Abstract
Both Lactococcus lactis and Lactobacillus plantarum contain a single alr gene, encoding an alanine racemase (EC 5.1.1.1), which catalyzes the interconversion of d-alanine and l-alanine. The alr genes of these lactic acid bacteria were investigated for their application as food-grade selection markers in a heterologous complementation approach. Since isogenic mutants of both species carrying an alr deletion (Δalr) showed auxotrophy for d-alanine, plasmids carrying a heterologous alr were constructed and could be selected, since they complemented d-alanine auxotrophy in the L. plantarum Δalr and L. lactis Δalr strains. Selection was found to be highly stringent, and plasmids were stably maintained over 200 generations of culturing. Moreover, the plasmids carrying the heterologous alr genes could be stably maintained in wild-type strains of L. plantarum and L. lactis by selection for resistance to d-cycloserine, a competitive inhibitor of Alr (600 and 200 μg/ml, respectively). In addition, a plasmid carrying the L. plantarum alr gene under control of the regulated nisA promoter was constructed to demonstrate that d-cycloserine resistance of L. lactis is linearly correlated to the alr expression level. Finally, the L. lactis alr gene controlled by the nisA promoter, together with the nisin-regulatory genes nisRK, were integrated into the chromosome of L. plantarum Δalr. The resulting strain could grow in the absence of d-alanine only when expression of the alr gene was induced with nisin.
Alanine racemases are pyridoxal 5′-phosphate-dependent enzymes involved in the interconversion of d-alanine (d-ala) and l-alanine (l-ala). Most well-studied alanine racemases originate from bacteria, including Escherichia coli (52), Pseudomonas putida (41), Salmonella enterica serovar Typhimurium (51), and several Bacillus species (21, 30, 39, 55). More recently, eukaryotic alanine racemases were investigated in more detail, e.g., in the fungus Tolypocladium niveum, where this enzyme is involved in the biosynthesis of cyclosporin A (27). Since d-Ala is involved in the cross-linking of cell wall peptidoglycan layers in many bacteria, this component is essential for their growth.
In E. coli, the dadX gene is involved in l-Ala catabolism. It encodes an alanine racemase and is situated in an operon together with the dadA gene, encoding a d-Ala dehydrogenase. The transcription of dadX and dadA was found to be repressed by glucose but induced by the presence of l-Ala (53). The dadX gene is responsible for 85% of the total alanine racemase activity in E. coli, and a second gene named alr is present, which is constitutively expressed (52, 53). Only the alr dadX double mutant is auxotrophic for d-Ala (53).
In Bacillus subtilis the alanine racemase gene (dal) is involved in alanine conversion and a dal deletion mutant was dependent on d-Ala supplementation when grown in rich media. However, in minimal medium, growth of the dal mutant was affected only after addition of l-Ala. Hence, it was suggested that B. subtilis possesses a second, l-Ala-repressible alanine racemase (21). The complete genome sequence of B. subtilis confirms the existence of a second gene (yncD) whose product shows high homology to alanine racemases (35). Furthermore, alanine racemases from Bacillus species are postulated to be involved in spore formation, since racemase activity is generally higher in spores than in vegetative cells (39, 40, 46). Spore alanine racemase converts the germinant l-Ala into its competitive inhibitor d-Ala and may regulate spore germination (54). The most extensively studied alanine racemase originates from Bacillus stearothermophilus and includes sequence analysis of the gene and protein, determination of the catalytic-site residues, and characterization of the biochemical properties of the protein (30, 44, 47). Moreover, the three-dimensional structure of the alanine racemase of this organism was determined by X-ray crystallography (43).
Several allosteric inhibitors of alanine racemases have been described, including d-cycloserine, hydroxylamine, and β-chloroalanine (1, 32, 36, 50). Inactivation of alrA, encoding alanine racemase, resulted in a 30-fold-lower MIC of d-cycloserine for Mycobacterium smegmatis (9). Furthermore, a d-cycloserine- resistant mutant of M. smegmatis was shown to display elevated alanine racemase activity, caused by a promoter-up mutation. Similarly, increasing the alrA gene dosage by cloning on a multicopy plasmid resulted in increased d-cycloserine resistance in M. intracellulare and M. bovis (6).
In the lactic acid bacteria (LAB) Lactobacillus plantarum and Lactococcus lactis, alanine racemase activity is encoded by homologous alr genes. Disruption of alr in both LAB resulted in auxotrophy for d-Ala on rich media (28, 29). Additionally, no growth of the L. plantarum deletion mutant was observed on minimal medium with or without l-Ala, indicating that l-Ala does not suppress another putative alanine racemase. In contrast to B. subtilis, alr appears to be the sole gene coding for alanine racemase activity in L. plantarum (28). LAB are important organisms for the production of fermented foods and feeds. Moreover, they are used as probiotics and have great potential to serve as delivery vehicles of health-promoting compounds to the human gastrointestinal tract (23, 24, 45). To further optimize these microbes for industrial exploitation, genetic modification approaches have been used (14). However, the application of genetically modified microorganisms in food products requires safe and sustainable genetic tools. Therefore, sophisticated food-grade marker systems are being developed that circumvent the addition of undesirable components like antibiotics to industrial fermentation processes. Several food-grade marker systems have been developed for the selection of a plasmid in LAB, of both the dominant and complementation type (14). Recently, a third type of food-grade selection markers was described, using a two-plasmid system for food-grade selection in lactococci (17).
d-Ala is not a regular constituent of industrial fermentation media, suggesting that the alanine racemase-encoding gene of LAB could be exploited as a food-grade complementation marker. Previously, a similar approach in B. subtilis showed that the dal gene, encoding alanine racemase, could be used as a functional and stringent complementation marker in rich media (21). Here, we describe the use of the alr gene as a heterologous food-grade selection marker in both L. plantarum and L. lactis. Plasmids expressing alr were introduced into alr mutants of both organisms and into their wild-type parent strains, using d-cycloserine resistance for selection of transformants in the latter strains. Selection appeared highly stringent, and plasmids were stably maintained during culturing. Moreover, alr expression levels in L. lactis could be correlated with d-cycloserine resistance levels, and a nisin-dependent alr mutant strain of L. plantarum was constructed that showed a conditional lethal phenotype.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
L. lactis MG1363 (22) and L. plantarum NCIMB8826 (26) were used in this study, and their derivatives and the plasmids and primers used are listed in Table 1. E. coli MC1061 (8) was used as the intermediate cloning host and was handled as described by Sambrook et al. (42). L. plantarum was grown at 30°C in MRS broth (Difco, West Molesey, United Kingdom) without aeration. L. lactis was grown at 30°C without aeration in M17 broth (Merck, Darmstadt, Germany) supplemented with 0.5% (wt/vol) glucose. D-Ala (200 μg/ml) was added to these media when indicated. When appropriate, antibiotics were added to the different media; for E. coli, ampicillin (50 μg/ml), erythromycin (250 μg/ml), and chloramphenicol (20 μg/ml); for L. lactis, chloramphenicol (10 μg/ml); for L. plantarum, erythromycin (5 μg/ml) plus lincomycin (10 μg/ml), and chloramphenicol (10 μg/ml).
TABLE 1.
Strains, plasmids and primers used in this studya
| Strain, plasmid, or primer | Relevant featuresb | Source of reference |
|---|---|---|
| Strains | ||
| L. plantaruma | ||
| MD007 | L. plantarum NCIMB8826 Δalr,d-Ala auxotroph | M. Deghorain, unpublished data |
| MD007Int6 | Emr, nisRK, pMEC10 chromosomal integration in MD007 at tRNASer | This work |
| MD007::pGIP014 | Emr, nisRK, pGIP014 chromosomal integration in MD007 at tRNASer | This work |
| L. lactis | ||
| NZ3900 | MG1363 with a chromosomal insertion of nisRK | 12 |
| PH3960 | NZ3900 Δalr, nisRK | 29 |
| Plasmids | ||
| pJDC9 | Emr | 10 |
| pGIP009 | Emr, pJDC9 derivative containing L. lactis alr gene | This work |
| pGIT032 | Emr Cmr, PldhL | 18 |
| pNZ2650 | Cmr, PnisA | 29 |
| pGIP011 | Emr Cmr, pGIT032 derivative containing L. lactis alr downstream of PldhL | This work |
| pGIP012 | Cmr, pNZ2650 derivative containing L. lactis alr downstream of PnisA | This work |
| pNG8048-Cre | Cmr | 7 |
| pUC-NcoI | Apr, pUC18 derivative containing NcoI and ribosome-binding site of prtP | 20 |
| pNZ7110 | Apr, pUC derivative containing cre and TpepN | This work |
| pNZ7115 | Apr, pNZ7110 with cre replaced by L. plantarum alr PCR product | This work |
| PNZ8020 | Cmr, PnisA | 11 |
| pGIP013 | pNZ8020 derivative containing L. plantarum alr from pNZ7115 | This work |
| pGEMT-easy | Apr | Promega |
| pGIP010 | Apr, pGEMT-easy derivative containing L. lactis alr PCR product | This work |
| pMEC10 | Emr, nisRK | 38 |
| pGIP014 | Emr, nisRK, pMEC10 derivative containing alr fragment of pGIP010 | This work |
| Primers | ||
| LLALR-5 | 5′-CGAGGATCCGCATAGTAATTTAGAAGCTGTTGC-3′ | This work |
| LLALR-11 | 5′-ACGCGTCGACATTTGTAAAGGCTTTTATGAGAT-3′ | This work |
| LLALR-12 | 5′-GGGGTACCATTTGTAAAGGCTTTTATGAGAT-3′ | This work |
| LLALR-13 | 5′-CGGAATTCATTACAGCTCCAAGACTAGTC-3′ | This work |
| TRNA | 5′-GCGAACCGGCTAATACCGGC-3′ | This work |
| 409301L1 | 5′-AACAGAAGGTGGGACAGTAG-3′ | This work |
| LplAlr_F | 5′-AGGCAATTTGCCATGGTTGTAATTGG-3′ | This work |
| LplAlr_R | 5′-ACGTTCTAGATTAATCTATATAAACTCTCGG-3′ | This work |
Strain NCIMB8826 was obtained from the National Collections of Industrial, Food and Marine Bacteria, Aberdeen, Scotland.
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Emr, erythromycin resistant. Underscored base pairs in primers indicate introduced restriction sites.
DNA manipulations.
Plasmid DNA was isolated from E. coli on a small scale using the alkaline-lysis method (2, 42). Large-scale plasmid DNA isolations were performed using Jetstar columns as specified by the manufacturer (Genomed GmbH, Bad Oberhausen, Germany). DNA isolation and transformation in L. lactis and L. plantarum were performed as described previously (15, 19, 31). Standard procedures were applied for DNA manipulations in E. coli (42). Restriction endonucleases, Taq and Pwo polymerase, T4 DNA ligase, and Klenow polymerase were used as recommended by the manufacturer (Promega, Leiden, The Netherlands, and Boehringer GmbH, Mannheim, Germany). Primers were purchased from Pharmacia Biotech (Roosendaal, the Netherlands) or Genset Oligos (Paris, France).
Plasmid constructions.
L. lactis MG1363 genomic DNA (22) was used as a template to amplify the alr gene, using primers LLALR-5 and LLALR-11, which were designed on the basis of the L. lactis IL1403 genome sequence (3). Sequence analysis of the amplicons obtained revealed 98% identity between the alr genes of L. lactis IL1403 and L. lactis MG1363. The 1.3-kb PCR product was digested with BamHI and SalI (restriction sites were introduced by the primers) and inserted into similarly digested pJDC9, resulting in pGIP009. To drive expression of the L. lactis alr gene by the ldhL promoter of L. plantarum (18), pGIP009 was digested with HindIII and SmaI and then ligated to pGIT032, previously digested with HindIII and HpaI. The resulting plasmid was designated pGIP011 and was introduced into L. plantarum NCIMB8826 and MD007. Similarly, to place alr expression under control of the nisA promoter of L. lactis (11), pGIP009 was digested with HindIII and BamHI and subsequently ligated to similarly digested pNZ2650, yielding pGIP012, which was introduced into L. plantarum MD007Int6 carrying the L. lactis nisRK genes on the chromosome.
The strategy described above was also used to construct plasmids expressing the L. plantarum alr gene for transformation and selection in L. lactis. To remove the HindIII site from pNG8048-cre (7), this plasmid was digested with HindIII, blunt ended with Klenow DNA polymerase, and backligated. The cre-TpepN fragment in this vector was removed with NcoI and XhoI and ligated to pUC-NcoI (20) digested with NcoI and SalI. Finally, digestion of this vector with EcoRI and HindIII, followed by Klenow DNA polymerase treatment, yielded a fragment containing RBS-cre-TpepN, which was inserted into a pUC18 derivative previously digested with SmaI. The resulting plasmid was designated pNZ7110. The alr gene was obtained by a PCR amplification using primers LplAlr_F and LplAlr_R and with L. plantarum NCIMB8826 chromosomal DNA as template. The 1.1-kb PCR product was digested with NcoI and XbaI and ligated to pNZ7110 digested with NcoI and NheI, in order to exchange cre for alr. The resulting plasmid was designated pNZ7115. Finally, RBS-alr-TpepN was digested from pNZ7115 as a BamHI-EcoRI fragment and cloned downstream of the nisA promoter in similarly digested pNZ8020, yielding pGIP013. This plasmid was used for transformation of NZ3900 and strain PH3960.
The alr mutant strains of L. plantarum NCIMB8826 and L. lactis NZ3900, designated MD007 and PH3960 (Table 1), respectively, carry a stable deletion of an internal fragment of 100 and 30 bp in the alr gene, resulting in auxotrophy for d-Ala in both organisms (28, 29). Since a large part of the alr genes in MD007 and PH3960 remain present in the chromosome, a heterologous complementation strategy was used here to avoid integration of the plasmid into the chromosome. Therefore, pGIP011 and pGIP012, containing the lactococcal alr gene under control of the ldhL and nisA promoter, were used for complementation of the L. plantarum Δalr strains MD007 and MD007Int6, respectively. Similarly, pGIP013, containing the L. plantarum alr gene under control of the nisA promoter, was used for complementation of the L. lactis Δalr strain PH3960.
To obtain a conditional alr mutant using the NICE system (12), a plasmid was designed for integration of the L. lactis alr gene into the chromosome of L. plantarum MD007. pGIP012 was used as template to amplify the nisA promoter fused to the alr gene, using primers LLALR-12 and LLALR-13 for the PCR. The 1.6-kb PCR product was cloned in pGEMT-easy (Promega Biotech), resulting in a vector designated pGIP010. This vector was digested with SacI and KpnI, and the resulting fragment containing PnisA and alr was subcloned into similarly digested pMEC10 (38), yielding the vector pGIP014. This vector was used for integration into the genome of L. plantarum MD007.
Alanine racemase assay.
L. lactis strain PH3960 harboring pGIP013 was cultured overnight at 30°C in GM17 with chloramphenicol. The culture was used to inoculate fresh medium (starting optical density at 600 nm [OD 600], 0.25) at 30°C. Growth was continued to an OD600 of 0.6. Nisin was then added to the cultures at concentrations of 0, 0.001, 0.01, 0.1, or 1.0 ng/ml, and growth was continued for 2.5 h. Cultures were harvested by centrifugation, and the pellets were washed with 2 ml of distilled water and recentrifuged. Subsequently, the cells were resuspended in distilled water to an OD600 of 20, 1 g of zirconium beads (48) was added to the cell suspension, and the cells were mechanically disrupted. After centrifugation, the cell extract was transferred to fresh tubes and stored at 4°C. Alanine racemase activity was measured by determining d-Ala conversion into l-Ala by spectrophotometrically monitoring the production of NADH in a coupled reaction as the addition of l-alanine dehydrogenase (l-ADH) from B. sphaericus converts l-Ala into pyruvate and ammonia. l-ADH was obtained from L. lactis NZ3900 harboring plasmid pNZ2650 (29). After induction with 1 ng of nisin per ml, a cell extract was prepared and used as source of l-ADH. The assay mixture used here contained 0.001 to 0.015 U of alanine racemase depending on the nisin induction level, 0.025 U of l-ADH, 100 mM sodium phosphate buffer (pH 8.0), 10 mM NAD +, and 50 mM d-alanine in a total reaction volume of 1 ml. After addition of the sample containing Alr activity, the OD340 at 25°C was monitored for at least 5 min and enzyme activities were correlated to the total protein content of the cell extracts as described previously (4).
Measurement of d-cycloserine resistance in cultures.
PH3960 harboring pGIP013 was grown overnight in GM17 containing chloramphenicol and d-Ala. Cultures were diluted 1:20 and grown in the same medium to an OD600 of 1.3. Afterwards, 1:8 dilutions were cultured for 1 h in the same medium with nisin concentrations ranging from 0 to 1 ng/ml. Then the cultures were diluted 1:10 in the same medium with d-cycloserine ranging from 0 to 1 mg/ml. OD600 values were measured, growth was continued for 2 h, and OD600 values were measured again. The OD increase in the cultures grown in the presence of d-cycloserine was compared to that observed in the cultures grown without added d-cycloserine.
The nucleotide sequences of the alr genes of L. plantarum NCIMB8826 and L. lactis MG1363 are accessible through the GenBank database under accession numbers Y08941 and Y18148, respectively.
RESULTS
Heterologous complementation of MD007 and PH3960.
To evaluate the potential of the alanine racemase-encoding gene, alr, as a heterologous complementation marker, pGIP011 (carrying L. lactis alr) was introduced into the alr deletion variant of L. plantarum NCIMB8826 (MD007). After introduction of pGIP011 into MD007, the cells were plated on MRS or MRS containing erythromycin, lincomycin, and d-Ala. After approximately 16 h of incubation, the MRS plates contained full-grown colonies selected by alanine racemase production, while after approximately 40 h, similar numbers of colonies were obtained on the antibiotic resistance selection plate. In addition, colonies selected by alr expression appeared more homogenous in size. These results indicate that in trans complementation by the L. lactis alr gene under control of the ldhL promoter results in regeneration of growth of the Δalr strain of L. plantarum without d-Ala supplementation. Similar results were obtained using pGIP012 for transformation into the L. plantarum Δalr strain MD007int6. Colonies appeared faster and were more homogenous in size when grown on MRS plates than when grown on MRS plates containing chloramphenicol and d-Ala. Similar observations were made when pGIP013 was introduced into the L. lactis Δalr strain PH3960 and subsequently plated on GM17 or GM17 containing chloramphenicol and d-Ala. Although tightly regulated, the high-copy NICE vector containing alr apparently provides sufficient alr transcription to fully complement the alr phenotype in both hosts, even without the addition of nisin, suggesting that only relatively low expression levels are required for growth.
Plasmid integrity was investigated in pGIP011 and pGIP013 transformants of L. plantarum MD007 and L. lactis PH3960, respectively. Twenty of these L. plantarum and L. lactis colonies were inoculated in medium containing erythromycin or chloramphenicol, respectively. All cultures were fully grown overnight. Following isolation of plasmids pGIP011 and pGIP013, restriction analyses revealed that the plasmids were apparently intact (data not shown), indicating that selection of the transformants is equally stringent on the basis of alr and on the basis of the antibiotic markers.
To evaluate the stability of these food-grade selectable plasmids, MD007Int6 colonies harboring pGIP012 and PH3960 colonies harboring pGIP013 were pregrown for 200 generations in MRS or GM17 medium, respectively, with or without d-Ala. After four successive cycles of growth for 50 generations, the cells were plated on medium containing d-Ala and the presence of the plasmid in the resulting colonies was assessed through the Alr phenotype by replica plating to plates with or without d-Ala (Table 2). After 200 generations of culturing in the presence of d-Ala, plasmid pGIP012 was undetectable in MD007Int6. However, all cells that were cultured without d-Ala still contained pGIP012 after 200 generations. This indicates that the alr marker strongly contributes to the stability of the plasmid in L. plantarum. In contrast, all tested cells of strain PH3960 contain plasmid pGIP013 after 200 generations, independent of the presence or absence of selective pressure. Plasmid DNA isolation of pGIP012 from MD007Int6 and pGIP013 from PH3960 suggests that the copy number of the lactococcal plasmid is 5 to 10 times higher (data not shown), possibly explaining the higher stability of pGIP013 than of pGIP012. Fermentation media do not usually contain d-Ala, making alr a stable, food-grade complementation marker under industrial conditions for both L. lactis and L. plantarum.
TABLE 2.
Stability of alr-carrying plasmids pGIP012 and pGIP013 in L. plantarum MD007Int6 and L. lactis PH3960a
| No. of generations of growth with d-ala | % of colonies with plasmids
|
|
|---|---|---|
| MD007Int6 Alr + | PH3960 Alr + | |
| 50 | 100 | 100 |
| 100 | 34 | 100 |
| 150 | 2 | 100 |
| 200 | 2 | 100 |
Cells were pregrown with or without d-Ala for multiples of 50 generations, and the presence of the plasmids was judged by the ability of the cells to grow without d-Ala. After culturing under selection pressure (without d-Ala), both L. plantarum and L. lactis have stably maintained their plasmids in all tested colonies. The results for the cultures grown without selection pressure (with D-ala) are presented in the table.
Selection of pGIP011 and pGIP013 in wild-type strains.
Several competitive inhibitors for Alr have previously been described (1, 32, 36, 50). In M. smegmatis, resistance to d-cycloserine could be correlated with the expression level of alr (6). This suggests that d-cycloserine resistance can be used for the dominant selection of transformants harboring a plasmid expressing alr. To evaluate this possibility in LAB, pGIP011 and pGIP013 were used for selection in the wild-type L. plantarum NCIMB8826 and L. lactis NZ3900. Plasmid pGIP011 was introduced into L. plantarum NCIMB8826, and the cells were plated on MRS with increasing concentrations of d-cycloserine. After approximately 96 h of growth at 30°C, colonies appeared on the plates with d-cycloserine concentrations up to 600 μg/ml (Table 3). The presence of pGIP011 in these colonies was confirmed by replica plating to MRS plates containing erythromycin and lincomycin (Table 3). The vast majority (96%) of the colonies originating from the MRS plate containing 600 μg of d-cycloserine per ml harbored pGIP011. Similarly, the possibility of selecting pGIP013 transformants of L. lactis strain NZ3900 was evaluated. Since pGIP013 contains the L. plantarum alr gene under control of the nisA promoter, dominant d-cycloserine-based selection was evaluated on plates containing 1 ng of nisin per ml. Addition of increasing concentrations of d-cycloserine resulted in a decrease of CFU appearing after approximately 96 h. Replica plating proved that all tested colonies from the plate containing 200 μg of d-cycloserine per ml possessed the Cmr phenotype, indicating that pGIP013 is present (Table 3). These results indicate that alr can be used as a dominant selection marker in wild-type variants of these LAB.
TABLE 3.
d-Cycloserine-based dominant selection of plasmids pGIP011 and pGIP013 in wild-type strains of L. plantarum and L. lactisa
| Organism | d-Cycloserine concn (μg/ml) | CFU | Antibiotic resistance (%) |
|---|---|---|---|
| L. plantarum | 0 (+Ery) | 746 | 100 |
| 450 | 442 | 11 | |
| 500 | 264 | 43 | |
| 550 | 139 | 75 | |
| 600 | 109 | 96 | |
| L. lactis | 0 (+Cm) | 326 | 100 |
| 100 | >1,000 | 25 | |
| 150 | 769 | 96 | |
| 200 | 418 | 100 |
Numbers of primary transformants are indicated (CFU). Selection stringency depends on the concentration of d-cycloserine present in the media, as judged by replica plating to determine the antibiotic resistance phenotype of the colonies growing at different d-cycloserine concentrations (percent antibiotic resistance). Control plates without d-cycloserine contain the appropriate antibiotic. Data are representative of three independent experiments. Ery, erythromycin; Cm, chloramphenicol.
Nisin inducible d-cycloserine resistance in PH3960.
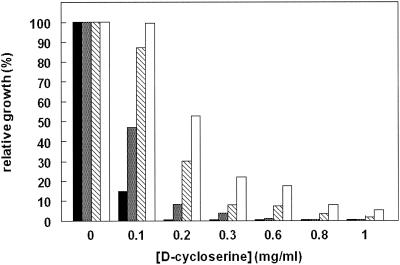
The correlation between d-cycloserine resistance and alr expression in L. lactis was further investigated. First, strain PH3960 harboring pGIP013 was used in a nisin induction experiment. Cell extracts were prepared from cultures grown in the presence of different concentrations of nisin and used for determination of the specific alanine racemase activity. Results from this experiment proved that induction with nisin led to overexpression of the alanine racemase enzyme, as indicated by the higher levels of alanine racemase activity (Table 4). Subsequently, strain PH3960 harboring pGIP013 was used for determination of d-cycloserine resistance levels at different nisin induction levels. The data clearly indicated that at higher nisin induction levels, the elevated Alr levels resulted in higher d-cycloserine resistance levels, as could be judged from culture growth (Fig. 1). Subsequently, an analysis was performed to determine if this correlation between nisin and d-cycloserine resistance could be detected on plates. Cells were grown to an OD600 of 0.5 and plated on GM17 containing chloramphenicol and different concentrations of nisin and d-cycloserine. After growth at 30°C, colonies appeared on the plates with d-cycloserine concentrations up to 300 μg/ml. A similar correlation between nisin and d-cycloserine resistance was found, as judged by the larger number of colonies formed after higher levels of nisin induction (data not shown). This experiment demonstrated that stepwise increase of alr expression led to a stepwise increase of the d-cycloserine resistance level in L. lactis.
TABLE 4.
Nisin induction experiment for the overexpression of L. plantarum alr in L. lactisa
| Nisin concn (ng/ml) | Sp act (U/g protein) |
|---|---|
| 0 | 3.4 |
| 0.001 | 5.6 |
| 0.01 | 10.7 |
| 0.1 | 36.8 |
| 1 | 70.7 |
Alr activity was measured in PH3960 harboring pGIP013 after induction with nisin concentrations ranging from 0 to 1 ng/ml.
FIG. 1.
Correlation between the level of Alr and the level of d-cycloserine resistance in L. lactis. PH3960 harboring pGIP013 was cultured with different concentrations of nisin and d-cycloserine. After 2 h, the growth rate was determined by OD600 measurement. Cultures were induced with 0, 0.01, 0.1, or 1 ng of nisin per ml (black, gray, hatched, and white bars, respectively). Note that growth without d-cycloserine was similar for all nisin concentrations and is taken as 100%.
Conditional alr mutant of L. plantarum.
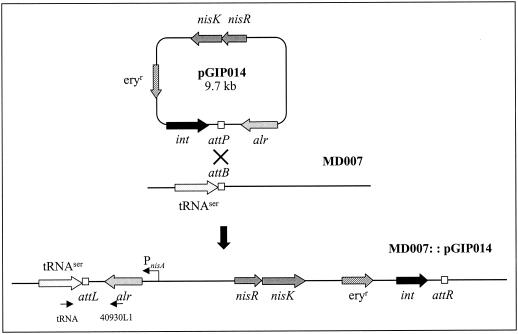
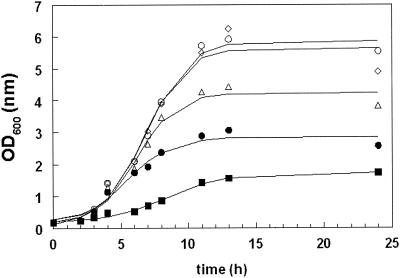
In addition to the use of plasmid pGIP013 for food-grade selection in L. lactis, the possibility of using alr as a single-copy chromosomal marker was investigated. For this purpose, pGIP014 was constructed and integrated into the chromosome of L. plantarum MD007. This vector contains the lactococcal alr gene under control of the nisA promoter. Other important features of the construct are the nisRK genes that are required for functional implementation of the NICE system in L. plantarum NCIMB8826. The alr fusion was cloned in the opposite orientation relative to the tRNASer to minimize readthrough from upstream promoters (Fig. 2). Finally, pGIP014 carries the int gene, which is involved in the integration of the plasmid into the chromosome via site-specific recombination between the attP and attB sites. This plasmid was transformed to the L. plantarum Δalr strain MD007, and the anticipated genomic organization after correct integration was verified by PCR (data not shown). A single colony possessing the correct genotype was designated MD007::pGIP014, and its growth characteristics were analyzed in relation to its d-Ala requirement under conditions in which the nisin concentration varied. These experiments revealed that without addition of nisin, this strain was unable to grow for at least 24 h in the absence of d-Ala in the plates. After pregrowth in liquid MRS medium with d-Ala, the strain was grown in MRS medium without d-Ala and with increasing concentrations of nisin (0.1 to 20 ng/ml). For 24 h, the growth of the cultures was monitored by measuring the OD600 (Fig. 3). These measurements revealed that addition of 20 ng of nisin per ml led to growth very similar to that of a control culture grown in the presence of d-Ala. Moreover, gradual induction of alr expression using the NICE system leads to a gradual increase in the growth rate. These experiments show that the alr gene can be used for single-copy selection on the chromosome.
FIG. 2.
Integration of pGIP014 into the chromosome of MD007 at the tRNASer locus by site-specific integration. pGIP014 harbors the attP site, which is identical to the chromosomally localized attB site. The int gene product catalyzes the recombination event between the attB and attP sites. The indicated primers tRNA and 40930L1 were used to identify the recombination event. The final strain was designated MD007::pGIP014. In this strain, the alr gene is orientated opposite to all neighboring genes, preventing readthrough expression from surrounding genes.
FIG. 3.
Growth of strain MD007::pGIP014 depends on nisin addition. An overnight culture of MD007::pGIP014 in MRS containing d-Ala was used to inoculate MRS containing nisin concentrations of 0.1 (solid squares), 1 (solid circles), 3 (triangles), and 20 (diamond) ng/ml. Open circles represent a control culture grown with d-Ala. Growth was monitored for 24 h by OD600 measurement. Data are representative of three independent experiments.
DISCUSSION
In the present study, we exploitated the alanine racemase-encoding gene as a safe, food-grade complementation marker for L. lactis and L. plantarum. Selection based on alr expression appeared to be faster and equally stringent compared to selection based on antibiotic resistance. Several food-grade selection markers, of both the dominant and complementation type, have been described for various bacteria, including the use of alr in B. subtilis (21). The experiments presented here demonstrate that the strategy used for B. subtilis could readily be extended and used for LAB. The plasmids used here were highly stable during culturing under selective pressure, making alr a valuable, sustainable, food-grade marker in these LAB, since d-Ala is absent in most industrial fermentation media. Additionally, some LAB are considered suitable candidate microorganisms for the delivery of health-promoting factors to the human gastrointestinal tract (23, 24, 45). The alr gene thus provides a valuable tool for stable maintenance of plasmids. Historically, both complementation and dominant markers have been reported that use the capacity to ferment a given sugar as the selection criterion (14). This includes, for lactobacilli, the use of the lacLM genes as a complementation marker in L. helveticus (25). However, the latter marker only functions in media containing lactose as the sole carbon source. Hence, to our knowledge, the alr marker presented here is the first food-grade marker in a Lactobacillus species that can be used independently of the carbon source available in the medium. Moreover, this is the first complementation marker described in L. plantarum. Finally, we have shown that alr could be selected both as a multicopy plasmid-based marker and as a single-copy chromosomal marker.
d-Cycloserine, a competitive inhibitor of alanine racemase activity, could be used for selection of plasmids expressing alr in the wild-type strains of L. plantarum and L. lactis. Essentially, the results prove that alr can be used as both a complementation and dominant marker, which to our knowledge is a novel feature among the food-grade marker systems described to date. Since several other compounds inhibiting alanine racemase activity are known (1, 32, 36, 50), replacement of d-cycloserine by any of the other inhibitors seems possible in this dominant selection system. Even though application possibilities with alr using dominant selection are limited, it is a functional tool in the laboratory, especially in L. plantarum and other LAB, since the number of functional markers in these organisms is limited. Hence, alr-based selection could provide a useful expansion of the genetic tools available for these LAB. Application of the alr gene as a food-grade complementation marker can probably be extended to many other LAB, including industrial strains. Construction of genetically modified Alr-deficient variants of such strains might not be required, since it seems feasible to select natural alr mutants of LAB by screening for the d-Ala requirement for growth. Although this strategy would require a negative screening procedure, the high-throughout screening methods that have become available might allow such an approach.
We have demonstrated how the stepwise overexpression of alr by the NICE system leads to a commensurate stepwise increase in the levels of d-cycloserine resistance in L. lactis, in both liquid and solid media. These results show that the activity of the nisA promoter and the cognate Alr expression levels in the cell can be correlated with d-cycloserine resistance levels. Tools for screening promoter libraries for conditionally active promoters have been described previously for LAB, e.g., use of the promoterless cat gene (49). However, correlation of the level of chloramphenicol resistance with promoter strength was possible only in a narrow range of promoter activities, and subtle differences in promoter strength probably remain undetected using this system. Using the NICE system (12), our experiments suggest that an alr-based system enables selection on basis of differences in promoter strength, in a wide dynamic range.
Thus far, the NICE system has been mainly used for overexpression of proteins (29, 37, 38). However, the NICE system has recently also been applied to more tailor-made expression strategies (13). Here we describe the introduction of the alr gene into the chromosome of L. plantarum under control of the NICE system, resulting in a nisin-controlled conditional mutant; hence, nisin acts as the trigger for the change in growth phenotype. To our knowledge, this is the first report describing a conditional Lactobacillus species mutant with a mutation in an essential gene using the NICE system. A similar essential-gene disruption strategy could also be used in the original host, L. lactis (34; I. C. Boels, unpublished data). However, NICE can be implemented in many gram-positive hosts (5, 16, 33, 38), generating similar regulatory characteristics, including promoter silence under noninducing conditions. These findings suggest that a broad application of this system allows us to study essential gene mutation phenotypes in a wide range of microorganisms.
Overall, we expanded the set of food-grade markers available in LAB for the expression of heterologous proteins with our alr selection system. Furthermore, our system will be helpful in the development of delivery vehicles, since cell lysis can be easily manipulated. Currently we are investigating the potential use of alr as a qualitative and quantitative reporter that can be applied in conditional promoter and promoter strength selection procedures.
Acknowledgments
We thank Steve Kotsonis for critically reviewing the manuscript.
This work was supported by the EU projects LABVAC (BIO4-CT96-0542) and LABDEL (EU-QLRT-2000-00340)
REFERENCES
- 1.Badet, B., and C. Walsh. 1985. Purification of an alanine racemase from Streptococcus faecalis and analysis of its inactivation by (1-aminoethyl)phosphonic acid enantiomers. Biochemistry 24:1333-1341. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, N. E., N. B. Harris, J. F. Wellehan, Z. Feng, V. Kapur, and R. G. Barletta. 1997. Overexpression of the D-alanine racemase gene confers resistance to d-cycloserine in Mycobacterium smegmatis. J. Bacteriol. 179:5046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campo, N., M. L. Daveran-Mingot, K. Leenhouts, P. Ritzenthaler, and P. Le Bourgeois. 2002. Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis. Appl. Environ. Microbiol. 68:2359-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 9.Chacon, O., Z. Feng, N. B. Harris, N. E. Caceres, L. G. Adams, and R. G. Barletta. 2002. Mycobacterium smegmatis d-alanine racemase mutants are not dependent on d-alanine for growth. Antimicrob. Agents. Chemother. 46:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155-164. [DOI] [PubMed] [Google Scholar]
- 11.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food- grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ruyter, P. G., O. P. Kuipers, W. C. Meijer, and W. M. de Vos. 1997. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat. Biotechnol. 15:976-979. [DOI] [PubMed] [Google Scholar]
- 14.de Vos, W. M. 1999. Safe and sustainable systems for food-grade fermentations by genetically modified lactic acid bacteria. Int. Dairy J. 9:3-10. [Google Scholar]
- 15.de Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis ssp. cremoris SK 11 gene encoding an extracellular serine protease. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 16.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emond, E., R. Lavall Ee, G. Drolet, S. Moineau, and G. LaPointe. 2001. Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl. Environ. Microbiol. 67:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferain, T., D. Garmyn, N. Bernard, P. Hols, and J. Delcour. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferain, T., J. N. Hobbs, Jr., J. Richardson, N. Bernard, D. Garmyn, P. Hols, N. E. Allen, and J. Delcour. 1996. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J. Bacteriol. 178:5431-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrari, E., D. J. Henner, and M. Y. Yang. 1985. Isolation of an alanine racemase gene from Bacillus subtilis and its use for plasmid maintenance in B. subtilis. Bio/Technology 3:1003-1007. [Google Scholar]
- 22.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, C., K. Robinson, R. W. Le Page, and J. M. Wells. 2000. Heterologous expression of an immunogenic pneumococcal type 3 capsular polysaccharide in Lactococcus lactis. Infect. Immun. 68:3251-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grangette, C., H. Muller-Alouf, D. Goudercourt, M. C. Geoffroy, M. Turneer, and A. Mercenier. 2001. Mucosal immune responses and protection against tetanus toxin after intranasal immunization with recombinant Lactobacillus plantarum. Infect. Immun. 69:1547-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashiba, H., R. Takiguchi, K. Jyoho, and K. Aoyama. 1992. Establishment of a host-vector system in Lactobacillus helveticus with beta-galactosidase activity as a selection marker. Biosci. Biotechnol. Biochem. 56:190-194. [DOI] [PubMed] [Google Scholar]
- 26.Hayward, A. C., and G. H. G. Davis. 1956. The isolation and classification of Lactobacillus strains from italian saliva samples. Br. Dent. J. 101:2733-2741. [Google Scholar]
- 27.Hoffmann, K., E. Schneider-Scherzer, H. Kleinkauf, and R. Zocher. 1994. Purification and characterization of eucaryotic alanine racemase acting as key enzyme in cyclosporin biosynthesis. J. Biol. Chem. 269:12710-12714. [PubMed] [Google Scholar]
- 28.Hols, P., C. Defrenne, T. Ferain, S. Derzelle, B. Delplace, and J. Delcour. 1997. The alanine racemase gene is essential for growth of Lactobacillus plantarum. J. Bacteriol. 179:3804-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hols, P., M. Kleerebezem, A. N. Schanck, T. Ferain, J. Hugenholtz, J. Delcour, and W. M. de Vos. 1999. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 17:588-592. [DOI] [PubMed] [Google Scholar]
- 30.Inagaki, K., K. Tanizawa, B. Badet, C. T. Walsh, H. Tanaka, and K. Soda. 1986. Thermostable alanine racemase from Bacillus stearothermophilus: molecular cloning of the gene, enzyme purification, and characterization. Biochemistry 25:3268-3274. [DOI] [PubMed] [Google Scholar]
- 31.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9-20. [DOI] [PubMed] [Google Scholar]
- 32.Kanda-Nambu, K., Y. Yasuda, and K. Tochikubo. 2000. Isozymic nature of spore coat-associated alanine racemase of Bacillus subtilis. Amino Acids 18:375-387. [DOI] [PubMed] [Google Scholar]
- 33.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koebmann, B. J., D. Nilsson, O. P. Kuipers, and P. R. Jensen. 2000. The membrane-bound H(+)-ATPase complex is essential for growth of Lactococcus lactis. J. Bacteriol. 182:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-56. [DOI] [PubMed] [Google Scholar]
- 36.Lambert, M. P., and F. C. Neuhaus. 1972. Mechanism of d-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 110:978-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez de Felipe, F., M. Kleerebezem, W. M. de Vos, and J. Hugenholtz. 1998. Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J. Bacteriol. 180:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavan, S., P. Hols, J. Deleour, M. C. Geoffroy, C. Grangette, M. Kleerebezem, and A. Mercenier. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston, R. A., and H. A. Douthit. 1984. Germination of Bacillus cereus spores: critical control by DL-alanine racemase. J. Gen. Microbiol. 130:3123-3133. [DOI] [PubMed] [Google Scholar]
- 40.Reusch, V. M., Jr., S. G. Hale, and B. J. Hurly. 1982. Levels of cell wall enzymes in endospores and vegetative cells of Bacillus subtilis. J. Bacteriol. 152:1147-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosso, G., K. Takashima, and E. Adams. 1969. Coenzyme content of purified alanine racemase from Pseudomonas. Biochem. Biophys. Res. Commun. 34:134-140. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Shaw, J. P., G. A. Petsko, and D. Ringe. 1997. Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution. Biochemistry 36:1329-1342. [DOI] [PubMed] [Google Scholar]
- 44.Soda, K., and K. Tanizawa. 1990. Thermostable alanine racemase. Its structural stability. Ann. N. Y. Acad. Sci. 585:386-393. [DOI] [PubMed] [Google Scholar]
- 45.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352-1355. [DOI] [PubMed] [Google Scholar]
- 46.Stewart, B. T., and H. O. Halvorson. 1953. Studies on the spores of aerobic bacteria. I. The occurence of alanine racemases. J. Bacteriol. 65:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanizawa, K., A. Ohshima, A. Scheidegger, K. Inagaki, H. Tanaka, and K. Soda. 1988. Thermostable alanine racemase from Bacillus stearothermophilus: DNA and protein sequence determination and secondary structure prediction. Biochemistry 27:1311-1316. [DOI] [PubMed] [Google Scholar]
- 48.van der Meer, J. R., J. Polman, M. M. Beerthuyzen, R. J. Siezen, O. P. Kuipers, and W. M. De Vos. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Vossen, J. M., J. Kok, and G. Venema. 1985. Construction of cloning, promoter-screening, and terminator-screening shuttle vectors for Bacillus subtilis and Streptococcus lactis. Appl. Environ. Microbiol. 50:540-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, E., and C. Walsh. 1978. Suicide substrates for the alanine racemase of Escherichia coli B. Biochemistry 17:1313-1321. [DOI] [PubMed] [Google Scholar]
- 51.Wasserman, S. A., C. T. Walsh, and D. Botstein. 1983. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 153:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wijsman, H. J. 1972. The characterization of an alanine racemase mutant of Escherichia coli. Genet. Res. 20:269-277. [DOI] [PubMed] [Google Scholar]
- 53.Wild, J., J. Hennig, M. Lobocka, W. Walczak, and T. Klopotowski. 1985. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol. Gen. Genet. 198:315-322. [DOI] [PubMed] [Google Scholar]
- 54.Yasuda, Y., K. Kanda, S. Nishiola, Y. Tanimoto, C. Kato, A. Saito, S. Fukuchi, and Y. Nakanishi. 1993. Regulation of l-alanine-initiated germination of Bacillus subtilis spores by alanine racemase. Amino Acids 4:89-99. [DOI] [PubMed] [Google Scholar]
- 55.Yonaha, K., T. Yorifuji, T. Yamamoto, and K. Soda. 1975. Alanine racemase of Bacillus subtilis var. atterimus. J. Ferment. Technol. 53:579-587. [Google Scholar]