Abstract
Meiotic recombination is not random along chromosomes; rather, there are preferred regions for initiation called hotspots. Although the general properties of meiotic hotspots are known, the requirements at the DNA sequence level for the determination of hotspot activity are still unclear. The sequence of six known hotspots in Saccharomyces cerevisiae was compared to identify a common homology region (CoHR). They reported that the locations of CoHR sequences correspond to mapped double-strand break (DSB) sites along three chromosomes (I, III, VI). We report here that a deletion of CoHR at HIS2, a hotspot used to identify the motif, has no significant effect on recombination. In the absence of CoHR, DSB formation occurs at a high frequency and at the same sequences as in wild-type strains. In cases where the deletion of sequences containing the CoHR motif has been shown to reduce recombination, we propose that it may be a reflection of the location of the deletion, rather than the loss of CoHR, per se.
Introduction
Genetic recombination has long been demonstrated to be an essential feature of meiosis (Baker et al, 1976; Lichten, 2001). It is important for the creation of genetic variation and is required for proper segregation of homologous chromosomes during the reductional division in almost all eukaryotes (Baker et al, 1976; Lichten, 2001). A feature of meiotic recombination is that it does not occur randomly along chromosomes; rather, it occurs more frequently in preferred regions dubbed hotspots. In Saccharomyces cerevisiae, hotspots are associated with high levels of recombination initiation via the formation of double-strand breaks (DSBs). Several meiotic recombination hotspots have been extensively studied in S. cerevisiae (e.g., Nicolas et al, 1989; White et al, 1991; Cherest & Surdin-Kerjan, 1992; Malone et al, 1992). These hotspots have many similar properties (e.g., high frequencies of DSBs and gene conversion (GC)); however, one property not shared is an easily identifiable DNA sequence required for DSB formation.
In an attempt to define a DNA motif important for recombination initiation, Blumental-Perry et al (2000) examined DNA sequences within 1 kilobase pair (kbp) of six well-characterized recombination hotspots in S. cerevisiae. A moderately degenerate motif, called CoHR (common homology region), was identified by sequence alignment (Fig 1A). Comparative sequence analysis was subsequently used to identify CoHR sites on chromosomes I, III and VI. The authors reported a correlation between the locations of CoHR sequences (Blumental-Perry et al, 2000) and mapped sites of DSB formation (Game, 1992; Zenvirth et al, 1992; Klein et al, 1996; Baudat & Nicolas, 1997). Blumental-Perry et al (2000) proposed that the CoHR sequence might provide the basis for understanding meiosis-induced chromatin changes that enable DSBs to occur at defined chromosomal sites. To substantiate the importance of the CoHR sequence, the authors examined published data from the ARG4 hotspot (de Massy & Nicolas, 1993). Most deletions at ARG4 that encompass the CoHR sequence also reduce conversion and DSBs.
Figure 1.
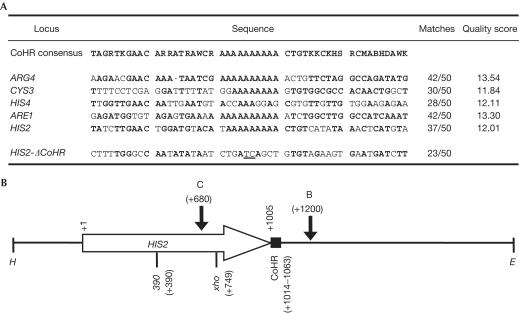
CoHR sequences and features at HIS2. (A) Comparison of the CoHR sequences of five known meiotic recombination hotspots (modified from Fig 1 of Blumental-Perry et al, 2000) and the sequence created by deleting CoHR at HIS2. Sequences and base designations are as in Fig 1 from Blumental-Perry et al (2000), except that the poly(A) region has been corrected from 12 to 10 bp. Matches (in bold) represent base pairs matching the CoHR consensus. The quality score has been taken from Table 1 of Blumental-Perry et al (2000). A comparison of the fusion created by ΔCoHR with the CoHR consensus is shown. Underlined bases represent the fusion junction created by ΔCoHR. (B) Important features at HIS2. The 2.6 kbp HindIII–EcoRI fragment containing HIS2 is shown. The direction of transcription of HIS2 is designated by the horizontal arrow. DSB sites are shown by vertical arrows, with C and B representing DSB-C and DSB-B, respectively. The location of CoHR is designated by the solid box at the end of the HIS2 coding region. Alleles used to measure gene conversion are designated by 390 and xho (Malone et al, 1992, 1994). E=EcoRI; H=HindIII.
One of the hotspots used to identify the CoHR motif was HIS2, which contains two DSB sites: DSB-C is located in the coding region (at +680 relative to the start of the coding region) and DSB-B is located about 200 bp downstream (at +1200) (Bullard et al, 1996; Fig 1B). Blumental-Perry et al (2000) reported that the 5′ region of HIS2 was used for identification of CoHR; however, the CoHR motif is actually located at the 3′ end of HIS2, 10 bp downstream from the end of the coding region (+1014 to +1063). Unlike the CoHR motif at some other hotspots, the CoHR motif at HIS2 is located neither at the 5′ end of the gene nor at the sites of DSB formation.
Studies of the HIS2 hotspot (Haring et al, 2003) and the ARG4 hotspot (Nicolas et al, 1989; de Massy & Nicolas, 1993) have demonstrated that sequences at the DSB sites can profoundly affect the frequency of DSB formation. Since the CoHR site at ARG4 is within the region of DSB formation, it is not surprising that a deletion encompassing this CoHR motif has an effect on hotspot activity; the DSB site is altered as well. It is difficult to determine the contribution of CoHR to hotspot activity by studying hotspots like ARG4 (Liu et al, 1995), ARE1 (YCR048W) (Liu et al, 1995) or CYS3 (de Massy et al, 1995), because CoHR sites lie within sequences at DSB sites. How can one determine the contribution of CoHR to hotspot activity, when removing CoHR alters the DSB site? Since CoHR is separable from the locations of DSB formation at HIS2, this hotspot allows us to test the importance of the CoHR motif; CoHR can be altered without altering sequences at the DSB sites.
Results
Comparison of CoHR sites and DSB sites on chromosome III
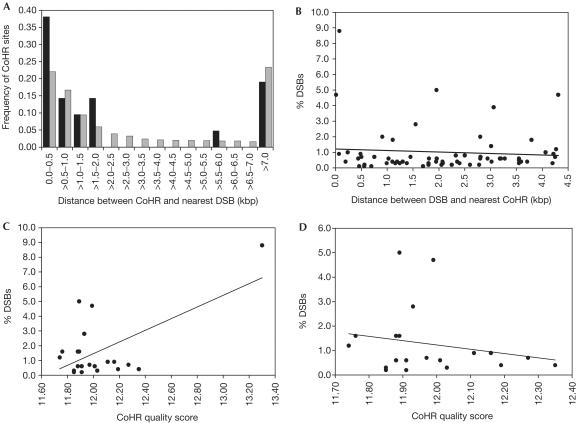
It was reported that the location of CoHR sites correlates with DSB sites on chromosomes I, III and VI (Blumental-Perry et al, 2000). The precise correlation of CoHR and DSB sites on chromosomes I and VI is difficult to determine, because the breaks on these chromosomes were mapped by pulse-field gel electrophoresis (Zenvirth et al, 1992; Klein et al, 1996) and therefore have lower resolution. However, Baudat & Nicolas (1997) mapped DSB formation on chromosome III to an average resolution of ±167 bp. We therefore re-examined the relationship between CoHR and DSB sites on this chromosome. Analysis of all 75 DSB sites (⩾0.1% breaks) on chromosome III revealed that 6.7% (5/75) are located at positions indistinguishable from CoHR sites (including the ARE1 and HIS4 sites used to define CoHR). We then examined the distance between DSB sites and CoHR sites by asking how far from each CoHR the nearest DSB site is located. If CoHR plays a role in the determination of hotspots, one might expect a DSB site relatively close to each CoHR site. We find that 76% (16/21) of chromosome III CoHR sites have a measurable DSB site within 2 kbp. The distribution of distances between CoHR sites and the nearest DSB site is shown in Fig 2A (black bars); the mean distance between a CoHR site and its closest DSB is 4.36±6.79 kbp. However, given that there are 75 DSB sites along the 317 kbp of chromosome III, it is not clear whether these data are different from what would be expected if CoHR sites were distributed randomly along chromosome III. The calculated distribution of distances from CoHR sites to the nearest DSB site for a random distribution (see Methods) along chromosome III is also shown in Fig 2A (grey bars); the mean distance is 4.66 kbp with a 95% confidence interval of 2.03–7.59 kbp. We note that the observed distance distribution does not significantly differ from the random distribution (χ2=9.31, df=14, P=0.81). Since the observed mean distance falls easily within the 95% confidence interval of the mean for a random distribution, the mean values are also not significantly different. It appears that there is no significant association of CoHR sequences with DSB sites on chromosome III.
Figure 2.
Relationships between CoHR sites and DSB sites on chromosome III. (A) Distribution of the distance between CoHR sites and the nearest DSB sites. The actual distribution, determined by combining data from Baudat & Nicolas (1997) and Blumental-Perry et al (2000), is represented by black bars. The random distribution determined by 1,000 independent trials (see Methods) is represented by grey bars. (B) Plot of DSB strength versus distance to nearest CoHR. All detectable DSBs within 4.36 kbp of a CoHR motif were examined. (C) Plot of DSB strength nearest to each CoHR versus CoHR quality score. (D) Plot of DSB strength nearest to each CoHR versus CoHR quality score, as in (C), excluding the ARE1 data point.
In the simplest hypothesis, the closer a DSB site is to a CoHR motif, the stronger the DSB site should be. The frequency of breaks for each DSB site within 4.36 kbp (the mean distance observed in Fig 2A) of a CoHR was plotted against the distance from the CoHR. Fig 2B demonstrates that there is no correlation (r=−0.08; P=0.52) between the strength of a DSB site and its distance from a CoHR site. Owing to the high level of degeneracy of the putative CoHR sequence, CoHR sites were identified based on a quality score system; only CoHR sites scoring above 11.7 were considered significant (for an explanation, see Blumental-Perry et al, 2000). Blumental-Perry et al (2000) calculated the quality scores of CoHR sites present in various constructs at ARG4 (de Massy & Nicolas, 1993) and argued that they might be responsible for the alterations in the frequency of DSBs observed. There is a significant correlation (r=0.61; P=0.0031) between the CoHR quality score and the strength (i.e., frequency) of the DSB nearest each CoHR (Fig 2C) when all 21 CoHR sites on chromosome III are examined. However, examination of the data (Fig 2C) suggested to us that most of the correlation came from the ARE1 data point (8.8% DSBs; quality score=13.30). ARE1 was, of course, one of the loci used to define CoHR. If we remove the ARE1 data point (Fig 2D), there is no correlation between the strength of DSB and quality score (r=−0.21; P=0.38). If we exclude both the sites on chromosome III used to define CoHR (HIS4 and ARE1), there is also no significant correlation (r=−0.20; P=0.41).
The CoHR motif at HIS2 has no effect on recombination
The data from chromosome III suggest that CoHR might not be important for the creation of DSBs. To test directly the role of CoHR, we precisely deleted (ΔCoHR) the entire 50 bp sequence described in Blumental-Perry et al (2000) (Fig 1A). GC was measured in diploids homozygous for ΔCoHR. In the absence of the CoHR motif, conversion of the his2-390 allele is 10.0% (LJL1-1; Table 1). This is not significantly different (G-test; P=0.11) from its isogenic counterpart containing an intact CoHR sequence (RM193; 12.9%). Conversion was also analysed in an isogenic ΔCoHR diploid (LJL1-2) containing the his2-xho allele. The conversion of this marker was 14.9% compared to 14.1% conversion in its CoHR-containing counterpart (RM169; Table 1); there is no statistical difference (G-test; P=0.67). GC remains very high at HIS2 in the absence of CoHR.
Table 1.
GC analysis of strains containing and lacking the CoHR motif at HIS2
| Diploid | Relevant genotype | Total GC | Total tetrads | GC (%) |
|---|---|---|---|---|
| RM193a |
his2-390/HIS2 |
80 |
622 |
12.9 |
| LJL1-1 |
his2-390-ΔCoHR/HIS2-ΔCoHR |
33 |
329 |
10.0 |
| RM169a |
his2-xho/HIS2 |
55 |
391 |
14.1 |
| LJL1-2 | his2-xho-ΔCoHR/HIS2-ΔCoHR | 47 | 315 | 14.9 |
aData taken from Malone et al (1994) and Haring et al (2003).
HIS2 retains a high frequency of DSBs in absence of CoHR
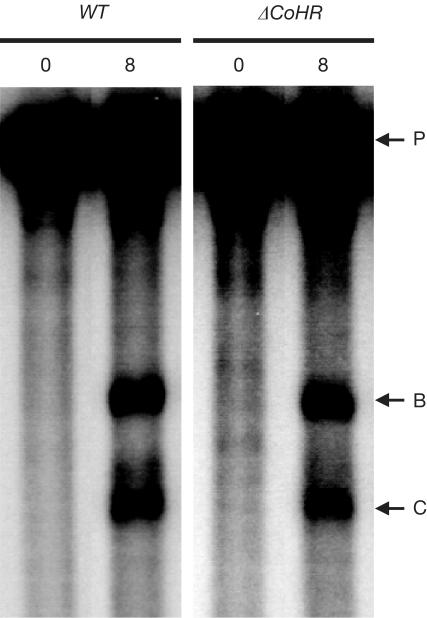
The persistence of high recombination levels at HIS2 in the absence of CoHR implies a high frequency of DSB formation. The measurement of DSBs at HIS2 in ΔCoHR diploids demonstrated that breaks still occur at high frequency (Table 2). The sites of DSB formation in ΔCoHR diploids were indistinguishable from the normal DSB-B and DSB-C sites (Fig 3). Quantitation of the breaks at DSB-B and DSB-C revealed that DSB formation is slightly reduced at both sites (Table 2). DSB-B is reduced from 2.8±1.1 to 1.7±0.2%; DSB-C is reduced from 1.9±0.9 to 0.9±0.3%. However, neither of these differences is significant at the 95% confidence level (Mann–Whitney test; P=0.21 for DSB-B; P=0.07 for DSB-C).
Table 2.
DSB analysis of strains containing and lacking the CoHR motif at HIS2
| Diploidsa | Relevant genotypeb | Isogenic RAD50 derivatives | No. of exp. | DSB-B (%) | DSB-C (%) | Total (%) | |
|---|---|---|---|---|---|---|---|
| SJH5-0 |
his2/HIS2 |
rad50S/rad50S |
RM193 |
4 |
2.8±1.1 |
1.9±0.9 |
4.7±2.0 |
| LJL1-5 |
|
|
RM169 |
|
|
|
|
| LJL1-3 |
his2-ΔCoHR/HIS2-ΔCoHR |
rad50S/rad50S |
LJL1-1 |
8 |
1.7±0.2 |
0.9±0.3 |
2.6±0.5 |
| LJL1-4 | LJL1-2 | ||||||
aThe data from diploids SJH5-0 and LJL1-5 were combined, and the data from LJL1-3 and LJL1-4 were combined. All these strains are isogenic.
bThe rad50S mutation was used, because DSBs form and accumulate in the presence of this allele.
Figure 3.
DSB formation at HIS2 in the presence and absence of the CoHR site. WT refers to a diploid in which CoHR is intact at HIS2; ΔCoHR refers to a diploid in which CoHR is deleted at HIS2. The numbers above each lane represent hours in sporulation. DSB-B, DSB-C and the parental (unbroken) fragments are designated by horizontal arrows labelled B, C and P, respectively.
Medium resolution mapping of HIS2 DSB sites
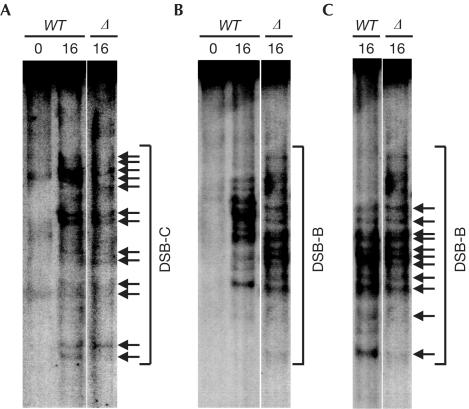
The CoHR deletion has no significant effect on GC or DSB formation at HIS2. To examine whether CoHR affects the precise locations of DSBs, the technique of medium-resolution mapping was employed (Liu et al, 1995; Diaz et al, 2002; Haring et al, 2003). Examination of the break pattern revealed that most (if not all) of the breaks present in diploids with an intact CoHR motif also occur when CoHR is absent. The pattern of breaks at DSB-C demonstrates that the positions of at least 13 break sites are indistinguishable (Fig 4A). At first glance, the pattern of breaks at DSB-B appears to be altered (Fig 4B), because ΔCoHR reduces all band sizes by 50 bp. To account for this, the wild-type lane was resized to mimic the electrophoresis of fragments 50 bp smaller. This lane could then be aligned with the lane containing breaks from a ΔCoHR diploid; at least 11 breaks occur at the same sequence positions (Fig 4C).
Figure 4.
Medium-resolution mapping of DSBs at HIS2 in the presence and absence of the CoHR site. DSBs at indistinguishable sequences in wild-type or ΔCoHR strains are designated by horizontal arrows. WT refers to a wild-type diploid with CoHR intact at HIS2; Δ refers to a diploid with CoHR deleted at HIS2. Numbers above lanes represent hours in sporulation. (A) Mapping of the break pattern at DSB-C. (B, C) Mapping of the break pattern at DSB-B. The pattern of DSB-B in (B) appears altered in a ΔCoHR strain, because the DSB fragments also contain the ΔCoHR (50 bp) deletion. To account for the 50 bp size difference due to ΔCoHR, the WT lane from (B) was stretched in (C) to mimic the running of 50 bp shorter fragments on a gel.
Discussion
The contribution that DNA sequence, per se, makes to the initiation of meiotic recombination has been difficult to resolve. Unlike replication origins (Deshpande & Newlon, 1992) or transcriptional promoters (Kollmar & Farnham, 1993), there is no obvious recombination initiation sequence. At least two types of sequences contribute to hotspot activity: sequences at DSB sites and ‘surrounding sequences' (e.g., Wu & Lichten, 1995; Haring et al, 2003). Sequences at DSB sites affect breaks at that particular site, whereas surrounding sequences are important for hotspot activity within a region (Haring et al, 2003).
The extent of surrounding sequences sufficient to maintain hotspot activity has yet to be determined. Wu & Lichten (1995) and Borde et al (1999) showed that an 8.5 kbp URA3::ARG4::pBR322 construct has different levels of recombination and DSB formation, depending on its position on chromosome III. Examination of these constructs by Petes & Merker (2002) showed that the recombination activity of inserted sequences can be correlated with the GC content of large surrounding chromosomal regions (30–100 kbp). More than 10 kbp are required to maintain 50% of the recombination activity of a hotspot (Ross et al, 1992; Haring et al, 2003) when moved to a different location. In all these constructs, DSB formation occurs at the same sites regardless of the location of the construct; only the frequency is affected. Although long-range surrounding sequences are important for hotspot activity in a region, it is less clear as to what local sequences determine the location and frequency of DSBs.
Our analysis of chromosome III demonstrates no correlation between the amount of DSB formation and the distance to, or quality score of, the nearest CoHR. Furthermore, the distribution of CoHR sites along chromosome III is not significantly different from that expected if CoHR sites were randomly distributed. Taken together, the data for chromosome III suggest that CoHR is not important for DSB formation.
The data clearly indicate that the CoHR sequence at HIS2 contributes little to hotspot activity. Deleting CoHR does not decrease recombination. To argue that CoHR is important, one would have to propose that there is another CoHR site nearby, or that the fusion resulting from ΔCoHR creates a new CoHR site allowing hotspot activity at HIS2 to remain. However, the next closest CoHR site to HIS2 is 6.5 kbp away (Blumental-Perry et al, 2000). In addition, the fusion sequence created by the deletion does not resemble the CoHR motif; only 23/50 bases match, and there is no poly(A) tract (Fig 1A). Not only do DSB breaks form at high frequency in a strain with the CoHR sequence deleted, but the locations of the individual DSBs appear unchanged as well. Since both DSB formation and GC remain high when CoHR is absent, we conclude that CoHR is not required for HIS2 hotspot activity.
Speculation
Since CoHR appears to be unnecessary for hotspot activity at HIS2, we suspect that CoHR may not be required for other hotspots. In those situations (e.g., ARG4) where the deletion of CoHR has been shown to affect recombination, we posit that it was due to loss of the DSB sites themselves or important sequences in the promoter region rather than CoHR. We note that CoHR located at HIS4 (another hotspot used to define the consensus sequence) is not at the site of DSB formation; it is located in the coding region a few hundred bases away. We predict that a deletion of CoHR at HIS4, or of other CoHR sites not located at DSB sites, would have a minimal effect on meiotic recombination. Although the data at HIS2 indicate that CoHR is not important for hotspot activity, studies of CoHR at other hotspots are necessary to confirm the generality of our results.
Methods
Strains. Isogenic derivatives of haploids RM96-15A and RM182-55C were crossed to create all diploids (Malone et al, 1994). Haploids containing the ΔCoHR mutation at HIS2 were created by two-step gene replacement (Rothstein, 1991) using pLJL2. Relevant genotypes of diploids are as follows: RM193 (his2-390/HIS2), RM169 (his2-xho/HIS2), LJL1-1 (his2-390-ΔCoHR/HIS2-ΔCoHR) and LJL1-2 (his2-xho-ΔCoHR/HIS2-ΔCoHR). Diploids used for DSB analysis are isogenic, except homozygous for rad50KI81::URA3 (rad50S; Alani et al, 1990).
Plasmids. To create the ΔCoHR mutation at HIS2, plasmid pSJH1 (Haring et al, 2003) was digested with SmaI and NotI. The digested ends were treated with Klenow and ligated to create pLJL1, eliminating the BamHI site in the multiple cloning site of pSJH1. Primers 362 (5′-ATCCCTTTTGGGCCAATATATAATCTGATCAGCTGTGTAG AAGTGAATGATCTTTCC-3′) and 375 (5′-GGAAAGATCATTCACTTCTACACAGCTGATCAGATTATAT ATTGGCCCAAAAGGGAT-3′) and pLJL1 were used with the QuickChange Site-Directed Mutagenesis Kit (Stratagene) to create pLJL2, containing a precise 50 bp deletion of the CoHR motif downstream of HIS2 (confirmed by DNA sequencing). pLJL2 was targeted for integration by BamHI digestion.
Chromosome III comparisons. All comparisons of DSB sites and CoHR sites on chromosome III were made by combining data from Baudat & Nicolas (1997) (http://www.yeastgenome.org/DSB_table.shtml) and Blumental-Perry et al (2000). Data were graphed using Microsoft Excel, and correlation coefficients were determined using Microsoft Excel and GraphPad Instat.
Generating a random distribution between CoHR and DSB sites. Computer simulations were used to investigate the distance between CoHR sites to the nearest DSB site when CoHR sites are randomly distributed along a chromosome. This process mimics a chromosome 316,613 bp long (i.e., chromosome III) with 75 DSB sites assigned to their known physical positions (Baudat & Nicolas, 1997). A total of 21 CoHR sites are then randomly distributed along the chromosome, and the average distance between each CoHR site and its nearest DSB site is recorded. After 1,000 independent replicates of this process, a distribution of the average distance between CoHR and DSB sites is generated and can be used to obtain confidence intervals and the mean distance. The program used for this analysis is available from J.M.C. upon request.
DSB analysis and medium-resolution mapping. DSB analysis and medium-resolution mapping were carried out as in Haring et al (2003). We estimate the precision of medium-resolution mapping to be ±8 bp.
Acknowledgments
We thank Jan Fassler, Joe Frankel, Kelley Foreman, Morgan Pansegrau and Demelza Houdek for comments on this manuscript. We also thank Jan Fassler and Greg Gingerich for experimental assistance. During part of this research, L.J.L. was supported by a Howard Hughes Undergraduate Research Fellowship. This research was funded initially by NSF Grant MCB97-28557 and subsequently by NSF Grant MCB00-83816, both to R.E.M.
References
- Alani E, Padmore R, Kleckner N (1990) Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436 [DOI] [PubMed] [Google Scholar]
- Baker BS, Carpenter AT, Esposito MS, Esposito RE, Sandler L (1976) The genetic control of meiosis. Annu Rev Genet 10: 53–134 [DOI] [PubMed] [Google Scholar]
- Baudat F, Nicolas A (1997) Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA 94: 5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumental-Perry A, Zenvirth D, Klein S, Onn I, Simchen G (2000) DNA motif associated with meiotic double-strand break regions in Saccharomyces cerevisiae. EMBO Rep 1: 232–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Wu TC, Lichten M (1999) Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol 19: 4832–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard SA, Kim S, Galbraith AM, Malone RE (1996) Double strand breaks at the HIS2 recombination hot spot in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93: 13054–13059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherest H, Surdin-Kerjan Y (1992) Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics 130: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B, Nicolas A (1993) The control in cis of the position and the amount of the ARG4 meiotic double-strand break of Saccharomyces cerevisiae. EMBO J 12: 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B, Rocco V, Nicolas A (1995) The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J 14: 4589–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS (1992) The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol Cell Biol 12: 4305–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz RL, Alcid AD, Berger JM, Keeney S (2002) Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol Cell Biol 22: 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC (1992) Pulsed-field gel analysis of the pattern of DNA double-strand breaks in the Saccharomyces genome during meiosis. Dev Genet 13: 485–497 [DOI] [PubMed] [Google Scholar]
- Haring SJ, Halley GR, Jones AJ, Malone RE (2003) Properties of natural double-strand break sites at a recombination hotspot in Saccharomyces cerevisiae. Genetics 165: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Zenvirth D, Dror V, Barton AB, Kaback DB, Simchen G (1996) Patterns of meiotic double-strand breakage on native and artificial yeast chromosomes. Chromosoma 105: 276–284 [DOI] [PubMed] [Google Scholar]
- Kollmar R, Farnham PJ (1993) Site-specific initiation of transcription by RNA polymerase II. Proc Soc Exp Biol Med 203: 127–139 [DOI] [PubMed] [Google Scholar]
- Lichten M (2001) Meiotic recombination: breaking the genome to save it. Curr Biol 11: R253–R256 [DOI] [PubMed] [Google Scholar]
- Liu J, Wu TC, Lichten M (1995) The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA–protein intermediate. EMBO J 14: 4599–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RE, Bullard S, Lundquist S, Kim S, Tarkowski T (1992) A meiotic gene conversion gradient opposite to the direction of transcription. Nature 359: 154–155 [DOI] [PubMed] [Google Scholar]
- Malone RE, Kim S, Bullard SA, Lundquist S, Hutchings-Crow L, Cramton S, Lutfiyya L, Lee J (1994) Analysis of a recombination hotspot for gene conversion occurring at the HIS2 gene of Saccharomyces cerevisiae. Genetics 137: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A, Treco D, Schultes NP, Szostak JW (1989) An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338: 35–39 [DOI] [PubMed] [Google Scholar]
- Petes TD, Merker JD (2002) Context dependence of meiotic recombination hotspots in yeast. The relationship between recombination activity of a reporter construct and base composition. Genetics 162: 2049–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross LO, Treco D, Nicolas A, Szostak JW, Dawson D (1992) Meiotic recombination on artificial chromosomes in yeast. Genetics 131: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R (1991) Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol 194: 281–301 [DOI] [PubMed] [Google Scholar]
- White MA, Wierdl M, Detloff P, Petes TD (1991) DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc Natl Acad Sci USA 88: 9755–9759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TC, Lichten M (1995) Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics 140: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G (1992) Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J 11: 3441–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]