpaE and podJ∷Tn5(NTD) (SC1119) strains have been described (5, 13, 17). The podJ∷Tn5(CTD) mutant was isolated in the same mini-Tn5lacZ2 screen that identified the cpaF∷Tn5 strain (5). The strain with an in-frame deletion of pleC (ΔpleC) was a gift from Urs Jenal (Biozentrum, Basel University, Basel). The ΔpodJ strain was made by creating an in-frame deletion of the sequence encoding residues 42–959 of PodJ and confirmed by complementation with podJ in trans. The yfp-podJ strain was constructed by fusing a version of yfp lacking the start and stop codon after the podJ start codon as has been described for yfp-cpaE (4). The yfp-podJ strain is chemotactic and sensitive to infection by bacteriophage ΦCbK, indicating that yfp-podJ is functional. For the construction of the ΔpilA-cpaF∷Ωaac3 strain, the sequence between codon 5 of pilA and codon 462 of cpaF was replaced with the Ωaac3 cassette from pHP45Ωaac3 that confers resistance to apramycin at 8 μg/ml (18). The pleC-gfp and yfp-cpaE strains were made as described by using pHPV290 and pHPV227, respectively (4).
Sequence of podJ.
An updated sequence of podJ replacing the previous one (17), which contains frameshifts that translate into a truncated version of PodJ (CC 2045) with a molecular mass of 50 rather than 103 kDa, was deposited in the Third Party Annotation Section of GenBank under accession number TPA: BK000536.
Antibody Production, Immunoblots, and Fractionation of Cell Lysates.
Hexa-histidine-tagged PodJ N-terminal domain (NTD) and C-terminal domain (CTD) were overexpressed in Escherichia coli by using pET28 (Novagen) and purified by using standard conditions (Qiagen, Valencia, CA). A total of 2 mg of each purified protein was used to immunize two rabbits (Covance, Richmond, CA) for the production of polyclonal antibodies. Immunoblots were performed as described (4, 19). The crude PodJ NTD and CTD serum was diluted 1/20,000 and 1/30,000, respectively. Fractionation of cell lysates into soluble and insoluble fractions was performed as described by Sandkvist et al. (20) except that cells were lysed completely by sonication.
Fluorescence Microscopy.
GFP/yellow fluorescent protein (YFP)-mediated fluorescence microscopy, immunofluorescence microscopy (IFM), and antibody affinity purification were performed as described (4, 21). The PodJ antibodies were used at 1:2,500 (NTD) and 1:1,000 dilution (CTD), respectively.
Results
podJ Mutants Fail to Accumulate PilA and a Modified Form of CpaC, CpaC*.
The podJ gene was identified in a Tn5 mutagenesis screen for mutants that display defects in polar organelle development and confer resistance to infection by bacteriophage ΦCbK (13). The observation that ΦCbK binds to pili as the first step in the infection process (5) suggests that podJ loss-of-function mutants are resistant to ΦCbK because of their inability to assemble pili. To test this possibility, immunoblots of cell extracts from wild-type and a podJ deletion strain were probed with antibodies raised against structural and regulatory factors required for pilus formation (Fig. 1B): the pilin subunit (PilA), the pilus outer membrane channel subunit (CpaC), and its assembly factor (CpaE) as well as antibodies against two regulatory proteins required for the accumulation of PilA, the histidine kinase PleC and the response regulator CtrA (4, 5). The ΔpodJ strain showed significantly reduced steady-state levels of PilA, which could account for the loss of pili as bacteriophage ΦCbK receptors. Although CpaE, CpaC, and PleC are present in the ΔpodJ mutant, CpaC*, a modified form of the CpaC monomer with a slower mobility on SDS/PAGE (4), fails to accumulate (Fig. 1B). We have shown previously that, unlike CpaC, which is present throughout the cell cycle, CpaC* transiently accumulates only in the stalked cell and predivisional cell (4). This requirement of PodJ for the accumulation of CpaC* suggests that PodJ may mediate an event at the cell pole required for CpaC function.
Polar Localization of CpaC and CpaE Depends on PodJ.
We demonstrated previously that both CpaC and CpaE are localized to the flagellated cell pole (4). Furthermore, ΔcpaE cells are unable to localize CpaC to the pole and are devoid of CpaC* (4). The absence of CpaC* in the ΔpodJ mutant cell extracts suggests that PodJ, similar to CpaE, is also required for the polar localization of CpaC. IFM using an affinity-purified CpaC antibody (4) showed that ΔpodJ mutant cells were devoid of the polar CpaC foci (Fig. 2A). Thus, PodJ, similar to CpaE, is in the pathway controlling CpaC modification and localization. To determine whether PodJ acts upstream of CpaE, we used fluorescence microscopy to observe the intracellular location of YFP–CpaE in a ΔpodJ strain in which the chromosomal cpaE gene is replaced with yfp-cpaE (4). As shown in Fig. 2B, ΔpodJ mutant cells were devoid of polar YFP–CpaE foci. From these results, together with the finding that CpaE is present in the ΔpodJ mutant (Fig. 1B), we conclude that PodJ provides the positional information for the polar localization of CpaE. In addition, these results suggest that the presence of CpaE is necessary but not sufficient for the modification of CpaC and its polar localization. Instead, CpaE function needs to be exerted at the correct location within the cell.
Figure 2.
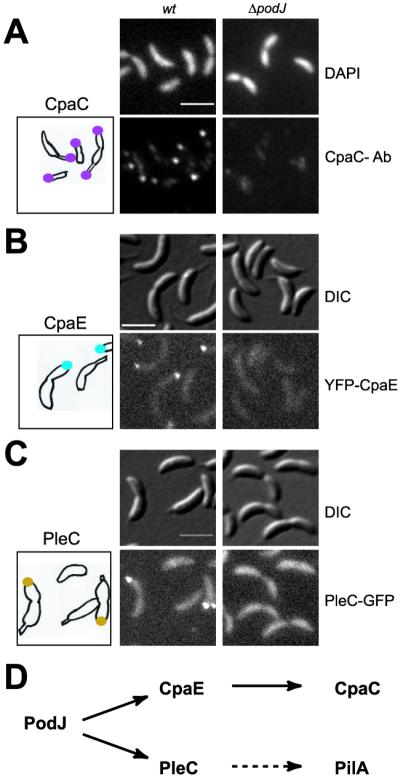
PodJ is required for the polar localization of the CpaC secretin, the CpaE pilus assembly factor, and the PleC histidine kinase. The location of each protein was examined in wild-type (wt) and ΔpodJ mutant cells. (A) Indirect IFM using affinity-purified CpaC antibody (Ab). Shown are images of 4′,6-diamidino-2-phenylindole (DAPI)-stained chromosomal DNA (Upper), IFM images of CpaC (Lower), and a drawing with the location of CpaC (purple dots) in wild-type cells. (B) CpaE location observed in cells expressing YFP–CpaE. Shown are differential interference contrast (DIC) images (Upper), fluorescence images (Lower), and a drawing with the location of YFP–CpaE (blue dots) in the podJ+ yfp-cpaE strain. (C) PleC location observed in cells expressing PleC–GFP. Shown are DIC images (Upper), fluorescence images (Lower), and a drawing with the location of PleC-GFP (gold dots) in the podJ+ pleC-gfp strain. (D) The parallel pathways of PodJ localized proteins. (Scale bars, 2 μm.)
PodJ Controls the Polar Localization of the PleC Histidine Kinase.
Fluorescence microscopy of a functional PleC–GFP fusion showed that polar PleC–GFP foci observed in podJ+ cells are absent in ΔpodJ cells (Fig. 2C). Instead, PleC–GFP is dispersed evenly throughout the cell membrane, indicating that PleC lacks the positional information for polar localization in the absence of PodJ.
The ΔpodJ mutant is motile, forms stalks, and is not completely devoid of PilA (Fig. 1B), unlike the ΔpleC and pleC(H610A) loss-of-function mutants (4). Thus, delocalized PleC has retained activity. To determine whether podJ acts upstream of pleC, the ΔpodJ mutation was introduced into ΔpleC and pleC(H610A) mutants, and the resulting strains were analyzed by differential interference contrast light microscopy for motility and by immunoblotting for PilA levels. Both double-mutant strains were nonmotile and completely devoid of PilA (data not shown), indicating that the pleC loss-of-function mutations are epistatic to the ΔpodJ mutation. Although PodJ is not required for PleC accumulation (Fig. 1B), it is required for its polar positioning, suggesting that mislocalization of PleC is responsible for the reduced PilA levels observed in the ΔpodJ mutant. These results, together with the PleC localization experiments, strongly support a model in which PodJ modulates PleC activity with respect to PilA accumulation by directing PleC to the polar site of pili biogenesis.
To determine whether PleC localization depends on CpaE, the pleC-gfp fusion was introduced into a strain (ΔpilA-cpaF∷Ω) with a deletion of the locus encompassing the pilus assembly genes pilA-cpaF. Fluorescence microscopy of this strain revealed PleC–GFP foci at the swarmer cell pole (data not shown), suggesting that PleC polar localization does not depend on CpaE. Thus, by directing the polar localization of the CpaE pilus assembly factor and the PleC histidine kinase, PodJ controls two parallel pathways that culminate in pili biogenesis at the cell pole (Fig. 2D).
podJ Encodes Two Distinct Isoforms, PodJS and PodJL.
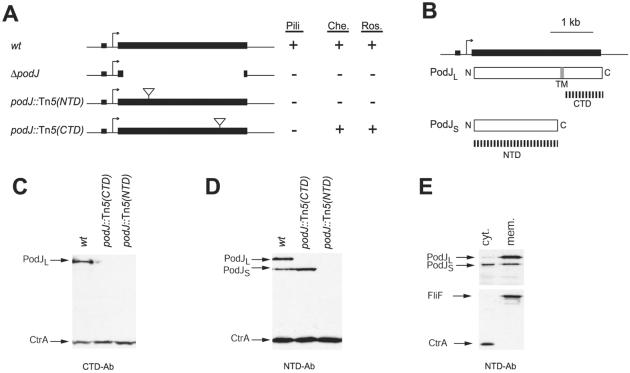
A podJ deletion mutant and a mutant harboring a Tn5 insertion near the 5′ end of podJ [podJ∷Tn5(NTD)] are impaired in pili biogenesis and are also nonchemotactic and unable to carry out holdfast-mediated rosette formation (Fig. 3A; refs 13 and 17). A mutant with a Tn5 insertion near the 3′ end of podJ [podJ∷Tn5(CTD)], however, lacks pili but is chemotactic and forms rosettes (Fig. 3A). The podJ gene encodes a predicted protein with a molecular mass of 103 kDa (974 residues) and a TM domain at residues 639–662 (Fig. 3B). A stretch of positively charged residues (RPRSRRRAWARP) immediately preceding the TM suggests that, according to the “positive-inside” rule (22), the TM separates PodJ into a cytoplasmic NTD (residues 1–638) and a periplasmic CTD (residues 663–974). The site of the Tn5 insertion in the podJ∷Tn5(CTD) mutant maps to the region encoding the CTD of PodJ, whereas that of podJ∷Tn5(NTD) is located within the NTD, suggesting that the CTD is specifically required for pilus assembly, and that the NTD is necessary and sufficient for chemotaxis and rosette formation.
Figure 3.
podJ encodes two isoforms, a full-length protein, PodJL, and a C-terminal truncated form, PodJS, with distinct biological properties. (A) Schematic of the podJ gene (black bar) in wild-type (wt) and podJ mutant strains and the effects of the mutations on the presence (+) or absence (−) of pili assayed by ΦCbK sensitivity, chemotaxis (Che.) and holdfast-mediated rosette formation (Ros.). Rosette formation was assayed in derivatives of CB15 wild type, and the other phenotypes were scored in strains derived from CB15N wild type. Triangles show the site of Tn5 insertions in podJ∷Tn5(NTD) and podJ∷Tn5(CTD). An arrow mediates the podJ transcriptional start site, and a black box represents a CtrA-binding motif in the promoter (17). (B) The podJ gene showing the predicted PodJ isoforms with the N and C termini and the putative transmembrane (TM) domain. Dashed black lines represent the NTD and the CTD that served as antigens for the generation of two PodJ antibodies. (C) Immunoblots of cell extracts from wild-type and podJ mutants using the antibody raised against the PodJ CTD (CTD-Ab). PodJL was the only protein detected by the CTD antibody in wild-type cells. In C and D, CtrA antibody serves as a control. (D) Immunoblots of cell extracts from wild-type and podJ mutants using the PodJ NTD antibody (NTD-Ab). Both PodJS and PodJL were detected by the NTD antibody in wild-type cells. (E) Immunoblots, using the antibody to PodJ NTD, of cell extracts from wild-type cells fractionated into a cytoplasmic fraction (cyt.) and a membrane fraction (mem.). CtrA and FliF serve as controls for cytoplasmic and membrane proteins, respectively.
Immunoblots using antibodies raised against recombinant CTD and NTD (Fig. 3B) revealed that PodJ is composed of two physically separable domains. As expected, the CTD-specific antibody recognizes a single protein with an apparent molecular mass of ≈110 kDa (Fig. 3C). The antiserum raised against the NTD detects two proteins, a longer form with the same mobility as the one detected by the CTD-specific antibody (designated PodJL) and a shorter form that migrates at ≈90 kDa (designated PodJS). The ΔpodJ (data not shown) and the podJ∷Tn5(NTD) mutant (Fig. 3D) are devoid of both PodJL and PodJS. However, PodJL, but not PodJS, was absent from the chemotaxis- and rosette-proficient podJ∷Tn5(CTD) mutant (Fig. 3D), suggesting that PodJS comprises the NTD required for chemotaxis and rosette formation. Moreover, the observation that PodJS does not react with the CTD-specific antibody provides evidence that PodJS represents a C-terminal truncated form of PodJL.
To ascertain that PodJs lacks the TM domain, wild-type cell extracts were separated into soluble (cytoplasmic) and insoluble (membrane) fractions (Fig. 3E) and probed for the presence of the PodJ isoforms by immunoanalysis using the NTD-specific antibody. Approximately half of the cellular content of PodJS is soluble, suggesting that it is at least partially cytoplasmic, whereas the other half seems to be associated with the cytoplasmic membrane. Unlike PodJS, PodJL is found exclusively in the membrane fraction, suggesting that the predicted TM provides PodJL with features similar to those of integral membrane proteins such as FliF (Fig. 3E; ref. 23).
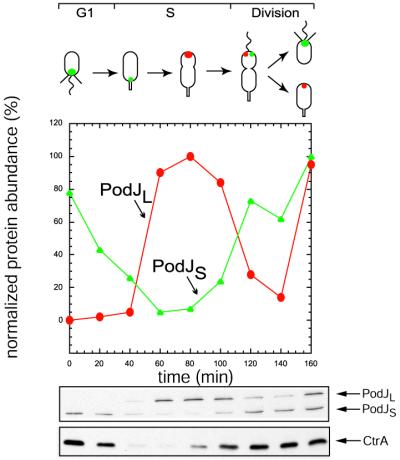
Steady-State Levels of PodJS and PodJL Peak at Different Stages of the Cell Cycle.
Immunoblots probed with the PodJ NTD antibody were used to determine the steady-state levels of both PodJ isoforms during the cell cycle (Fig. 4). These experiments revealed that the two PodJ isoforms are present in mutually opposing cell types. The full-length translation product, PodJL, accumulates in the stalked cell. PodJS, on the other hand, appears in the late predivisional cell at a time when PodJL steady-state levels decrease. After cell division, PodJL appears in the stalked cell progeny, while PodJS is in the swarmer cell progeny. The accumulation of PodJL in the stalked cell and early predivisional cell coincides with the onset of podJ transcription (17). In the late predivisional cell, PodJL is likely to be converted to the truncated form, PodJS, by proteolytic cleavage.
Figure 4.
PodJL and PodJS steady-state levels peak at different times during the cell cycle. Swarmer cells were isolated by Ludox gradient centrifugation and released into M2G medium. Samples of equal volume were collected from the synchronous culture every 20 min and subjected to immunoblotting by using the PodJ NTD and CTD antibodies, with antibodies to CtrA as a control. (Top) The subcellular location of PodJL (red) and PodJS (green) during the cell cycle inferred from the results shown below and those shown in Fig. 5. (Middle) The relative levels of PodJL (red) and PodJS (green) steady-state levels during the cell cycle, determined from the immunoblots shown (Bottom).
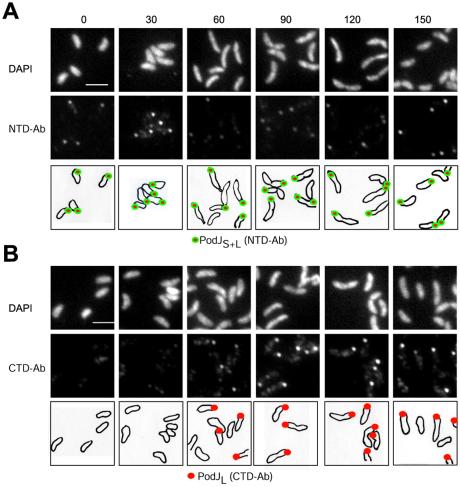
PodJS and PodJL Localize to the Incipient Swarmer Cell Pole.
To explore the subcellular location of PodJL and PodJS, IFM using the PodJ NTD- and CTD-specific antibodies was performed. Both antibodies revealed strong polar foci in wild-type cells that were not present in the ΔpodJ mutant (data not shown). To distinguish between the localization pattern of PodJL and that of PodJS, synchronized wild-type cells were collected at different stages of the cell cycle and subjected to IFM by using either NTD- or CTD-specific antibodies. By using the PodJ NTD-specific antibody, polar foci were detected in wild-type swarmer cells (Fig. 5A) which do not contain PodJL (Fig. 4). Similar results were obtained by observing the location of YFP–PodJ in a yfp-podJ strain by fluorescence microscopy and immunoblotting (data not shown). Thus, PodJS is located at the swarmer cell pole. The antibody to PodJ CTD, which only detects PodJL, yielded fluorescent foci at the cell pole of the predivisional cells when PodJL first accumulates. Fluorescence microscopy using the yfp-podJ strain revealed that YFP–PodJ was localized to the pole opposite the stalked pole in predivisional cells (data not shown). These results show that the localization of PodJL, the isoform required for pilus formation, occurs at the same site as and precedes that of PleC and CpaE localization. The decrease in PodJL steady-state levels in the late predivisional cell is followed by the appearance of PodJS at the new swarmer cell pole, the site previously occupied by PodJL. Thus, both PodJ isoforms, PodJL and PodJS, sequentially appear at the incipient swarmer cell pole of the predivisional cell, but only PodJS is retained at the pole of the progeny swarmer cell.
Figure 5.
The subcellular location of PodJL+S and PodJL in synchronized wild-type cells determined by indirect IFM using the PodJ NTD (A) and PodJ CTD (B) antibody. As swarmer cells progressed through the cell cycle, samples were taken at 0, 30, 60, 90, and 120 min, the cells were fixed and treated with lysozyme, and fluorescent foci were observed. A and B show 4′,6-diamidino-2-phenylindole (DAPI)-stained images (Top), IFM images (Middle), and drawings of the location of PodJL+S (green circles filled red) or PodJL (red dots) during the cell cycle. (Scale bars, 2 μm.)
Discussion
The asymmetric polar location of pilus assembly proteins relies on two mechanisms acting in concert. The first mechanism, described in this study, utilizes a localization factor, PodJ, to position a pili assembly protein, CpaE, and a pili regulatory protein, the PleC histidine kinase, at the cell pole. The second mechanism mediates polar asymmetry by restricting the location of these proteins to only one of the two cell poles by using PleC∼P to signal the release of itself and CpaE from the pole of the swarmer cell at the swarmer-to-stalked cell transition (4).
The podJ gene encodes two isoforms: PodJL, required for pilus biogenesis, and PodJS, responsible for chemotaxis and the formation of holdfast-mediated rosettes. PodJL provides the positional information for the localization of the CpaE assembly protein and the PleC histidine kinase to the incipient swarmer pole of the predivisional cell. PodJS may act in a manner similar to PodJL in serving as a localization factor or it may affect chemotaxis and holdfast formation in some other way. Consistent with their distinct roles in polar organelle development, the PodJ isoforms are located at the cell poles at different times in the cell cycle (Fig. 4). Because the podJ gene is transcribed in the stalked cell and early predivisional cell, coincident with the accumulation of the full length PodJL isoform, it is likely that the shorter PodJS isoform is a proteolytic product of PodJL. After the onset of the suspected proteolytic cleavage of PodJL just before cell division, PodJS remains at the pole of the progeny swarmer cell, and PodJL is lost from this pole. Thus, a temporally controlled proteolytic event is likely to mediate polar development during the cell cycle.
The CpaC secretion channel may perform a function at the swarmer cell pole and at the tip of the stalk. Unlike CpaE and PleC, which are lost at the swarmer-to-stalked cell transition, CpaC remains at its polar position after pili and the flagellum are lost (4). As a consequence, CpaC is retained at the tip of the elongating stalk (Fig. 1A). In the absence of PodJ, CpaC is unable to localize to the cell pole and, consequently, is also absent from the tip of the stalk later in the cell cycle. We propose that, like for CpaC, PodJ may control the localization of a holdfast biogenesis factor by directing it to the incipient swarmer cell pole of the predivisional cell, where it may persist in an inactive state until after the swarmer-to-stalked cell transition, when it mediates holdfast biogenesis at the tip of the nascent stalk. The lack of positional information for its polar location in the absence of PodJ may result in the inability to form holdfasts. In light of this hypothesis, it is noteworthy that the hfaA gene, encoding a holdfast attachment protein, is transcribed in the swarmer cell compartment of the predivisional cell rather than at the time when the holdfast forms (3).
We have shown that several two-component signal transduction proteins including DivJ, DivK, CckA, and PleC are dynamically localized to Caulobacter cell poles during the course of the cell cycle (23–25). Thus far, it has not been possible to determine whether both the activity of a protein and its location within the cell are required for the same downstream signaling pathway. Using the accumulation of PilA as a readout of the PleC phosphorelay, we provide evidence that the ability of PleC to localize to the cell pole is necessary for the regulation of PilA accumulation. In a ΔpodJ mutant, active PleC is delocalized, and PilA accumulation is impaired. The complete loss of PleC activity not only fails to support PilA accumulation but also impedes stalk biogenesis, motility (4), and holdfast-mediated rosette formation (13). Although the polar localization of PleC is important for pili and perhaps holdfast biogenesis, it is not required for stalk biogenesis or motility. Active but delocalized PleC (PleC∼P) is present in the cell, because the ΔpodJ strain only exhibits the PilA and holdfast phenotypes. Thus, for at least one branch of the PleC signal transduction pathway, impaired signaling is probably a direct consequence of the inability to localize the PleC histidine kinase to the cell pole. The local activity of PleC at the swarmer cell pole is likely to be critical for the activation of a target response regulator, like the DivK single domain response regulator, that also is localized to this pole (25). The possibility that only fully active PleC localizes to the pole and PodJ serves primarily to stimulate PleC activity can be excluded based on the result that an inactive version of PleC, lacking the site of autophosphorylation, retains its ability to localize to the cell pole (4).
The CpaE pilus assembly protein provides another example of the necessity of polar localization for protein function. Strains lacking CpaE activity fail to localize the CpaC pilus secretion channel (4). In a ΔpodJ strain, CpaE is present but not localized, and consequently CpaC is not positioned at the cell pole. Thus, CpaE function is impaired in the ΔpodJ strain. These observations are consistent with the view that CpaE must perform its function at the correct subcellular location, the incipient swarmer cell pole, and that the mere presence of CpaE is not sufficient for the formation of the polar CpaC pilus secretion channel.
Acknowledgments
We are indebted to Jeff Skerker for the isolation of the podJ∷Tn5(CTD) strain. We thank Urs Jenal and Bert Ely for providing strains and plasmids and members of the Shapiro Lab for critical reading of the manuscript. This work was supported by the National Institutes of Health Grant GM 32506/5120M2 and Defense Advanced Research Planning Agency Grant MDA972-00-0032. P.H.V. was funded in part by a grant from the Swiss National Science Foundation and the Roche Research Foundation.
Abbreviations
- NTD
N-terminal domain
- CTD
C-terminal domain
- YFP
yellow fluorescent protein
- TM
transmembrane
- IFM
immunofluorescence microscopy
Note Added in Proof.
While this work was in progress, the laboratory of Yves Brun (Indiana University, Bloomington) also identified the localization pattern and function of PodJ (personal communication).
Footnotes
Data deposition: The podJ nucleotide sequence reported in this paper has been deposited in the Third Party Annotation Section of the GenBank database (accession no. TPA: BK000536).
References
- 1.Jensen R B, Wang S C, Shapiro L. Nat Rev Mol Cell Biol. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- 2.Alley M R, Maddock J R, Shapiro L. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 3.Janakiraman R S, Brun Y V. J Bacteriol. 1999;181:1118–1125. doi: 10.1128/jb.181.4.1118-1125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viollier P H, Sternheim N, Shapiro L. EMBO J. 2002;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skerker J M, Shapiro L. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wall D, Kaiser D. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 7.Soto G E, Hultgren S J. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russel M. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 9.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley A P. Proc Natl Acad Sci USA. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laub M T, McAdams H H, Feldblyum T, Fraser C M, Shapiro L. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 11.Ohta N, Lane T, Ninfa E G, Sommer J M, Newton A. Proc Natl Acad Sci USA. 1992;89:10297–10301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecht G B, Lane T, Ohta N, Sommer J M, Newton A. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S P, Sharma P L, Schoenlein P V, Ely B. Proc Natl Acad Sci USA. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laub M T, Chen S L, Shapiro L, McAdams H H. Proc Natl Acad Sci USA. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ely B. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 16.Evinger M, Agabian N. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crymes W B, Jr, Zhang D, Ely B. J Bacteriol. 1999;181:3967–3973. doi: 10.1128/jb.181.13.3967-3973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blondelet-Rouault M H, Weiser J, Lebrihi A, Branny P, Pernodet J L. Gene. 1997;190:315–317. doi: 10.1016/s0378-1119(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 19.Jenal U, Shapiro L. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- 20.Sandkvist M, Bagdasarian M, Howard S P, DiRita V J. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen R B, Wang S C, Shapiro L. EMBO J. 2001;20:4952–4963. doi: 10.1093/emboj/20.17.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Heijne G, Gavel Y. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 24.Wheeler R T, Shapiro L. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs C, Hung D, Shapiro L. Proc Natl Acad Sci USA. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]