Abstract
Many enzymes of the bacteriochlorophyll and chlorophyll biosynthesis pathways have been conserved throughout evolution, but the molecular mechanisms of the key steps remain unclear. The magnesium chelatase reaction is one of these steps, and it requires the proteins BchI, BchD, and BchH to catalyze the insertion of Mg2+ into protoporphyrin IX upon ATP hydrolysis. Structural analyses have shown that BchI forms hexamers and belongs to the ATPases associated with various cellular activities (AAA+) family of proteins. AAA+ proteins are Mg2+-dependent ATPases that normally form oligomeric ring structures in the presence of ATP. By using ATPase-deficient BchI subunits, we demonstrate that binding of ATP is sufficient to form BchI oligomers. Further, ATPase-deficient BchI proteins can form mixed oligomers with WT BchI. The formation of BchI oligomers is not sufficient for magnesium chelatase activity when combined with BchD and BchH. Combining WT BchI with ATPase-deficient BchI in an assay disrupts the chelatase reaction, but the presence of deficient BchI does not inhibit ATPase activity of the WT BchI. Thus, the ATPase of every WT segment of the hexamer is autonomous, but all segments of the hexamer must be capable of ATP hydrolysis for magnesium chelatase activity. We suggest that ATP hydrolysis of each BchI within the hexamer causes a conformational change of the hexamer as a whole. However, hexamers containing ATPase-deficient BchI are unable to perform this ATP-dependent conformational change, and the magnesium chelatase reaction is stalled in an early stage.
Keywords: ATPase‖chlorophyll biosynthesis‖integrin‖metallation‖sensor arginine
The insertion of Mg2+ into protoporphyrin IX is the first unique step in chlorophyll biosynthesis. This reaction is catalyzed by the enzyme magnesium chelatase, which consists of three distinct subunits. In Rhodobacter capsulatus and other bacteria, these subunits are named BchI, BchD, and BchH whereas in plants, algae, and cyanobacteria, the homologous proteins generally are named ChlI, ChlD, and ChlH with average molecular masses of 40, 70, and 140 kDa, respectively (1). There appear to be two ATP-dependent processes in the magnesium chelatase reaction (2, 3). One involves the BchH subunit and its binding of protoporphyrin IX, which is enhanced by ATP and Mg2+ (4, 5). The other is the ATP- and Mg2+-dependent activation of BchI and BchD (1), which associate in a complex upon ATP binding (6–9). Both the BchI and the BchH subunits have ATPase activity, although it is unclear whether ATP hydrolysis is required for the two processes (7, 10, 11). The noncovalent binding of protoporphyrin IX to BchH implies that it plays the role of a catalytic subunit in the metallation reaction (1). Recently, it was found that the smallest subunit, BchI, belongs to the large family of AAA+ proteins (ATPases associated with various cellular activities) (12, 13) and that the C terminus of the intermediate subunit, BchD, is similar to an integrin I domain (12).
AAA+ proteins are Mg2+-dependent ATPases, and they usually are found in various multimeric states (14, 15). They play essential roles in a broad range of cellular activities, including proteolysis, membrane fusion, cytoskeletal regulation, protein folding, DNA replication (13–15), and porphyrin metallation (12). It has been shown for several AAA+ proteins that a significant change of conformation occurs during the ATP hydrolysis cycle, and it has been suggested that this may be a general feature of these proteins (15–20). Integrins are known to participate in cell–matrix and cell–cell interactions (21, 22). They are involved in signaling to and from cells in various physiological processes, including morphogenesis, cell migration, immunity, and wound healing (23, 24). The connection between magnesium chelatase, AAA+ proteins, and integrins has become especially exciting since an Arabidopsis thaliana gun5 mutant, deficient in chloroplast-to-nucleus signal transduction, recently was found to be deficient in the 140-kDa magnesium chelatase subunit (25). Thus, these recent findings suggest magnesium chelatase and Mg2+ chelation as key components in chloroplast development (26–28).
Among 357 lethal barley mutants deficient in chlorophyll biosynthesis and chloroplast development, 20 are deficient in magnesium chelatase activity (29). Among the seven mutant alleles of the Xantha-h gene encoding the smallest subunit of barley magnesium chelatase (corresponding to BchI), four are recessive and three are semidominant. The homozygous mutants are all yellow because of lack of chlorophyll. The heterozygotes carrying the recessive allele are green and indistinguishable from WT, whereas the heterozygotes carrying the semidominant allele are pale green. The recessive mutations prevent transcription of the gene (30), and the semidominant alleles are missense mutations leading to changes in single amino acid residues (31). The amino acid changes in the three semidominant mutants named Xantha-hclo 125, -hclo 157, and -hclo 161 are D207N, R289K, and L111F, respectively (sequence numbers of R. capsulatus BchI). The pale green phenotype of the semidominant heterozygous barley mutants were considered to be due to a competitive binding of the mutated and WT 40-kDa subunits to the other subunits, resulting in an inhibition of the magnesium chelatase reaction (31). The competitive inhibition could affect the interaction between the 40- and 70-kDa subunits, which are known to undergo an ATP-dependent activation step (6, 8, 9). However, the recently solved structure of the R. capsulatus BchI reveals that 40-kDa subunits interact with each other in a hexameric ring complex (12). In the BchI structure, an N-terminal domain containing the Walker A and B motif (32) is connected with a C-terminal four-helix bundle by a long helical region. All three altered amino acid residues are located in the interface between two neighboring subunits in the AAA+ hexamer and close to the areas forming the ATP-binding site (Fig. 1). L111 is located on one side whereas D207 and R289 are on the opposite side of the wedge-shaped structure. The three mutations provide a unique possibility to study the interaction between subunits in an AAA+ hexamer. In contrast to the situation for the barley magnesium chelatase, a heterologous expression system for the R. capsulatus magnesium chelatase exists (34). Therefore, the three mutations were made in the R. capsulatus bchI gene encoding the BchI protein.
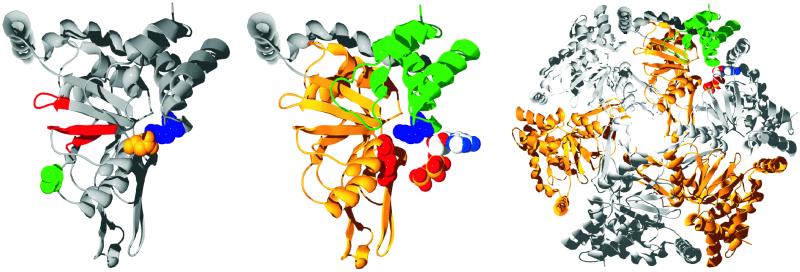
Figure 1.
(Left) The structure of R. capsulatus BchI is shown as a ribbon diagram. The Walker AB motif and the sensor 1 region are colored red, and the residues that are changed in the mutants are shown as space-filled residues, with L111 colored green, D207 colored gold, and R289 colored blue. (Center) R. capsulatus BchI folds into two domains connected by a long helical region colored in gray. The N-terminal is gold, and the C-terminal domain is green. Two important residues in AAA+ proteins, the arginine finger and the sensor arginine, are shown as space-filled residues and are colored red and blue, respectively. An ATP from a neighboring subunit is modeled next to BchI. (Right) From the BchI hexamer, it is clearly seen that the mutated residues lie at the interface between two neighboring subunits. All images were made with swiss-pdbviewer (33), rendered with pov-ray (Persistence of Vision, Indianapolis), and colored by corel draw (Corel, Ottawa).
Several AAA+ proteins are known to form ring-closed hexamers, bind and hydrolyze ATP, and undergo conformational changes (14–17, 35). To elucidate which of these properties determines the activity of BchI, and thereby the activity of magnesium chelatase, we modified BchI by site-directed mutagenesis. The modified BchI subunits are deficient in ATP hydrolysis and magnesium chelatase activity. We show that modified BchI is able to mix with WT BchI in oligomeric complexes. The oligomerization is ATP-dependent but results in inactive complexes. These observations provide a model for the dysfunction of the semidominant barley mutants Xantha-hclo 125, -hclo 157, and -hclo 161.
Materials and Methods
Magnesium Chelatase from Barley Mutant Seedlings.
Grains of barley, both WT and Xantha-hclo 157, were planted in moist vermiculite and grown in 12-h light/12-h dark cycles at 20°C. After 7 days, the seedlings were harvested. The yellow homozygous Xantha-hclo 157 seedlings were separated from the dark- and light-green seedlings. Soluble protein extracts were prepared as described by Kannangara et al. (36). Total soluble protein concentrations for WT and homozygous Xantha-hclo 157 were determined by Bio-Rad Protein Assay based on the Bradford dye-binding procedure (37). The magnesium chelatase activity assays were performed as described below. When assayed individually, 100 μg of soluble protein extract was used. In assays with combined WT and homozygous Xantha-hclo 157 extracts, 100 μg from each preparation was used.
Mutagenesis, Expression, and Purification of BchI Proteins.
To study the missense mutations identified in the barley Xantha-h gene (31), the corresponding mutations were made in the R. capsulatus bchI gene. The amino acid changes L111F, D207N, and R298K were introduced by site-directed mutagenesis with the QuikChange method (Stratagene) by using Vent DNA polymerase (New England Biolabs) and a PTC-200 DNA Engine (MJ Research, Cambridge, MA). Each mutation altered a restriction enzyme site, and positive clones were identified by agarose gel electrophoresis after digestion with the relevant restriction enzyme. The following primers were used: L111Fu 5′-CGTCGATCTGCCGtTCGGCGTGTCGG-3′, L111Fl 5′-CCGACACGCCGAaCGGCAGATCGACG-3′, D207Nu 5′-GGCCGCAGCTTCTGaACCGTTTCGGCC-3′, D207Nl 5′-GCCGAAACGGTtCAGAAGCTGCGGCCG-3′, R289Ku 5′-GGTCCGACGGTtTaaaGGGCGAGCTGACGC-3′, and R289Kl 5′-GTCAGCTCGCCCtttAaACCGTCGGACCCC-3′. Introduced mismatches are in lowercase, and restriction sites are in bold. L111F eliminated a BsrBI site whereas D207N and R289K introduced a XmnI and DraI site, respectively. One positive clone from each mutation was sequenced to confirm that no other mutations had been generated. Expression and purification of BchI, BchD, and BchH were performed as described by Willows and Beale (34).
Magnesium Chelatase Assay.
All magnesium chelatase activity assays were performed in a 100-μl reaction mixture. The protein content in each 100 μl was 31 μg BchH, 6 μg BchD, and 2.5–5 μg BchI for the R. capsulatus system. The assay contained 4 mM ATP, 10 mM phosphocreatine (Sigma), 0.025 unit/μl creatine phosphokinase (Sigma), 8% glycerol, 15 mM MgCl2, 1 μM protoporphyrin IX, 4 mM DTT, and 50 mM Tricine⋅NaOH, pH 8.5. After incubation at 32°C, the assays were stopped by adding 900 μl of alkaline acetone [14 M NH4OH/acetone/H2O (1:80:20 vol/vol/vol)], and the precipitate was removed by centrifugation. A fluorescence spectrum of the supernatant was taken from 550 to 650 nm with excitation at 418 nm by using a FluoroMax-2 (ISA-SPEX, Edison, NJ) for R. capsulatus or a LS 50 B (Perkin–Elmer) for barley.
ATP Hydrolysis Assays.
ATPase activity was measured by hydrolysis of [8-14C]ATP (specific activity, 2.1 GBq/mmol; Perkin–Elmer). Each assay contained 50 mM Tricine⋅NaOH, pH 8.5, 8% glycerol, 15 mM MgCl2, 4 mM DTT, 80 μM unlabeled ATP, 20 μM radioactive ATP, and 0.2 μM BchI in a total volume of 10 μl. During incubation at 32°C, samples (1 μl) were taken out at different times and quenched by the addition of 1 μl of 2 M formic acid. The ATP, ADP, and AMP components then were separated by TLC (polyethyleneimine-cellulose F; Merck), using 1 M formic acid and 0.7 M LiCl as the mobile phase. A STORM phosphorimager system (Amersham Pharmacia) was used for detection and quantification.
Biomolecular Interaction Analysis by Surface Plasmon Resonance.
All surface plasmon resonance experiments were performed on a Biacore 2000 biosensor (Biacore AB, Stockholm) at 28°C and a continuous flow of 10 μl/min. The running buffer, with or without ATP (1 mM), containing 50 mM Tricine⋅NaOH, pH 8.0, 15 mM MgCl2, 1 mM DTT, and 0.005% P20 (Biacore), was filtered and degassed before use. WT BchI was biotinylated with the biotin linker EZ-Link Sulfo-NHS-LC-Biotin (Pierce) or EZ-Link 5-(Biotinamido) pentylamine (Pierce). Electrospray MS indicated that a minimum of 80% of the BchI was derivatized with a single biotin linker, with the remainder having two biotins per molecule. The biotinylated BchI was immobilized in channel 2 of a streptavidin-coated sensor chip SA. Channel 1 served as an internal reference, and the signal from this channel was subtracted from channel 2. During analysis, BchI protein samples (WT, L111F, D207N, and R289K) with a concentration of 10 μM were injected over the chip at 20 μl/min for 5 min. After protein injection, the dissociation of interacting protein was monitored for an additional 150 s. In the end of each sample cycle, the sensor chip was stripped from interacting protein by washing with running buffer without ATP followed by regeneration with 1 M NaCl before additional samples were injected. All experiments were conducted in triplicate.
Gel Filtration.
Aliquots (10 μl) of WT, L111F, D207N, or R289K BchI (3 μg/μl) were injected to a Superose 12 column connected to a SMART system (Amersham Pharmacia). The running buffer, with or without ATP (1 mM), containing 50 mM Tricine⋅NaOH, pH 8.0, 15 mM MgCl2, and 1 mM DTT, was filtered and degassed before use. Each protein was passed through the column both in the presence and absence of ATP. When ATP was included in the running buffer, the proteins were preincubated on ice for 15–20 min with 1 mM ATP. The relative protein content in each elution fraction was determined by SDS/PAGE (38) and stained with colloidal Coomassie brilliant blue (39), and the bands were quantified with a Personal Densitometer SI (Amersham Pharmacia).
Results
Magnesium Chelatase Assays with Mutants.
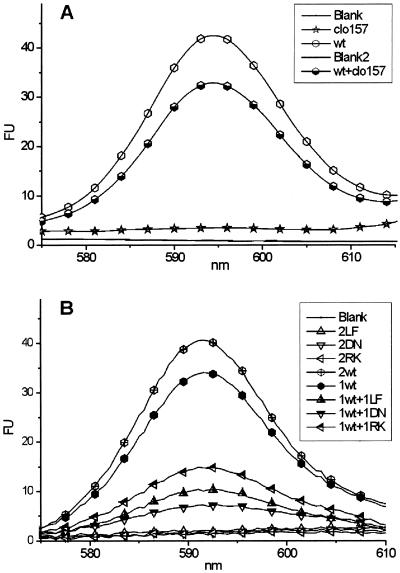
To study the effect of the semidominant mutant alleles in vitro, cleared cell extracts were prepared from WT and homozygous Xantha-hclo 157 barley, corresponding to the R. capsulatus R289K BchI. When the two preparations were assayed individually for magnesium chelatase activity, WT had a high activity whereas Xantha-hclo 157 had no activity. In an assay in which the two extracts were combined, the total activity was lower than for the WT alone (Fig. 2A). This finding indicated that the Xantha-hclo 157-encoded protein disrupted the activity of magnesium chelatase and is consistent with the semidominant chlorina phenotype. To study the observation more thoroughly, R. capsulatus BchI with the modifications D207N, R289K, and L111F corresponding to Xantha-hclo 125, Xantha-hclo 157, and Xantha-hclo 161, respectively, was made by site-directed mutagenesis of the R. capsulatus bchI gene. A standard magnesium chelatase assay was used to analyze the functionality of the modified BchI. WT BchI together with BchD and BchH catalyzed the insertion of Mg2+ into protoporphyrin IX. When WT BchI was replaced by either the L111F, D207N, or R289K BchI, no measurable magnesium chelatase activity was detected. Assays containing both WT and modified BchI had a lower activity than the WT alone, confirming the observation with barley crude cell extract and showing that L111F, D207N, and R289K BchI disrupted magnesium chelatase activity (Fig. 2B).
Figure 2.
(A) Crude cell extracts from barley, both WT and Xantha-hclo 157, were assayed for magnesium chelatase activity. Xantha-hclo 157 extracts alone had no activity, and they inhibited the activity of the WT when they were mixed. (B) Purified magnesium chelatase subunits from R. capsulatus were assayed for magnesium chelatase activity. WT BchH and BchD subunits were in all assays with various combinations of WT and modified BchI. The numbers in the legend indicate the relative amount of BchI (1 corresponds to 2.5 μg, and 2 corresponds to 5 μg). Symbols were added to identify the curves.
ATPase Activity.
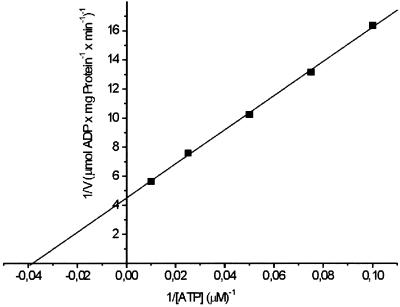
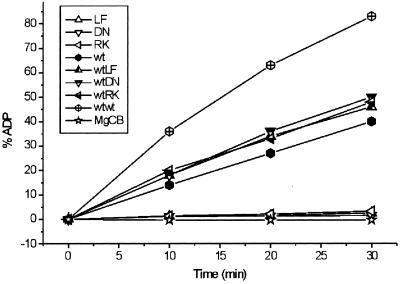
Purified BchI hydrolyzes ATP to ADP and phosphate. WT BchI ATPase activity, when measured in a continuous linear assay over 40 min, gave a Vmax of 220 nmol/(mg protein × min) with a Km of 26 μM and kcat of 0.14 s−1 (Fig. 3). L111F, D207N, and R289K BchI possess ≈5% of the WT activity when assayed under the same conditions as the WT (Fig. 4). To determine whether the modified BchI disrupted the ATPase activity of the WT as it disrupted the magnesium chelatase activity, assays with WT and modified BchI in equimolar ratio were set up. In these assays, the WT activity was not inhibited and the activity correlated well with the sum of WT and modified BchI assayed separately (Fig. 4).
Figure 3.
A Lineweaver–Burk plot of ATPase activity of WT R. capsulatus BchI. Vmax and Km were found to be 220 nmol (mg protein)−1 min−1 and 26 μM, respectively. Seventy-six nanograms of BchI was used in the assays.
Figure 4.
Different combinations of modified and WT R. capsulatus BchI were assayed for ATPase activity. Each of the three modified BchI proteins on its own possesses about 5% of WT activity. When L111F, D207N, or R289K BchI is added to WT BchI, the activity of the WT is not inhibited.
Protein–Protein Interaction Analyzed by Gel Filtration.
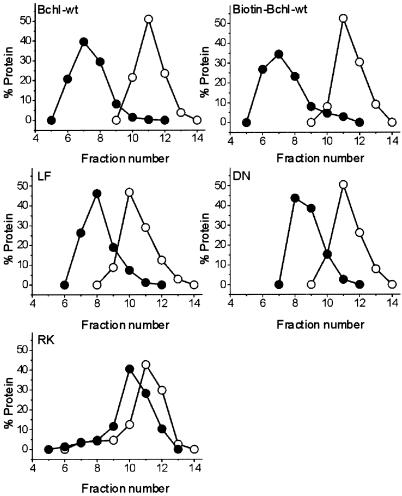
In gel-filtration experiments, a shift in elution volume toward that corresponding to a larger molecular size occurred for WT, biotinylated WT, L111F, D207N, and R289K BchI when ATP (1 mM) was included in the running buffer (Fig. 5). WT and biotinylated WT BchI were shifted the most followed by D207N, L111F, and R289K. After the shift, the proteins eluted at a volume corresponding to a molecular mass of 90–130 kDa. The expected molecular mass for a hexamer is 228 kDa. The peak of BchI eluted in the presence of ATP varied depending on the concentration of BchI injected, with higher concentrations giving higher apparent molecular masses. The disagreement in molecular mass thus can be explained by the equilibrium between monomeric and oligomeric BchI. As BchI passes through the column in the presence of ATP, it is in an equilibrium between monomer and hexamer and consequently is not eluted at the volume expected for a hexamer. In the absence of ATP, BchI migrates as a monomer.
Figure 5.
Mutant and WT BchI were passed through a Superose 12 gel-filtration column in the absence of ATP (○). When 1 mM ATP was included in the buffer, the proteins interacted and a shift toward an increase in size occurred (●). Elution profile of standard proteins: ribonuclease A, 13.7-kDa fraction 19; chymotrypsinogen A, 25-kDa fraction 15; ovalbumin, 43-kDa fraction 11; BSA, 67-kDa fractions 9–10; void, 2,000-kDa fraction 2.
Protein–Protein Interaction Analyzed by Surface Plasmon Resonance.
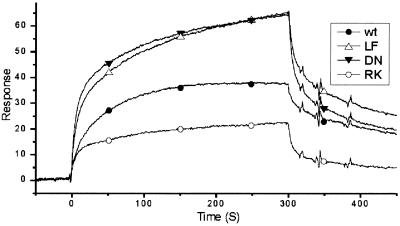
The capacity of WT and modified BchI to interact and form oligomers in the presence or absence of ATP also was analyzed with a Biacore 2000. WT BchI was biotinylated and confirmed to be active in magnesium chelatase activity assays, ATPase assays, and ATP-dependent complex formation in gel filtration before being immobilized on a Sensor Chip SA (Biacore) and used for all Biacore analysis. When ATP was omitted from the running buffers, no interactions were detected between the immobilized BchI on the sensor chip with either the WT or modified BchI proteins. However, when ATP (1 mM) was included in the running buffer of the Biacore, both WT and modified BchI proteins interacted with the biotinylated WT BchI protein immobilized on the surface of the SA chip (Fig. 6). D207N and L111F had association and dissociation rates higher than WT. R289K had a lower association rate but a higher dissociation rate than WT (Fig. 6). The WT BchI has a slower association rate than any of the modified BchI proteins but it forms oligomers that are more stable than those of modified BchI as indicated by the slower dissociation rate. The gel-filtration results correlate well with the Biacore observations and show that ATP is required for interaction and oligomerization of BchI.
Figure 6.
Biomolecular interactions were analyzed by using a Biacore 2000. The sensograms show how mutant and WT BchI interact with the immobilized WT BchI on the sensor chip in the presence of 1 mM ATP. Symbols were added to identify the curves.
Discussion
The requirement for ATP in the activation step of the magnesium chelatase reaction is well documented (1–3). ATP has been considered to be involved in the formation of a complex between the I and D subunits, which is created upon ATP binding and does not depend on ATP hydrolysis (2, 6, 7). Here, we present data showing that the binding, but not the hydrolysis, of ATP also is needed in a step involving only BchI, namely, the formation of oligomeric BchI. It has yet to be determined whether ATP also binds to BchD or whether BchI is the only ATP-binding subunit in the BchI-, BchD-, and ATP-dependent activation step.
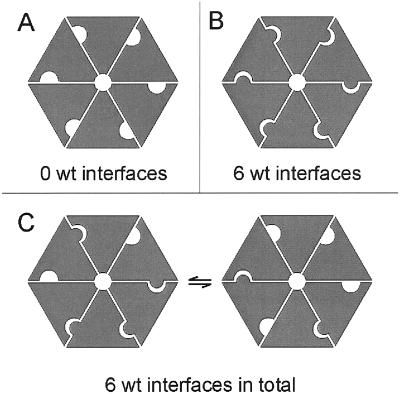
The magnesium chelatase activity measured in vitro was inhibited by the addition of L111F, D207N, and R289K BchI subunits. In contrast, the ATPase activity of WT BchI was not affected by the addition of mutant BchI. In BchI and other AAA+ proteins, the ATP-binding site is located at the interface between two neighboring subunits in the hexamer. Residues from one side of the wedge-shaped monomer, together with residues from the other side of a neighboring monomer, build up the ATP-binding site (14, 15, 18, 40). The mutants used in this study all give rise to changes of amino acid residues positioned in the interface between two neighboring subunits. The affected residue L111 is placed on the Walker side of the wedge whereas D207 and R289 are placed on the opposite side (Fig. 1). In a hexamer built up from any of the three L111F, D207N, or R289K mutant BchI proteins, it is not possible to obtain a WT interface, because one of the sides of the interface will always contain a modified residue. In a hexamer built up of a mixture of mutant and nonmutant BchI, depending on how a WT and a mutant subunit interact, the interface between the two subunits can be either WT or mutant. The total ATPase activity in an equimolar mixture of WT and modified BchI corresponded to the sum of WT and modified BchI assayed separately. This observation suggests that a pure WT hexamer is not required for ATPase activity. Rather, every interface within the hexamer acts autonomously. In an equimolar mixture of modified and WT BchI, the fraction of hexamers consisting purely of WT subunits is theoretically 0.56 = 0.016, provided that modified and WT proteins form a unit of a hexamer with equal ease. Thus, the expected activity from such a mixture would be 1.6% of WT activity if only the hexamers made up from WT subunits were active. Our value of 53% is much closer to the expected activity of 52.5% for a WT–mutant mixture when every subunit within the hexamer acts autonomously (2 × 0.52 × 1 + 2 × 0.52 × 0.05 = 0.525). The retained activity observed is explained by the number of WT interfaces remaining conserved even in the presence of modified BchI (Fig. 7).
Figure 7.
Model showing the formations of interfaces in mutant (A) and WT (B) hexamers and the conservation of the number of WT interfaces in an equimolar mixture (C). BchI monomers, gray wedges.
If there is a direct correlation between the ATPase activity of BchI and magnesium chelatase activity, an equimolar mixture of WT and modified BchI should yield 52.5% magnesium chelatase activity when combined with BchD and BchH. However, only 20–35% magnesium chelatase activity was observed depending on the modified BchI protein added. This observation suggests that modified BchI present in the hexamer abolishes the magnesium chelatase activity and supports the idea that the functional unit of BchI is the hexamer. The observed activities with equimolar mixture of WT and modified BchI suggest that one or two modified subunits in a hexamer still could allow activity. However, it is also likely that one modified subunit in a hexamer is enough to inactivate the complex, although such low activity (1.6%) was not observed. The reason could be that WT BchI has lower affinity for a modified subunit, which is supported by our Biacore and gel-filtration experiments. Therefore, it is possible that WT hexamers are overrepresented in an equimolar mixture of WT and modified BchI subunits. Our results strongly suggest that the semidominance of the Xantha-hclo 125, -hclo 157, and -hclo 161 mutations is due to the ability of the defective 40-kDa subunits to combine with WT 40-kDa subunits in leaves of heterozygous mutant plants. The small proportion of hexamers consisting only of WT 40-kDa subunits rescues the heterozygous plants but is not enough to provide sufficient magnesium chelatase activity to give WT levels of chlorophyll.
Several studies show that ATP binding but not ATP hydrolysis is important for the BchI–BchD activation step of the magnesium chelatase reaction (6, 7). However, ATPase activity of BchI alone has been demonstrated in several studies (7, 10, 41). This study provides strong evidence that the hydrolysis of ATP by BchI is essential for magnesium chelation. The modified proteins L111F, D207N, and R289K form oligomers in the mere presence of ATP but are deficient in ATPase activity. When assayed for magnesium chelatase activity, modified BchI together with BchD and BchH shows no activity. Therefore, oligomerization is insufficient to activate BchI in the magnesium chelatase reaction, which suggests that an additional event is needed for magnesium chelation to occur. This also is supported by a study with N-ethylmaleimide-modified magnesium chelatase subunits (42). The additional event may be a structural change on ATP hydrolysis. Structural changes have been observed in other AAA+ hexamers during the ATP-dependent hydrolysis cycle (16, 17, 19, 20). Two residues known to be involved in ATP-dependent conformational changes in AAA+ proteins are the two arginine residues normally referred to as the “arginine finger” and the “sensor arginine,” respectively (13, 15, 20). These arginines, placed in the interface between two subunits in the hexamer, interact with ATP and thereby induce a conformational change (13, 18, 20). In BchI, these residues are R208 and R289, respectively. R208 is located in the N-terminal domain, and R289 is located in the C-terminal domain (Fig. 1). We have shown that, despite the lack of the sensor arginine in R289K, BchI is still capable of binding ATP and forming oligomers. In D207N, it is likely that the arginine finger, R208, is affected structurally by the modification because the charge of the neighboring residue is changed from negative to neutral. The displacement of the arginine finger or the sensor arginine does not disrupt ATP binding and oligomerization but does disrupt ATP hydrolysis. We postulate that the absence of ATP hydrolysis prevents the N- and C-terminal domains from being brought into a functional conformation, and without this conformational change, the magnesium chelatase reaction stalls in an early state.
Acknowledgments
We are grateful to Simon Gough and Salam Al-Karadaghi for fruitful discussions. This work was made possible thanks to generous support from the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (FORMAS) and The Lund and Malmö City Foundation (to M.H.), The Lars Hierta Foundation and The Sven and Lilly Lawski Foundation (to A.H.), and the Australian Research Council (Grant A09905713) and an International Researcher Exchange Scheme Award (Grant X00001636) (to R.D.W.). We also acknowledge Macquarie University Centre for Analytical Biotechnology.
References
- 1.Willows R D, Gibson L C, Kannangara C G, Hunter C N, von Wettstein D. Eur J Biochem. 1996;235:438–443. doi: 10.1111/j.1432-1033.1996.00438.x. [DOI] [PubMed] [Google Scholar]
- 2.Walker C J, Weinstein J D. Biochem J. 1994;299:277–284. doi: 10.1042/bj2990277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker C J, Willows R D. Biochem J. 1997;327:321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karger G A, Reid J D, Hunter C N. Biochemistry. 2001;40:9291–9299. doi: 10.1021/bi010562a. [DOI] [PubMed] [Google Scholar]
- 5.Jensen P E, Gibson L C, Hunter C N. Biochem J. 1998;334:335–344. doi: 10.1042/bj3340335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson L C, Jensen P E, Hunter C N. Biochem J. 1999;337:243–251. [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen P E, Gibson L C, Hunter C N. Biochem J. 1999;339:127–134. [PMC free article] [PubMed] [Google Scholar]
- 8.Papenbrock J, Gräfe S, Kruse E, Hänel F, Grimm B. Plant J. 1997;12:981–990. doi: 10.1046/j.1365-313x.1997.12050981.x. [DOI] [PubMed] [Google Scholar]
- 9.Gräfe S, Saluz H P, Grimm B, Hänel F. Proc Natl Acad Sci USA. 1999;96:1941–1946. doi: 10.1073/pnas.96.5.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansson M, Kannangara C G. Proc Natl Acad Sci USA. 1997;94:13351–13356. doi: 10.1073/pnas.94.24.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen B L, Jensen P E, Gibson L C, Stummann B M, Hunter C N, Henningsen K W. J Bacteriol. 1998;180:699–704. doi: 10.1128/jb.180.3.699-704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodje M N, Hansson A, Hansson M, Olsen J G, Gough S, Willows R D, Al-Karadaghi S. J Mol Biol. 2001;311:111–122. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- 13.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 14.Vale R D. J Cell Biol. 2000;150:F13–F19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura T, Wilkinson A J. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 16.Hanson P I, Roth R, Morisaki H, Jahn R, Heuser J E. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 17.Rouiller I, Butel V M, Latterich M, Milligan R A, Wilson-Kubalek E M. Mol Cell. 2000;6:1485–1490. doi: 10.1016/s1097-2765(00)00144-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Shaw A, Bates P A, Newman R H, Gowen B, Orlova E, Gorman M A, Kondo H, Dokurno P, Lally J, et al. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 19.Lenzen C U, Steinmann D, Whiteheart S W, Weis W I. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 20.Song H K, Hartmann C, Ramachandran R, Bochtler M, Behrendt R, Moroder L, Huber R. Proc Natl Acad Sci USA. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 22.Emsley J, Knight C G, Farndale R W, Barnes M J, Liddington R C. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 23.Bella J, Berman H M. Struct Fold Des. 2000;8:R121–R126. [Google Scholar]
- 24.van der Flier A, Sonnenberg A. Cell Tissue Res. 2001;305:285–298. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki N, Brusslan J A, Larkin R, Nagatani A, Chory J. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kropat J, Oster U, Rudiger W, Beck C F. Plant J. 2000;24:523–531. doi: 10.1046/j.1365-313x.2000.00898.x. [DOI] [PubMed] [Google Scholar]
- 27.Jarvis P. Curr Biol. 2001;11:R307–R310. doi: 10.1016/s0960-9822(01)00171-3. [DOI] [PubMed] [Google Scholar]
- 28.Rodermel S. Trends Plant Sci. 2001;6:471–478. doi: 10.1016/s1360-1385(01)02085-4. [DOI] [PubMed] [Google Scholar]
- 29.Henningsen K W, Boynton J E, von Wettstein D. Dan Acad Sci Lett Biol Skr. 1993;42:1–349. [Google Scholar]
- 30.Jensen P E, Willows R D, Petersen B L, Vothknecht U C, Stummann B M, Kannangara C G, von Wettstein D, Henningsen K W. Mol Gen Genet. 1996;250:383–394. doi: 10.1007/BF02174026. [DOI] [PubMed] [Google Scholar]
- 31.Hansson A, Kannangara C G, von Wettstein D, Hansson M. Proc Natl Acad Sci USA. 1999;96:1744–1749. doi: 10.1073/pnas.96.4.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 34.Willows R D, Beale S I. J Biol Chem. 1998;273:34206–34213. doi: 10.1074/jbc.273.51.34206. [DOI] [PubMed] [Google Scholar]
- 35.Patel S, Latterich M. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- 36.Kannangara C G, Vothknecht U C, Hansson M, von Wettstein D. Mol Gen Genet. 1997;254:85–92. doi: 10.1007/s004380050394. [DOI] [PubMed] [Google Scholar]
- 37.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Fling S P, Gregerson D S. Anal Biochem. 1986;155:83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- 39.Neuhoff V, Arold N, Taube D, Ehrhardt W. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 40.Bochtler M, Hartmann C, Song H K, Bourenkov G P, Bartunik H D, Huber R. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 41.Petersen B L, Kannangara C G, Henningsen K W. Arch Microbiol. 1999;171:146–150. doi: 10.1007/s002030050692. [DOI] [PubMed] [Google Scholar]
- 42.Jensen P E, Reid J D, Hunter C N. Biochem J. 2000;352:435–441. [PMC free article] [PubMed] [Google Scholar]