Abstract
Finding new protein-coding genes is one of the most important goals of eukaryotic genome sequencing projects. However, genomic organization of novel eukaryotic genomes is diverse and ab initio gene finding tools tuned up for previously studied species are rarely suitable for efficacious gene hunting in DNA sequences of a new genome. Gene identification methods based on cDNA and expressed sequence tag (EST) mapping to genomic DNA or those using alignments to closely related genomes rely either on existence of abundant cDNA and EST data and/or availability on reference genomes. Conventional statistical ab initio methods require large training sets of validated genes for estimating gene model parameters. In practice, neither one of these types of data may be available in sufficient amount until rather late stages of the novel genome sequencing. Nevertheless, we have shown that gene finding in eukaryotic genomes could be carried out in parallel with statistical models estimation directly from yet anonymous genomic DNA. The suggested method of parallelization of gene prediction with the model parameters estimation follows the path of the iterative Viterbi training. Rounds of genomic sequence labeling into coding and non-coding regions are followed by the rounds of model parameters estimation. Several dynamically changing restrictions on the possible range of model parameters are added to filter out fluctuations in the initial steps of the algorithm that could redirect the iteration process away from the biologically relevant point in parameter space. Tests on well-studied eukaryotic genomes have shown that the new method performs comparably or better than conventional methods where the supervised model training precedes the gene prediction step. Several novel genomes have been analyzed and biologically interesting findings are discussed. Thus, a self-training algorithm that had been assumed feasible only for prokaryotic genomes has now been developed for ab initio eukaryotic gene identification.
INTRODUCTION
The shear scale of current eukaryotic genomic sequencing is astounding. As of October 2005, 531 eukaryotic sequencing projects have been registered (www.genomesonline.org). All but 161 ‘expressed sequence tag (EST)-only’ projects generate contigs of genomic DNA and 49 genome projects have been already completed. While extracting information about protein-coding genes from this enormous and growing collection of DNA sequences is of primary importance, this goal still presents a significant challenge.
Gene annotation in new eukaryotic genomic sequence could be done either by intrinsic (ab initio) methods (1–6) or by the methods using extrinsic evidence (7–21). Developing ab initio gene prediction methods for eukaryotic genomes has commonly been considered a difficult task. This difficulty for novel genomes is aggravated by the absence of the sufficiently large and reliable training sets. Specialized gene finding methods using extrinsic information frequently involve mapping relevant cDNA, EST and protein sequences to genomic DNA (7–17). Yet another type of extrinsic evidence is provided by the alignment of genomic DNA in question to a reference genome to extract specific patterns of matching nucleotides correlating with aligned protein-coding regions (18–21).
The general drawback of the extrinsic approaches is that they are inherently database-dependent and may fall short of providing sufficient support for gene annotation in novel genomes. Majority of ESTs and cDNAs (if available at all) are related to highly expressed genes and frequently represent partial mRNA (22). Even if the reference sequence, EST, cDNA and so on are available, pin-pointing exon boundaries and delineation of short exons may present a challenge (22). Therefore, improvement of the ab initio gene finding could provide a critically important resource for annotation of novel genomes.
Parameters of statistical gene models have been traditionally derived by supervised training. For instance, for eukaryotic genomes that have been completed already some time ago such as Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens the statistical gene models for advanced gene finders, e.g. GenScan (1), Genefinder (http://ftp.genome.washington.edu/cgi-bin/genefinder_req.pl) and Genie (4), were determined from sets of genes validated by EST, cDNA or protein data. Supervised training requires a rather large set of experimentally validated genes, therefore sequencing of a novel eukaryotic genome should have been in progress for quite some time before the required set of genes validated by EST or cDNA data would be available. Here we show that accurate statistical gene models for novel genomes could be derived by another type of training which works straight with anonymous genomic data available at a rather early stage of the sequencing project.
We have designed a new eukaryotic gene finding algorithm using hidden Markov models (HMM) and employing the unsupervised training procedure. We had to address the problem of derivation of statistical gene models from data with missing features, i.e. DNA sequences whose nucleotides are not labeled as coding or non-coding. In the HMM theory similar problems are generally solved by the Baum–Welch algorithm (23,24) seeking to identify the vector of parameters possessing the maximum likelihood given the observed data. The convergence properties of the Baum–Welch algorithm are not predictable beyond the provable statement of its convergence to a point of a local maximum, which is common for any general algorithm of expectation-maximization (EM) type. Another alternative for the HMM parameter estimation, which is readily amenable for using flexible training strategies, is the Viterbi training. The Baum–Welch and Viterbi training algorithms have already been used to estimate HMM parameters for gene finding algorithms, e.g. the prokaryotic gene finders GeneMarkS (25) and EasyGene (26). Note that for non-HMM prokaryotic gene identification algorithms non-supervised procedures for estimation of gene model parameters have also been described (27–29). However, it was commonly assumed, that implementation of a non-supervised procedure producing high precision gene models is not feasible for eukaryotic genomes with more complex gene organization. These difficulties include the higher dimension of the parameter space and apparently more complex profile of the likelihood function owing to the high level of noise introduced within iterations by abundant chances of mislabeling of nucleotides situated in non-coding regions.
Generation of gene predictions for a novel eukaryotic genome, as described below, occurs in parallel with the unsupervised iterative estimation of gene model parameters by the Viterbi training. At each iteration, the algorithm takes genomic sequence labeled by the Viterbi algorithm at the previous iteration into coding and non-coding regions, re-estimates model parameters, and computes a new sequence parse and labeling. This general path of the iterative Viterbi training process is modified by addition of restrictions on possible changes of parameters to ensure convergence of the iteration process to the biologically relevant point. At the point of convergence the set of sequence labels is transformed into the list of gene predictions, the program output.
By using test sets generated for well-studied genomes A.thaliana, C.elegans and D.melanogaster, it was shown that the new self-training algorithm generates gene predictions with comparable or higher accuracy as compared with the algorithm using models estimated by a supervised training. Finally, the new program GeneMark.hmm ES-3.0 (E—eukaryotic; S—self-training; 3.0—the version number) was applied for gene prediction in draft genomes of Anopheles gambiae, Ciona intestinalis, Chlamydomonas reinhardtii and Toxoplasma gondii. This analysis revealed earlier unknown genes in each genome. Most interesting biological findings are discussed in the last section of the paper.
MATERIALS AND METHODS
Datasets
In this study we have used complete genomic sequences of A.thaliana, C.elegans, and D.melanogaster (GenBank, www.ncbi.nlm.nih.gov) as well as draft genomes of A.gambiae (Ensembl, www.ensembl.org), C.intestinalis (DOE Joint Genome Institute, www.jgi.doe.gov), C.reinhardtii (DOE Joint Genome Institute, www.jgi.doe.gov) and T.gondii (ToxoDB, toxodb.org). The G + C content of the genomes considered in this study varies from 35% (C.elegans) to 63% (C.reinhardtii).
Training sets
Annotations of the A.thaliana, C.elegans and D.melanogaster genomes given in the TIGR Arabidopsis database (www.tigr.org), WormBase (www.wormbase.org) and FlyBase (www.flybase.net), respectively, were used for compiling 1000 gene strong training datasets (with no overlap with the corresponding test sets), for each of the three species. In these sets we have included sequences containing genes either validated by the cDNA/EST mapping or confirmed by the RT–PCR technique. These sequence sets were used for deriving statistical gene models by supervised training.
Test sets
The following criteria were used for admission of gene containing sequences to the test sets: (i) a gene should possess ATG start codon and canonical acceptor/donor sites; (ii) intron/exon structure should be supported by EST/cDNA alignment (12); (iii) no alternative isoforms supported by EST/cDNA should be mentioned in annotation; and (iv) a gene should not overlap with any other annotated gene. Sequences containing multiple genes are preferable for the accuracy assessment (30). To include into the test set a region of genomic DNA with multiple validated genes situated adjacent to each other, we have tested annotated intergenic regions for genes missed in annotation by searching against databases of EST/cDNA sequences (12). However, even with these precautions we could not guarantee that no gene remained in intergenic regions of the test sequences that contained three or more adjacent validated genes. For A.gambiae, C.elegans, C.intestinalis, C.reinhardtii and T.gondii the above stated rules of admission to a test set did not produce many records with multiple genes. Therefore, these test sets contained mostly one validated gene per sequence. The sizes (in terms of number of genes) of the test sets are as follows: A.gambiae—144, A.thaliana—1026, C.elegans—183, C.intestinalis—314, C.reinhardtii—43, D.melanogaster—361 and T.gondii—65.
GeneMark.hmm E-3.0 for eukaryotic genomes, supervised model parameterization
The initial GeneMark.hmm algorithm was developed for gene finding in prokaryotic genomes (31) and later was extended for use of eukaryotic gene models GeneMark.hmm E-1.0 (A. Lukashin and M. Borodovsky, unpublished data). The next program version GeneMark.hmm E-2.0 (G. Tarasenko and M. Borodovsky, unpublished data) has been used for annotation of several plant genomes including A.thaliana (30,32) and Oryza sativa (33). Here we describe the latest program version, GeneMark.hmm E-3.0.
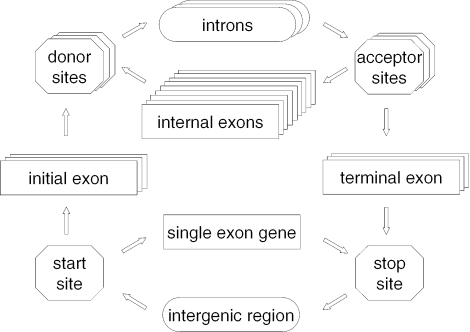
The statistical model of genomic sequence organization employed in the GeneMark.hmm algorithm is a HMM with duration (34) or a hidden semi-Markov model (HSMM). The HSMM architecture consists of hidden states for initial, internal and terminal exons, introns, intergenic regions and single exon genes (Figure 1). It also includes hidden states for start site (initiation site), stop site (termination site), and donor and acceptor splice sites. In what follows, we refer to such hidden states as site states.
Figure 1.
Diagram of hidden states of the HSMM employed in the eukaryotic GeneMark.hmm (E-3.0); only states emitting sequence of the direct DNA strand are shown, while the states generating sequence of the complementary strand (the mirror symmetrical part of the diagram with reversed arrows and horizontal symmetry line crossing ‘intergenic region’ state) are omitted.
The site states emit nucleotide sequences of fixed length modeled by positional (inhomogeneous) Markov chains (35,36). The length and parameters of these models are site type-dependent and determined from the sets of sequences of verified sites of a given type. Note that the models for sequences emitted by splice site states are also intron phase-dependent.
The protein-coding states (initial, internal, terminal exons and single exon gene) emit nucleotide sequences modeled by the three-periodic inhomogeneous Markov chains (1,37,38). Parameters of these models are chosen to be tied and are estimated from the sets of annotated protein-coding sequences (Datasets section). Orders of the Markov chains, up to the 5th order, are chosen depending on the total length of the training sequence.
The non-coding states (intron and intergenic region) emit sequences modeled by homogeneous Markov chains (1,37,38). Importantly, the parameters of the intron and intergenic region models may not be the same. Parameters of the intron models are estimated from the set of annotated intron sequences. Since a set of reliably annotated intergenic regions is not readily available, parameters of the models of intergenic regions are estimated from the set of direct and reverse complement of intron sequences.
Hidden state duration distributions are derived as approximations of observed in the training set length distributions of the sequences associated with a particular hidden state. For exon sequences this approximation is derived in two steps: (i) averaging the length frequencies over a period of three to eliminate the three-periodic component (this step is not needed for the of intron state duration approximation); and (ii) applying a smoothing algorithm, such as the nearest neighbor method (39) to get the final approximation. For adequate derivation of the distribution of duration of the sequence emitted by the state corresponding to the single exon gene we have to overcome a difficulty caused by the small sample effects, such as overfitting, as the set of single exon genes is commonly a rather small fraction of the supervised training set. It turned out that a reasonable approximation to the single exon gene length distribution is provided by the length distribution of annotated CDSs of all genes in the training set. Finally, in the absence of the reliable set of intergenic regions the uniform probability distribution is used for the duration of intergenic state.
Duration distributions are characterized by minimum and maximum values. The maximum duration of a sequence emitted from an exon state is set to the maximum ORF length observed in the given genome, while the minimum duration is 3 nt. The minimum and maximum durations of intron and intergenic sequences are set to 20 and 10 000 nt, respectively.
Initiation and termination of the trajectory of the hidden states of HSMM is allowed in either intron or intergenic state. Distribution of the length (duration) of the sequence emitted by the initial and terminal states differs from the duration distribution defined for the regular (internal) state. This distribution is determined as a convolution of a regular distribution of the state-specific duration with the uniform distribution. Note that the initialization and termination hidden states are not shown in the HSMM diagram (Figure 1).
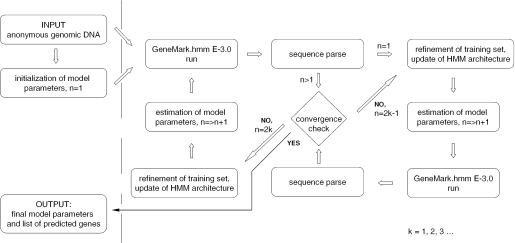
Outline of the unsupervised gene finding algorithm GeneMark.hmm ES-3.0
The algorithm of parallel unsupervised (automatic) training and gene prediction (Figure 2) consists of the following steps: (i) all parameters of the HSMM model with reduced architecture are initialized (as described below); (ii) GeneMark.hmm E-3.0 is run to determine a genomic sequence parse into ‘coding’ and ‘non-coding’ regions and the input genomic sequence is labeled with respect to this parse; and (iii) the subsets of the uniformly labeled fragments (selected as described below in the training set refinement procedure) are used for re-estimation of parameters of HSMM. Steps (ii) and (iii) are repeated until the convergence.
Figure 2.
The step-wise diagram of the iterative unsupervised parameterization of HSMM implemented in GeneMark.hmm ES-3.0.
Initial choice of the model structure and parameters
Initially, site state emissions are defined as follows: donor (acceptor) site states emit just two canonic GT (AG) dinucleotides; initiation (termination) site states emit canonic triplet sequence ATG (TGA, TAG, TAA). Sequences emitted by non-site states have uniform length distributions. Nucleotide sequences emitted by the non-coding states are described by the zero-order Markov model with parameters estimated with regard to the frequencies of nucleotides of the given genome. The sequence emitted by the protein-coding state is generated by one of the following initial models: (i) the second-order inhomogeneous Markov chain with heuristically defined parameters (40); (ii) the fifth-order inhomogeneous Markov chain with parameters derived from the sets of non-overlapping ORFs longer than 1000 nt; and (iii) homogeneous zero-order Markov model for a DNA sequence with G + C content elevated by 8% in comparison with the genome G + C content.
Training set refinement and update of parameter estimations
To reduce risk of including mislabeled genomic sequences into categorized datasets generated in the course of unsupervised training we filter out rather short predicted protein-coding regions. If a predicted exon–intron structure produces a whole protein-coding sequence (CDS) shorter than 800 nt the elements of this structure, both introns and exons, are not admitted to the training set at the next iteration. (Exception to this rule is described in the section ‘Minimum genome size required for automatic training’.)
We intentionally restrict the parameter space at the initial steps, with limitations becoming less stringent in the course of training. It has been shown that the behavior of HMMs is more sensitive to variations in the estimates of emission probabilities than to variations in the estimates of transition probabilities (41). Therefore, the self-training procedure starts with estimation of emission probabilities involved in the models for protein-coding and non-coding regions, extending the estimation process at the later steps to estimation of the distributions of duration as well as transition probabilities between hidden states.
At the initial iterations, the algorithm is allowed only to change the parameters of the Markov chain models of the sequences emitted by coding and non-coding states. After several iterations the parameters of the models of the sequences emitted by the site states are ‘unfrozen’ and later the length (duration) probability distribution for sequences emitted by coding and non-coding states is made free to change in correspondence with the statistics defined by the predicted gene structures. The emission probability values in the models of introns and intergenic regions are tied up until the point of convergence.
The lengths of sequences emitted by site states are related to the number of nucleotide positions that carry the site specific information. These lengths, constant within iteration, are allowed to change between iterations. The rules of change are illustrated by the following example. A particular position of non-coding sequence is assumed to carry specific information about the nearby splice site if the nucleotide frequency distribution in the given position differs from the (stationary) distribution of frequencies characteristic for the endogenous intronic region. We use the Kullback-Liebler (KL) distance as a measure of the difference between two distributions; and determine the value of KL-distance in positions of the sequence extending from the canonical splice site into the intron. The length at which the KL-distance decreases down to almost zero level, the one typical for positions situated well inside intron region, is accepted as the upstream (acceptor) or the downstream (donor) duration of the splice site model. This length is used in the next iteration as the length of the intronic part of the sequence emitted by the hidden state representing an acceptor (donor) site. The length of the exonic part of the sequence emitted by the acceptor (donor) site does not exceed 3 nt in any iteration.
The iterative update of the probability distribution of the single exon gene length could cause difficulty for the algorithm convergence owing to easy ‘autocatalytic’ amplification of the randomly occurred abundance of the single exon genes of a certain length. To regularize the procedure, in place of the length distribution of a single exon gene the length distribution of the whole set of predicted CDSs (both single and multiple exon genes) was used in iterations.
The predicted parse of genomic sequence into coding and non-coding regions provides information for further refinement of the sets of labeled sequences as described above and, finally, for updating the estimates of the model parameters to be used in the next iteration. Therefore, at a given iteration we could distinguish between the ‘training’ or input parse, the sequence parse obtained as a result of previous iteration, and the predicted or output parse, the actual new parse obtained as a result of the current iteration.
The difference between the training and predicted parses could be characterized by the nucleotide level sensitivity and specificity values (Sn and Sp), with ‘gene annotations’ defined by the training parse and ‘gene predictions’ defined by the predicted parse. The condition for termination of the iteration process is then defined in terms of the Sn and Sp values. The automatic training procedure is completed (reaching the convergence) as soon as both Sn and Sp rise above 97%. The final output parse defines the predicted exon–intron structures while the values of parameters of the models derived from the final training parse are considered to be the final parameter estimates.
Finally the gene finding algorithm with the models derived by the unsupervised training is applied to the test sets. We characterize the prediction accuracy by sensitivity and specificity values at the levels of nucleotides, internal exons and exon boundary sites (30,42).
RESULTS AND DISCUSSION
Comparison of supervised and unsupervised modes
The new version of GeneMark.hmm employing the newly developed unsupervised training procedure was applied to the genome sequences of A.thaliana, C.elegans, D.melanogaster, A.gambiae, C.intestinalis, C.reinhardtii and T.gondii. The accuracy of gene prediction was assessed using the test sets of validated genes. The differences in the numbers of the genes in the test sets are explained by the differences in the size of population of known cDNAs and ESTs for each species. For A.thaliana, C.elegans and D.melanogaster whose genomes are relatively well studied we were able to compile large enough training sets, not overlapping with the test sets, and compare performances of the GeneMark.hmm algorithm using both models estimated by unsupervised and supervised training (Table 1). It is seen that the unsupervised models outperform the supervised ones in 9 out of 12 categories for A.thaliana, 8 out of 12 for C.elegans and 4 out of 12 for D.melanogaster. Thus, from this data one could conclude that automatically derived models produced prediction accuracy in the range of slightly better to comparable with the accuracy produced by models derived from validated training sets. The quality of a supervised model depends slightly on the size of a training set. For instance, with tripling the size of the A.thaliana genome training set beyond currently used 1000 genes the Sn and Sp values improve one percent on average.
Table 1.
Values of several categories of sensitivity and specificity (Sn/Sp) and (Sn + Sp)/2 characterizing the accuracy of gene predictions produced for the group of ‘well-studied’ genomes by the eukaryotic GeneMark.hmm with models derived by both unsupervised and supervised training
| A.thaliana | C.elegans | D.melanogaster | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unsupervised | Supervised | Unsupervised | Supervised | Unsupervised | Supervised | |||||||
| Nucleotide | 97.7 | 96.3 | 97.2 | 95.8 | 99.1 | 96.4 | 97.8 | 96.7 | 97.9 | 95.4 | 98.1 | 95.6 |
| 94.8 | 94.3 | 93.6 | 95.5 | 92.9 | 93.1 | |||||||
| Internal exons | 91.2 | 89.5 | 91.2 | 89.9 | 94.0 | 92.7 | 90.9 | 90.9 | 91.3 | 90.5 | 87.2 | 88.7 |
| 87.8 | 88.5 | 91.3 | 90.8 | 89.7 | 90.2 | |||||||
| Initiation sites | 80.1 | 78.3 | 80.1 | 76.0 | 85.8 | 77.4 | 79.2 | 73.3 | 83.9 | 78.7 | 83.4 | 78.9 |
| 76.5 | 71.9 | 68.9 | 67.4 | 73.5 | 74.3 | |||||||
| Termination sites | 87.5 | 85.3 | 88.3 | 83.5 | 95.1 | 85.2 | 94.0 | 86.8 | 89.2 | 83.2 | 89.5 | 84.2 |
| 83.1 | 78.6 | 75.3 | 79.6 | 77.2 | 78.8 | |||||||
| Donor sites | 94.0 | 92.2 | 94.0 | 91.9 | 96.2 | 93.5 | 93.7 | 92.6 | 92.8 | 90.0 | 91.3 | 90.2 |
| 90.3 | 89.8 | 90.8 | 91.4 | 87.2 | 89.1 | |||||||
| Acceptor sites | 94.0 | 92.1 | 93.6 | 91.4 | 97.3 | 94.5 | 95.2 | 94.0 | 93.0 | 90.0 | 90.5 | 89.2 |
| 90.2 | 89.2 | 91.6 | 92.8 | 87.0 | 87.9 | |||||||
Boldface highlights the higher value in comparison of unsupervised and supervised modes (ES-3.0 versus E-3.0).
For novel genomes of A.gambiae, C.intestinalis, C.reinhardtii and T.gondii the large enough supervised training sets and subsequently the accurate supervised models were not available. The unsupervised training was the only viable option and Table 2 shows the values of Sn and Sp characterizing the accuracy of finding the elements of exon–intron structure by GeneMark.hmm using the ‘unsupervised’ models. The algorithm performs best for C.reinhardtii. One of the factors contributing to the better accuracy of C.reinhardtii gene recognition is the high genome G + C content built up as a result of mutation pressure toward G and C substitutions. Through the time of genome evolution this pressure has produced high G + C content in the third positions of codons signifying the highly biased codon usage pattern. In turn, this bias in triplet composition contributes to the higher discrimination power of the models of protein-coding regions and, eventually, more accurate exon–intron structure predictions. Relatively lower performance of gene prediction for T.gondii is apparently related to larger than usual fraction of non-canonic splice sites observed in this genome, estimated at ∼10% (Matthew Berriman, personal communication). Additionally, T.gondii has the longest among all considered genomes most probable intron size, 380 nt, and medium genome G + C content (thus lacking the augmented discrimination power of the gene models of high or low G + C content genomes).
Table 2.
Same as in Table 1, for the group of novel genomes and the unsupervised mode only (GeneMark.hmm ES-3.0)
| A.gambiae | C.intestinalis | C.reinhardtii | T.gondii | |||||
|---|---|---|---|---|---|---|---|---|
| Nucleotides | 96.0 | 90.5 | 98.3 | 94.2 | 97.4 | 97.4 | 89.6 | 88.4 |
| 85.0 | 90.0 | 97.4 | 87.1 | |||||
| Internal exons | 89.3 | 88.9 | 94.8 | 93.5 | 91.4 | 93.4 | 80.2 | 81.7 |
| 88.4 | 92.1 | 95.4 | 83.1 | |||||
| Initiation sites | 77.8 | 72.9 | 79.6 | 71.3 | 82.9 | 78.4 | 58.5 | 65.1 |
| 67.9 | 63.0 | 73.9 | 71.7 | |||||
| Termination sites | 86.1 | 78.9 | 85.4 | 75.9 | 92.7 | 87.7 | 66.2 | 73.7 |
| 71.7 | 66.3 | 82.6 | 81.1 | |||||
| Donor sites | 89.7 | 86.9 | 95.3 | 92.5 | 94.1 | 95.2 | 81.3 | 84.4 |
| 84.1 | 89.7 | 96.3 | 87.5 | |||||
| Acceptor sites | 92.3 | 88.5 | 96.3 | 93.3 | 93.5 | 94.6 | 82.0 | 85.2 |
| 84.7 | 90.3 | 95.7 | 88.3 | |||||
As can be seen from Tables 1 and 2 specificity values are lower than sensitivity ones for both supervised and unsupervised training procedures. This difference is reaching for some categories of accuracy measures up to 19% for genomes of A.gambiae and C.intestinalis. A general reason for elevation of the false positive error rate and lowering the specificity value is the ‘boundary effect’, which is related to cutting off the gene upstream and downstream sequences that carry information able to preclude false positive predictions at the 5′ and 3′ ends of the test sequence. Thus, the cited specificity values could be considered as a lower bound of the real values.
Initialization of unsupervised training
We have tested three different parameter initialization strategies (Materials and Methods) and observed convergence of the unsupervised training procedure to almost one and the same point in parameter space regardless of the chosen point of initialization (data not shown). The choice of an initialization point did affect the number of iterations necessary to reach convergence, though the difference itself did not exceed three iterations. It was quite surprising that the ‘weak’ initialization with the protein-coding sequence model defined as the homogeneous zero-order Markov model whose parameters were deduced just from the genome G + C content, could produce almost the same results as other more elaborate initialization strategies.
All the Sn and Sp values observed and cited here for the models derived by the unsupervised training procedure have been produced with initialization parameters of the protein-coding sequence model determined by the heuristic rules (40). On average, this type of initialization required the smallest number of iterations.
Dynamics of convergence of training iterations
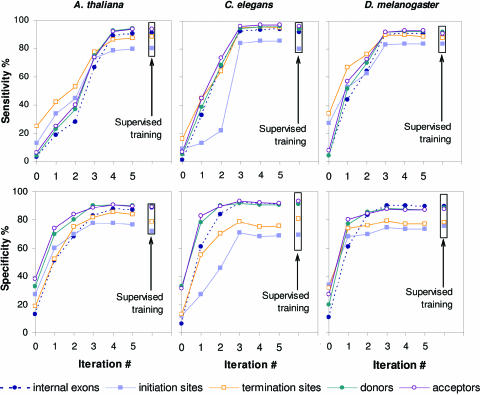
As the unsupervised training progresses through iterations, the characteristics of gene prediction accuracy, Sn and Sp, could be measured at each step by plugging the current models into GeneMark.hmm E-3.0 and running the program on a test set. The iteration index dependence of the Sn and Sp values for the test sets generated for A.thaliana, C.elegans and D.melanogaster is shown in Figure 3. The initial heuristically derived models produce predictions with the nucleotide level Sn and Sp values in the range between 5 and 40%. For each of the three species subsequent three to four iterations bring Sn and Sp values within the 5% vicinity of the steady-state level and the accuracy level of the algorithm using authentic ‘supervised’ models. Note that the observed growth of specificity values in the first two iterations is much faster than the growth of sensitivity values. This trend we consider as necessary condition of the right course of convergence. The reversed trend, with sensitivity growing while specificity remains low should end up in convergence to biologically irrelevant point as abundant false positive prediction would eventually bias the model parameters. Since distributions of durations remain uniform and unchanged in the first three iterations, the significant growth in accuracy in these initial iterations is due to improvement of estimates of emission probabilities. Naturally, the number of correctly predicted protein-coding regions grows from iteration to iteration. For example, in the A.thaliana genome, ∼17 000 exons are predicted in the first iteration, ∼23 000 after third iteration and ∼115 000 at convergence.
Figure 3.
Gene prediction accuracy parameters (Sn and Sp), as determined on the test sets for A.thaliana, C.elegans and D.melanogaster, are shown as functions of the iteration index. For gene predictions produced by models defined at initialization, the Sn and Sp values are shown at zero index value. Upon application of GeneMark.hmm ES-3.0 to genomes of A.gambiae, C.intestinalis, C.reinhardtii and T.gondii we observed similar dynamics of change of the Sn and Sp parameters measured on the relevant test sets (data not shown).
Essentially, the goal of the automatic training algorithm, GeneMark.hmm ES-3.0, is parameterization of the HSMM model from unlabeled (un-annotated) genomic sequence. To reach this goal we implement the Viterbi training algorithm, which generally does not guarantee finding the global maximum of the likelihood criterion. Hence, the convergence point in parameter space is not necessarily the point where the estimated parameters exactly match the true values or the accuracy of the predictions on a test set is the highest. The process of improving accuracy is not even monotonic. For instance, in the C.elegans case the specificity dropped by ∼2% in iterations four and five.
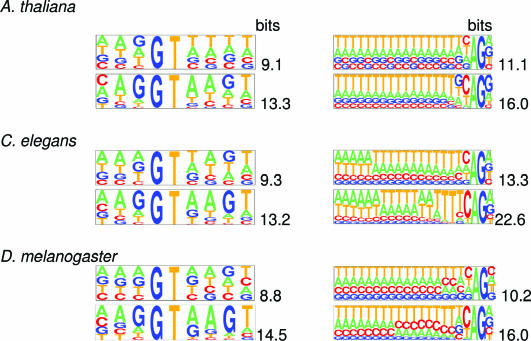
The decrease in iterations of the values of the KL-distance between the unsupervised donor (acceptor) site models and supervised donor (acceptor) models indicates that the algorithm brings the site model parameters close to ones of the supervised model, though not to the exactly same values. The set of sequences identified as emitted by the splice site state changes in iterations while accumulating more and more predicted donor (acceptor) sequences. These sequences and, hence, the splice site duration (with initial duration of two nucleotides) increase in length. For A.thaliana, C.elegans and D.melanogaster the pictograms in Figure 4 illustrate the patterns of nucleotide frequencies in the sets of sequences surrounding predicted donor site (9 nt long) and predicted acceptor sites (21 nt long). It is seen that these models gain a significant increase in information content through the iterations. The bit values of the information content at convergence are very close to the bit values of the information content for the ‘supervised’ site models.
Figure 4.
Pictograms of positional nucleotide frequency distributions observed around donor site (left column) and acceptor site (right column). Pictograms of the distributions derived after the first iteration are shown in the top panels of panel pairs, for the distributions derived at the algorithm convergence—in the bottom panels. Values (in bits) of the information content of the first-order positional Markov model derived from the aligned sequences are shown next to the pictograms. (The pictograms were drawn by the software utility available at genes.mit.edu/pictogram.html).
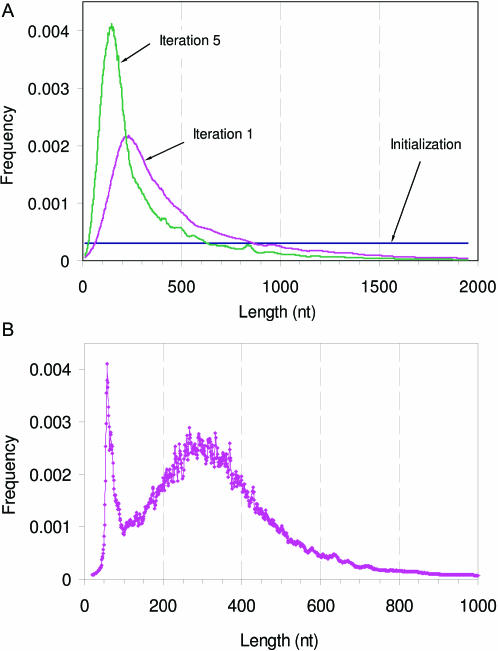
The exon length distribution transforms in iterations from the uniform one to the skewed bell shaped one (Figure 5A) close to the one observed in the supervised training set. Contrary to the unimodal distribution regularly observed in many species, the C.intestinalis intron length distribution derived from EST to DNA alignments has been reported to have two picks, the first and rather sharp one at 60 nt and the second one at 300 nt (43). Notably, the length distribution of introns predicted in the course of automatic training shows the shape matching the reported one (Figure 5B).
Figure 5.
(A) Change of the shape of predicted exon length distribution through iterations (D.melanogaster). Note that the GeneMark ES-3.0 algorithm continues to use uniform exon length distribution in the first three iterations. At convergence point the predicted exon length distribution coincides with the exon length distribution produced by the supervised training (not shown). (B) The shape of the C.intestinalis intron length distribution reached at iterations convergence.
It is worthwhile to note that the two peaks intron distribution may occur in the unsupervised training as an artifact if the genomic sequence in question has a low coverage and frame-shifts owing to sequence errors are probable to appear in protein-coding regions. In a simple experiment we randomly deleted a nucleotide at every 1000 nt stretch of the genomic sequence of A.thaliana, thus some frame-shifts were artificially introduced. Upon the self-training algorithm application this aberration in the sequence data affected the length distribution of introns, but not that of exons, by creating an extra peak in the short intron range (∼45 nt). The algorithm has found most of the coding region correctly, but at the frame-shift region it was typically forced to introduce a short intron.
With respect to the influence of the G+C content of a genome on the convergence process, the higher speed of convergence and the higher accuracy was observed in genomes with both high G + C content (C.reinhardtii) and low G + C content (A.thaliana and C.elegans). The largest number of iterations and lower gene prediction accuracy was observed in the T.gondii genome with medium G + C content. The observed trend of decrease of the discrimination power of the gene finding models in the sequences with medium G + C content is consistent with our earlier observations made for prokaryotic genomes (J. Besemer and A. Lomsadze, personal communication).
The performance of unsupervised training procedure could be influenced by the presence of repetitive sequences of transposable elements (TEs) that frequently carry genes required for their mobility. This effect increases with the increase of the fraction of the genome occupied by TEs, and is especially pronounced if the repeat population is dominated by large families (such as LINE and ERV). For the eukaryotic genomes analyzed in the current paper, fraction of the genome occupied by TEs has not exceeded 12% (as identified by RepeatMasker, available at repeatmasker.genome.washington.edu). Also, these genomes contain TEs belonging to diverse families, while the families having large number (thousands) of members are absent. The unsupervised training procedure implemented with or without prior repeat masking has produced the models that did not differ significantly in terms of gene prediction accuracy. The difference in (Sn + Sp)/2 value of internal exon prediction was below 0.5% for A.thaliana and C.intestinalis, 1% for C.elegans and C.reinhardtii, 1.8% for D.melanogaster and 3.8% for A.gambiae.
Minimum genome size required for automatic training
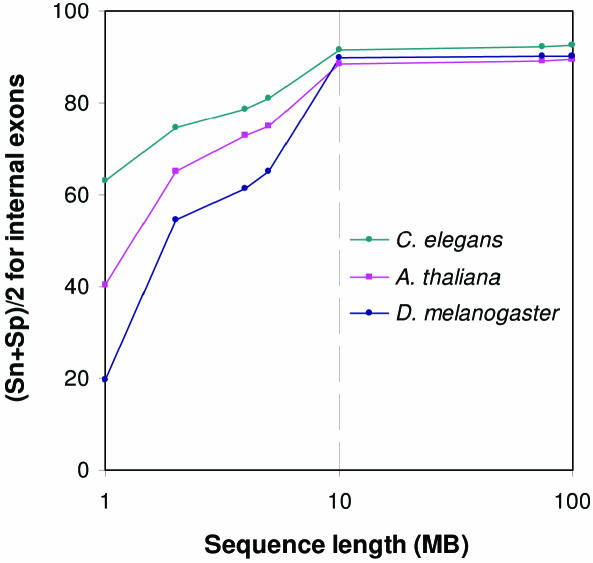
Model parameterization by unsupervised training makes robust and reliable gene prediction in eukaryotic genomic sequences feasible at rather early stages of genome sequencing. We have evaluated the dependence of the quality of models with parameters estimated by unsupervised training on the length of available genomic sequence. Genomic sequences of various sizes were used as inputs to the unsupervised training procedure. In these experiments we have met difficulty to consistently implement the restriction of removing predicted gene structures with CDS shorter than 800 nt from the updated training set. Therefore in the range of input sequence length below 10 MB, exon–intron structures with CDS longer than 300 nt (rather than 800 nt) predicted in the course of iterations were admitted to the updated training sets. Dependence of the average accuracy of prediction of internal exons, characterized by value of (Sn + Sp)/2, on the input sequence length is shown in Figure 6. For A.thaliana (87%), C.elegans (91%) and D.melanogaster (90%) the accuracy of prediction of internal exons reaches high enough levels for training sequence size as moderate as 10 Mb. However, for such a length of the input sequence, the number of iterations on average increases 2-fold. Another observation is that regardless of the type of genome considered, the growth of the training sequence beyond 10 Mb size did not produce a significant increase in accuracy. Therefore, our current results suggest that accumulation of 10 Mb of sequence in a eukaryotic genome sequencing project is sufficient for unsupervised parameterization of the statistical model (HSMM) employed in the gene finding algorithm. Still, there is a caveat stating that the minimal input sequence size depends on the gene density and a larger sequence might be needed for the genomes of higher organisms that are populated with TEs. However, there are many genomes of low eukaryotes, such as fungi, which may not be suitable targets for automatic training of gene models described here. The primary reason for that is the small number of introns. For genomes with very low intron population, such as yeast with slightly >200 introns per ∼6000 genes, it is practical to use HMMS's without intron related states, a simple modification of automatic prokaryotic gene finders (25,26).
Figure 6.
The internal exon prediction accuracy of GeneMark.hmm ES-3.0 characterized by (Sn + Sp)/2, as a function of the length of genomic sequence available for unsupervised training.
We emphasize that at the ‘early stage’ of genome sequencing, when the extrinsic evidence for a sufficiently large training set is unlikely to be found, the unsupervised training and prediction method, such as GeneMark.hmm ES-3.0, is arguably the only one to help analyze (predict genes in) available sequence data. This situation may change at a point when a large enough training set becomes available and the user of the gene finding tool is offered a choice of models either obtained by supervised or unsupervised training. However, it is difficult to decide a priori how to proceed with this dilemma. As we have seen above GeneMark.hmm ES-3.0 demonstrated superior of at least equal quality of gene prediction for the three well-studied genomes in comparison with GeneMark.hmm E-3.0 which is using the models derived by supervised training. We argue below that in general there are several reasons why the models obtained by unsupervised training could be still better than ones obtained by supervised training even if the large enough training set is available. Therefore, a selection of a particular method and model type (serial versus parallel training) requires direct comparison of performance of each method on a reliable test set.
The quality of the model of a protein-coding region derived by a supervised training is likely to be affected by bias of cDNA/EST data to highly expressed genes. The self-training method, however, would sample labeled sequences from the whole genomic space, and, as a result, would produce less biased models. Also, it is conventional to sample annotated intron sequences for the training set of the supervised model of an intergenic region. However, for some genomes (e.g. A.thaliana) the accurate model of intergenic region is different from the intron model.
Comparison with SNAP
The SNAP program (6) has been the only one developed so far with the goal to provide a gene finding tool for novel eukaryotic genomes with limited experimental data. For novel genomes SNAP offers the following technique of deriving so-called bootstrapped models. First, the user has to choose models for the new genome from the collection of readily available supervised models for well-known genomes. Second, a single model is plugged into SNAP to obtain predictions for the new genome. Third, these predictions are used as the training set for the bootstrapped model. Note that an additional option utilized in the SNAP paper is to choose several models, to run program with each model and to combine all the predictions into the training set.
In our comparison of the programs we had to accommodate the following restrictions. The bootstrapped models for SNAP have not been available for download. The accuracy of the SNAP gene predictions for bootstrapped models was only cited at a nucleotide level.
We have used two test sets. The first one, the set of sequences with one gene per sequence is available at the SNAP website (www.biomedcentral.com/content/supplementary/1471-2105-5-59-S1.gz). The second test set compiled for this study (Datasets section) is available at opal.biology.gatech.edu/GeneMark/eukset_080105.html.
The first test set allowed comparing programs in two avenues of competition (i) with use of supervised models and (ii) with use of unsupervised models (GeneMark.hmm) or bootstrapped models (recorded data for SNAP).
As explained above we were restricted to the use of Sn and Sp values characterizing the gene prediction accuracy on nucleotide level (Table 3). All the data for SNAP cited in Table 3 were given in the original paper (6). The SNAP paper also offers accuracy assessment for seven bootstrapped models for each of the genomes of A.thaliana, C.elegans and D.melanogaster.
Table 3.
Values of nucleotide level sensitivity and specificity (Sn/Sp) along with (Sn + Sp)/2 for gene predictions produced by GeneMark.hmm and SNAP for the group of ‘well-known’ genomes
| GeneMark.hmm unsupervised (ES-3.0) | SNAP bootstrap | GeneMark.hmm supervised (E-3.0) | SNAP supervised | GenScan supervised | Genefinder supervised | Augustus supervsed | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A.thaliana | 98.3 | 96.5 | 96.6 | 94.9 | 98.4 | 96.3 | 97.1 | 96.2 | 79.9 | 86.4 | – | – | – | – |
| 94.7 | 93.2 | 94.2 | 95.2 | 92.9 | – | – | ||||||||
| C.elegans | 99.1 | 97.1 | 96.7 | 93.9 | 97.7 | 97.0 | 97.6 | 95.9 | – | – | 98.1 | 96.7 | – | – |
| 95.1 | 91.1 | 96.2 | 94.2 | – | 95.3 | – | ||||||||
| D.melanogaster | 93.8 | 90.0 | 92.5 | 89.6 | 93.2 | 90.5 | 94.3 | 90.4 | – | – | – | – | 92.4 | 90.5 |
| 86.1 | 86.6 | 87.7 | 86.5 | – | – | 88.6 | ||||||||
The unsupervised mode of GeneMark.hmm (ES-3.0) is compared with the bootstrapped mode of SNAP (first two columns). The performance of the supervised modes of the two programs, GeneMark.hmm E-3 and SNAP, are also com pared (last five columns) with performance of GenScan (trained on A.thaliana), Genefinder (trained on C.elegans) and Augustus (trained on D.melanogaster). All figures in this table, except ones for GeneMark.hmm, are cited from (6). The accuracy of GeneMark.hmm was assessed on the same test sets downloaded from the SNAP website.
Comparison of the performance of the GeneMark.hmm ES-3.0 with automatically trained models against SNAP using the best for the given species (out of seven) bootstrapped model shows (the first two columns of Table 3) that the (Sn + Sp)/2 values are higher for GeneMark.hmm by 1.7% for A.thaliana (the training set for the bootstrapped model is generated from the C.elegans model predictions); by 3.3% for C.elegans (the bootstrapped model is based on the A.thaliana and O.sativa models predictions); and by 0.4% for D.melanogaster (the bootstrapped model is based on the O.sativa model predictions). Use of supervised models in both programs results in a very similar performance with marginally better accuracy of GeneMark.hmm (the last two columns).
Note that the results of the tests of GenScan, GeneFinder and Augustus trained respectively for A.thaliana, C.elegans and D.melanogaster have been reported for the first test (6) and are cited in the last three columns of Table 3. It is seen that comparison of GeneMark.hmm accuracy with the accuracy of these three programs shows equal or better results for GeneMark.hmm.
To work with the second test set the SNAP program was downloaded from homepage.mac.com/iankorf/snap-2005-07-27.tar.gz along with the supervised models for A.thaliana, A.gambiae, C.intestinalis, C.elegans and D.melanogaster. While the bootstrapped models for these genomes were not available, the results described in the original paper (6) showed that the SNAP supervised models uniformly outperform the bootstrapped models; therefore, the SNAP supervised models are providing a sufficient benchmark for our purposes.
Comparison of gene prediction accuracy on the second test set produced the following results (Table 4). On the nucleotide level the iteratively parameterized GeneMark.hmm models have shown consistently better performance in terms of (Sn + Sp)/2 values than the supervised models of SNAP. Further, we move to the accuracy measured by the specificity and sensitivity of the exact internal exon prediction. Note that the accuracy assessment based on exact exon prediction apparently has more relevance for practical purposes. For instance, massive mis-prediction of splice sites by 1 nt will significantly change predicted protein products. This event is easily detected if accuracy is assessed at the level of exact exon prediction, while it is almost unnoticeable at the nucleotide level. As it is seen from Table 4, on the internal exon level, the values of (Sn + Sp)/2 for GeneMark.hmm are higher by 6.4% for A.thaliana, by 3.5% for A.gambiae, by 23.7% for C.intestinalis, by 6.0% for C.elegans, and by 5.5% for D.melanogaster. The results of comparison with SNAP using supervised models provide lower bounds for the performance differences that would exist in comparison with SNAP using bootstrapped models. Therefore, the performance level of GeneMark.hmm ES-3.0 with iteratively trained models, would have even larger margin over the level of performance of SNAP with the bootstrapped models.
Table 4.
Values of sensitivity and specificity (Sn/Sp) along with (Sn + Sp)/2 for gene predictions produced by GeneMark.hmm and SNAP for the test sets described in the Dataset section
| A.thaliana | A.gambiae | C.intestinalis | C.elegans | D.melanogaster | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GeneMark.hmm unsupervised | SNAP supervised | GeneMark.hmm unsupervised | SNAP supervised | GeneMark.hmm unsupervised | SNAP supervised | GeneMark.hmm unsupervised | SNAP supervised | GeneMark.hmm unsupervised | SNAP supervised | |||||||||||
| Nucleotide | 97.7 | 96.3 | 93.6 | 94.5 | 96.0 | 90.5 | 87.6 | 84.5 | 98.3 | 94.2 | 90.1 | 82.2 | 99.1 | 96.4 | 97.2 | 95.7 | 97.9 | 95.4 | 94.8 | 93.9 |
| 94.8 | 95.3 | 85.0 | 81.4 | 90.0 | 74.3 | 93.6 | 94.1 | 92.9 | 92.9 | |||||||||||
| Internal | 91.2 | 89.5 | 79.7 | 83.8 | 89.3 | 88.9 | 81.7 | 84.7 | 94.8 | 93.5 | 80.9 | 71.3 | 94.0 | 92.7 | 87.1 | 87.1 | 91.3 | 90.5 | 85.8 | 85.5 |
| exons | 87.8 | 87.8 | 88.4 | 87.6 | 92.1 | 61.7 | 91.3 | 87.1 | 89.7 | 85.2 | ||||||||||
| Initiation | 80.1 | 78.3 | 75.6 | 75.0 | 77.8 | 72.9 | 65.2 | 66.2 | 79.6 | 71.3 | 61.0 | 52.2 | 85.8 | 77.4 | 73.2 | 67.4 | 83.9 | 78.7 | 78.1 | 77.7 |
| sites | 76.5 | 74.3 | 67.9 | 67.2 | 63.0 | 43.4 | 68.9 | 61.5 | 73.5 | 77.3 | ||||||||||
| Termination | 87.5 | 85.3 | 84.0 | 83.5 | 86.1 | 78.9 | 78.8 | 76.0 | 85.4 | 75.9 | 63.3 | 54.6 | 95.1 | 85.2 | 89.4 | 80.9 | 89.2 | 83.2 | 78.1 | 77.5 |
| sites | 83.1 | 82.9 | 71.7 | 73.2 | 66.3 | 45.9 | 75.3 | 72.4 | 77.2 | 76.8 | ||||||||||
| Donor sites | 94.0 | 92.2 | 83.7 | 86.9 | 89.7 | 86.9 | 80.8 | 83.1 | 95.3 | 92.5 | 82.7 | 72.6 | 96.2 | 93.5 | 90.1 | 89.0 | 92.8 | 90.0 | 86.9 | 86.6 |
| 90.3 | 90.0 | 84.1 | 85.3 | 89.7 | 62.4 | 90.8 | 87.8 | 87.2 | 86.2 | |||||||||||
| Acceptor | 94.0 | 92.1 | 84.6 | 87.8 | 92.3 | 88.5 | 83.3 | 83.7 | 96.3 | 93.3 | 83.6 | 73.5 | 97.3 | 94.5 | 93.5 | 92.1 | 93.0 | 90.0 | 87.7 | 87.3 |
| sites | 90.2 | 91.0 | 84.7 | 84.1 | 90.3 | 63.3 | 91.6 | 90.6 | 87.0 | 86.8 | ||||||||||
The unsupervised mode of Gene-Mark.hmm (ES-3.0) is compared with the supervised mode of SNAP which typically performs better than the SNAP bootstrapped mode (6). The supervised models from SNAP were downloaded from the SNAP website (see text).
Biological implications of new predictions
Ability to identify earlier unknown genes emphasizes the usefulness of the new method. Among the genes newly predicted (Supplementary Table S1 and Supplementary File S2 with corresponding sequences in Supplementary Data), several interesting examples are worth noting.
New ‘housekeeping’ or important metabolic genes
Presence of these genes in the genomes under study could be expected but has not been known previously. In this category, the genes coding for the following proteins could be mentioned.
In D.melanogaster: TTD-A subunit of the basal transcription complex TFIH (Supplementary Table S1, #1), an ortholog of transcription factor TBF5 of Gallus gallus). This protein is involved in general control of transcription and transcription-associated DNA repair, and possibly in cell cycle regulation. Defects of these processes lead to carcinogenesis. Identification of the ortholog of this protein in Drosophila, a model organism which is well studied genetically, paves the way for development of the genetic assays for the functional analysis of TTD-A in future.
In A.gambiae:
cytochrome c oxidase subunit VIc (#2), a component of the mitochondrial electron transport chain;
Nup84p (#54), evolutionary conserved component of the complex required for the nuclear pore biogenesis.
In C.intestinalis:
mitochondrial ribosomal protein L10 homolog (#15);
ortholog of the eukaryotic translation initiation factor 3, subunit 8 from rat or subunit p110 from human (#27); this protein also shows a similarity to the hypothetical protein LOC395052 from Xenopus, suggesting that a Xenopus protein may also play a role in translation;
ortholog of RNA polymerase I associated factor 53 from human, mouse and Xenopus (#23).
In C.reinhardtii:
homologs of the ribosomal proteins S21 (#6), S21e (#1), S9/S16 (#10);
nucleolar protein Nop10p (#2);
A homolog of Sec6 β-family protein (#4) that is a component of the Sec61 protein secretory system, studied previously in yeast and also found in humans and apes.
Genes having homologs in the phylogenetically closely related organisms
While presence of these genes in the genomes under study makes sense based on the phylogenetic positions of the given organisms, they have not been identified previously by other methods.
Example of such a gene is an A.gambiae homolog of the ‘royal jelly’ protein (#34) involved in control of cast differentiation in honey bee, and also homologous to Drosophila protein CG7463-PA. Presence of such a protein in Anopheles provides additional valuable information on its phylogeny among different groups of insects.
Another example from Anopheles is distant homolog of the mammalian male enhanced antigen 1, suggested to play an important role in the late stage of mammalian spermatogenesis (#22). It also shows a homology to Drosophila protein CG14341-PB, isoform B, and to E3 ubiquitin ligase URE-B1. Identification of this protein in Anopheles further confirms its broad evolutionary conservation, despite the possible divergence of its specific biological roles.
Unexpected genes
Genes whose presence in the genomes under study has not been known previously and could not be easily predicted from the general point of view. Discovery of these genes provides new useful information about the evolution of the specific gene families and/or biology of the specific organisms.
The genes coding for the following proteins could be mentioned in this category.
In A.gambiae: Homolog of the mammalian neurochondrin (#52). Neurochondrin is produced in mammalian chrondrocytes, bone-producing cells and some neurons, and is thought to play a specific role in regulating cell resorption. Presence of this protein in the insects, which have no internal skeleton, suggests that functions of neurochondrin could be broader than initially proposed. This protein also shows a homology to the Drosophila protein CG2330-PA. To our knowledge, similarity between that protein and neurochondrin has not been noticed previously.
In C.elegans: Tetracycline resistance protein of group C (#45) detected previously in prokaryotes such as Shigella and present in the transposon Tn10. Presence of such a protein in a eukaryotic organism is unexpected and may indicate a horizontal transmission. A possibility of horizontal transmission between bacteria and nematodes provides a new insight into biology of these organisms.
The reasons why the genes (listed in Supplementary Table S1) with protein products having similarity to known proteins have not been detected earlier by the DNA-to-protein searches (e.g. by the BLASTX application) seem to be as follows.
Selection of the threshold E-value in similarity searches using BLASTX-like program reflects the trade-off between sensitivity and specificity. It is natural to set up a relatively low E-value to avoid the flow of meaningless hits. Obviously, this may lead to missing some real genes. First, if similarity of the protein product to known proteins is weak, then even if the gene has one or more long exons, it might not be detected. Second, even if a similarity of the protein product to a known protein is quite high, but the gene structure consists of several short exons they might not be detected on their own. When these short exons are assembled into complete gene by a gene finding program, similarity is easier to detect. Many of the newly identified genes presented in the paper fall to one of these categories, thus, they were difficult to detect by the DNA-to-protein search.
CONCLUDING REMARKS
We have demonstrated that the iterative training algorithm, GeneMark.hmm ES-3.0, developed for novel eukaryotic genomes makes possible the automatic parameterization of high-performance gene models. When tested onwell-studied genomes, this method provides a higher or matching accuracy of gene finding in comparison with traditional use of models derived from training sets of validated genes. It was shown that the convergence of the proposed iterative training procedure is robust with regard to choice of initial parameters of the model. Some difficulties, however, could be anticipated for some genomes of both low and high eukaryotes. If the number of intron containing genes is too small, as observed in several low eukaryotic genomes (such as yeast), extracting informative models for splice sites and other conserved sequence patterns (like brunch point) becomes a cumbersome task for a designer of an ab initio method. On the other hand, in several genomes of high eukaryotes, such as human, mouse and rat, repetitive sequences may strongly bias the automatic model parameterization. For higher organisms with genomes populated with large families of TEs, repeat identification and elimination from the input sequence should be a standard additional step in the proposed algorithm. Also, if the genome under study has a significant variation in G + C content, sequence segmentation into contigs with more homogeneous G + C content, segment clusterization as well as multiple model initializations should be used. Still, for the eukaryotic genomes analyzed in this study difficulties have been significant as well. Therefore, even after presentation of the initial ideas on the unsupervised training procedure developed for GeneMark.hmm E-2.0 at the 2003 Gordon Bioinformatics conference at the Oxford University and at the 2003 International Bioinformatics Conference at Georgia Tech, we have spent quite a bit of time to further improve the gene finding algorithm and the automatic training procedure.
WWW and software resources
The new gene finding programs for novel eukaryotic genomes available for use via Internet: GeneMark.hmm E-3.0 at opal.biology.gatech.edu/GeneMark/eukhmm.cgi; GeneMark.hmm ES-3.0 at opal.biology.gatech.edu/GeneMark/gmseuk.cgi. The final gene predictions of GeneMark.hmm ES-3.0 are returned to the address provided by a submitter through e-mail. Note that for several eukaryotes GeneMark.hmm E-3.0 can use both models derived by supervised and unsupervised training. The source code of the new programs is freely available from the authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to John Besemer for valuable discussions. We thank Yuan Tian and Nataliya Shmeleva for help in the test sets preparation. This work was supported in part by grant HG00783 to M.B. and grant GM58763 to Y.O.C. from the US National Institutes of Health (NIH). Funding to pay the Open Access publication charges for this article was provided by NIH grants HG00783 and GM58763.
Conflict of interest statement. None declared.
REFERENCES
- 1.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 2.Krogh A. Two methods for improving performance of an HMM and their application for gene finding. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:179–186. [PubMed] [Google Scholar]
- 3.Parra G., Blanco E., Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reese M.G., Kulp D., Tammana H., Haussler D. Genie—gene finding in Drosophila melanogaster. Genome Res. 2000;10:529–538. doi: 10.1101/gr.10.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanke M., Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:II215–II225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 6.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gish W., States D.J. Identification of protein coding regions by database similarity search. Nature Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 8.Gelfand M.S., Mironov A.A., Pevzner P.A. Gene recognition via spliced sequence alignment. Proc. Natl Acad. Sci. USA. 1996;93:9061–9066. doi: 10.1073/pnas.93.17.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogozin I.B., Milanesi L., Kolchanov N.A. Gene structure prediction using information on homologous protein sequence. Comput. Appl. Biosci. 1996;12:161–170. doi: 10.1093/bioinformatics/12.3.161. [DOI] [PubMed] [Google Scholar]
- 10.Kulp D., Haussler D., Reese M.G., Eeckman F.H. Integrating database homology in a probabilistic gene structure model. Pac. Symp. Biocomput. 1997:232–244. [PubMed] [Google Scholar]
- 11.Laub M., Smith D. Finding intron/exon splice junctions using INFO, INterruption Finder and Organizer. J. Comput. Biol. 1998;5:307–321. doi: 10.1089/cmb.1998.5.307. [DOI] [PubMed] [Google Scholar]
- 12.Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pachter L., Batzoglou S., Spitkovsky V.I., Banks E., Lander E.S., Kleitman D.J., Berger B. A dictionary-based approach for gene annotation. J. Comput. Biol. 1999;6:419–430. doi: 10.1089/106652799318364. [DOI] [PubMed] [Google Scholar]
- 14.Mironov A.A., Roytberg M.A., Pevzner P.A., Gelfand M.S. Performance-guarantee gene predictions via spliced alignment. Genomics. 1998;51:332–339. doi: 10.1006/geno.1998.5251. [DOI] [PubMed] [Google Scholar]
- 15.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pachter L., Alexandersson M., Cawley S. Applications of generalized pair hidden Markov models to alignment and gene finding problems. J. Comput. Biol. 2002;9:389–399. doi: 10.1089/10665270252935520. [DOI] [PubMed] [Google Scholar]
- 17.Schlueter S.D., Dong Q., Brendel V. GeneSeqer@PlantGDB: gene structure prediction in plant genomes. Nucleic Acids Res. 2003;31:3597–3600. doi: 10.1093/nar/gkg533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer I., Durbin R. Comparative ab initio prediction of gene structures using pair HMMs. Bioinformatics. 2002;18:1309–1318. doi: 10.1093/bioinformatics/18.10.1309. [DOI] [PubMed] [Google Scholar]
- 19.Kent W.J., Zahler A.M. Conservation, regulation, synteny, and introns in a large-scale C.briggsae–C.elegans genomic alignment. Genome Res. 2000;10:1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- 20.Batzoglou S., Pachter L., Mesirov J.P., Berger B., Lander E.S. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgenstern B., Rinner O., Abdeddaim S., Haase D., Mayer K.F., Dress A.W., Mewes H.W. Exon discovery by genomic sequence alignment. Bioinformatics. 2002;18:777–787. doi: 10.1093/bioinformatics/18.6.777. [DOI] [PubMed] [Google Scholar]
- 22.Mathe C., Sagot M.F., Schiex T., Rouze P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002;30:4103–4117. doi: 10.1093/nar/gkf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dempster A.P., Laird N.M., Rubin D.B. Maximum likelihood from inclomplete data via the EM algorithm. J. R. Stat. Soc. 1977;39:31–38. [Google Scholar]
- 24.Baldi P. On the convergence of a clustering algorithm for protein-coding regions in microbial genomes. Bioinformatics. 2000;16:367–371. doi: 10.1093/bioinformatics/16.4.367. [DOI] [PubMed] [Google Scholar]
- 25.Besemer J., Lomsadze A., Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen T.S., Krogh A. EasyGene—a prokaryotic gene finder that ranks ORFs by statistical significance. BMC Bioinformatics. 2003;4:21. doi: 10.1186/1471-2105-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frishman D., Mironov A., Mewes H.W., Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Audic S., Claverie J.M. Self-identification of protein-coding regions in microbial genomes. Proc. Natl Acad. Sci. USA. 1998;95:10026–10031. doi: 10.1073/pnas.95.17.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes W.S., Borodovsky M. How to interpret an anonymous bacterial genome: machine learning approach to gene identification. Genome Res. 1998;8:1154–1171. doi: 10.1101/gr.8.11.1154. [DOI] [PubMed] [Google Scholar]
- 30.Pavy N., Rombauts S., Dehais P., Mathe C., Ramana D.V., Leroy P., Rouze P. Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences. Bioinformatics. 1999;15:887–899. doi: 10.1093/bioinformatics/15.11.887. [DOI] [PubMed] [Google Scholar]
- 31.Lukashin A.V., Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Initiative T.A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 33.Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 34.Rabiner L.R. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- 35.Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984;12:505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M.Q., Marr T.G. A weight array method for splicing signal analysis. Comput. Appl. Biosci. 1993;9:499–509. doi: 10.1093/bioinformatics/9.5.499. [DOI] [PubMed] [Google Scholar]
- 37.Borodovsky M.Y., Sprizhitskii Y.A., Golovanov E.I., Aleksandrov A.A. Statistical patterns in primary structures of functional regions in E.coli genome: III. Computer recognition of coding regions. Mol. Biol. 1986;20:1145–1150. [PubMed] [Google Scholar]
- 38.Borodovsky M.Y., McIninch J.D. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 1993;17:123–153. [Google Scholar]
- 39.Silverman B.W. Density Estimation for Statistics and Data Analysis. London, NY: Chapman and Hall; 1986. [Google Scholar]
- 40.Besemer J., Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitrophanov A.Y., Lomsadze A., Borodovsky M. Sensitivity of hidden Markov models. J. Appl. Probab. 2005;42:632–642. [Google Scholar]
- 42.Burset M., Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- 43.Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.