Abstract
Selenoprotein synthesis is conserved from bacteria to man. It involves the differential decoding of the UGA stop codon as selenocysteine. The proteomes of both prokaryotes and eukaryotes, with the exception of yeast, contain only few selenoproteins. This low number is explained by a counterselection of readily oxidized selenocysteine after the introduction of oxygen into the atmosphere and the need to conserve selenoenzymes that control redox homeostasis of cells. Lack of selenoprotein synthesis in vertebrates impairs the oxidative stress defence and causes lethality. Here we show that Drosophila mutants that lack the translation elongation factor SelB/eEFsec fail to decode the UGA codon as selenocysteine, but they are viable and fertile. Oxidative stress responses and the lifespan of these flies are not affected. Protecting cells from oxidative stress can therefore not account for the selection pressure that conserves selenoprotein biosynthesis during the course of evolution.
Keywords: Drosophila, eEFsec, SelB, selenoprotein, oxidative, stress
Introduction
Selenoproteins are implicated in oxidative stress defence and in a number of clinical syndromes including cancer in man (Moustafa et al, 2003). Their synthesis depends on the differential decoding of the UGA stop codon as selenocysteine (Atkins & Gesteland, 2000; Böck, 2000). In bacterial mRNA, this process is regulated by a special ‘stem–loop' structure, termed the SECIS element, next to the UGA codon. This element binds SelB, an EF-Tu-like elongation factor, which associates with selenocysteine-tRNA. Functional formation of the SECIS–SelB–tRNA complex ensures that the neighbouring UGA is read as selenocysteine instead of a stop codon (Böck, 2000). In higher organisms such as archaea, flies and man, the selenocysteine-coding machinery is conserved but modified. First, the SECIS element is located far from the UGA codon in the 3′ untranslated region of the mRNA (Low & Berry, 1996). Second, the SelB function is provided by two proteins, termed eEFsec and SECIS-binding protein 2 (SBP2) (Copeland et al, 2000; Fagegaltier et al, 2000; Tujebajeva et al, 2000). The available evidence therefore suggests that selenoprotein synthesis predates the diversion of bacteria, archaea and eukaryotes, and that the principle of synthesis has been conserved during the course of eukaryotic evolution (Low & Berry, 1996; Atkins & Gesteland, 2000; Hirosawa-Takamori et al, 2000; Driscoll & Copeland, 2003).
In mammals, a number of key enzymes involved in oxidative stress defence carry a UGA-coded selenocysteine in their enzymatic centres (Driscoll & Copeland, 2003). As both vertebrates and invertebrates have highly efficient oxidative stress defence systems (Melov, 2002), the need for selenoproteins for the control of redox homeostasis appears to be the likely selective force by which selenoprotein synthesis has been conserved throughout the animal kingdom. Here we report that the Drosophila translation elongation factor SelB/eEFsec is required for the differential decoding of the UGA codon. SelB/eEFsec mutations impair the differential UGA-decoding mechanism, but viability, lifespan and oxidative stress reactions of the mutants are not affected. This surprising finding challenges the view that a selenocysteine-based oxidative stress defence system is the key in conserving the selenoprotein biosynthesis system during the course of evolution.
Results and Discussion
In Drosophila, a genetically amenable invertebrate, three selenoprotein-coding genes were previously identified by homology cloning and whole-genome sequence analysis (Hirosawa-Takamori et al, 2000; Castellano et al, 2001; Martin-Romero et al, 2001; Kryukov et al, 2003). To assess the maximal effect of loss of selenoprotein biosynthesis in flies, we searched for the SelB/eEFsec homologue by in silico analysis of the genome, cloned the gene and generated a SelB/eEFsec knockout mutation.
Structure And Expression Of Drosophila SelB/eEFsec Gene
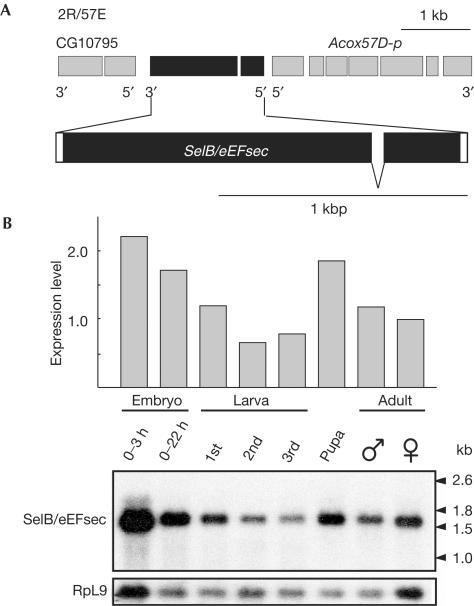
The structure of the Drosophila SelB/eEFsec gene, as revealed by sequence comparison of cDNA and corresponding genomic DNA, is shown in Fig 1A. It is located on the right arm of the second chromosome in section 57E and contains two exons that code for a single transcript of ∼1.6 kb (Fig 1A). Comparison of SelB/eEFsec of Drosophila and mouse (Fagegaltier et al, 2000; Tujebajeva et al, 2000) revealed 40% similarity. SelB/eEFsec mRNA could be detected during all stages of the Drosophila life cycle, with high levels during early embryogenesis, a decrease during larval stages and enrichment in pupae and adults (Fig 1B).
Figure 1.
Structure and expression of Drosophila SelB/eEFsec. (A) Genomic organization of Drosophila SelB/eEFsec showing that the gene is composed of two exons; coding sequences (black bars) and untranslated regions (open bars) are indicated. (B) Developmental expression of SelB/eEFsec transcripts as revealed by northern blot analysis using a SelB/eEFsec-specific probe; the amount of SelB/eEFsec mRNA was normalized against RpL9 transcripts serving as an internal control.
Generation Of SelB/eEFsec Mutant Alleles
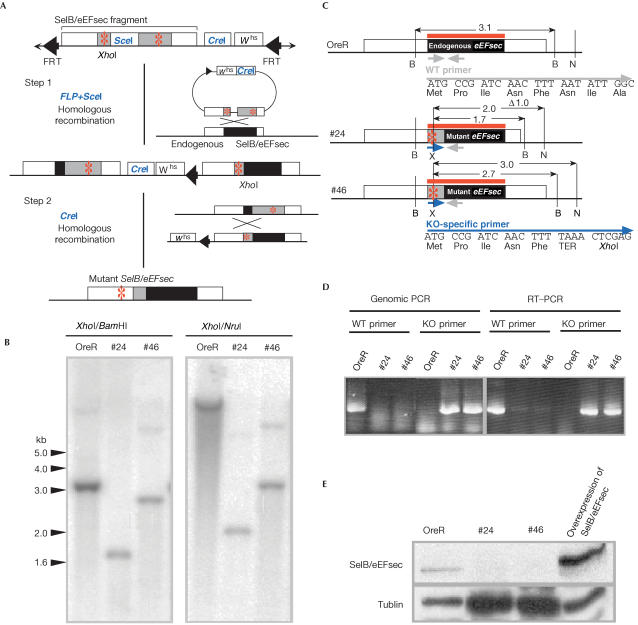
SelB/eEFsec knockout mutations were generated by homologous recombination (Rong & Golic, 2000; Rong et al, 2002). Briefly, we generated transgenic flies containing a nonfunctional SelB/eEFsec gene having the two TAA stop codons and a frameshift in the coding region (Fig 2A,C). We screened 1,000 individuals for a recombination event (Fig 2A) and identified two SelB/eEFsec mutants (KO#24 and KO#46) (Fig 2B) that carry deletions of ⩽1 kb and carry both TAA stop codons as well as the altered reading frame (Fig 2C; see Material and methods). RT–PCR showed that only mutant mRNA is expressed in the mutants (Fig 2D). Furthermore, antibodies directed against a C-terminal polypeptide detect SelB/eEFsec on western blots of protein extracts derived from both wild-type flies and flies that overexpress SelB/eEFsec from a cDNA-based transgene, but not in extracts from homozygous SelB/eEFsec individuals (Fig 2E). This result indicates that the SelB/eEFsec knockout mutants, which carry a deficiency corresponding to the essential N-terminal region of the protein, are null mutations.
Figure 2.
Schematic representation of SelB/eEFsec mutagenesis by homologous recombination. (A) The targeting DNA fragment excised by FLP from the donor construct is expected to recombine with endogenous SelB/eEFsec locus (step 1; for details, see Rong & Golic, 2000; Rong et al, 2002). In a second step, the tandem duplication was reduced to a single copy by a homologous recombination event due to a CreI-mediated double-strand break (step 2). Red asterisks: introduced TAA stop codon. (B) Southern blot with genomic DNA from wild-type and homozygous knockout mutant (KO#24 and KO#46) individuals. DNA was digested by XhoI, which is the newly generated site only in mutant DNA (see (A)), and by either BamHI or NruI. The SelB/eEFsec-specific DNA hybridization probe is indicated by red lines. (C) Gene structure of the wild-type (shown on top) and of SelB/eEFsec mutant alleles (#24 and #46). Restriction sites are as follows: B (BamHI), N (NruI) and X (XhoI). Size of DNA fragments in kilobase pairs (kb). (D) Genomic DNA and total RNA were prepared from wild-type (OreR) or homozygous knockout mutant flies (KO#24 and KO#46). Wild-type and knockout-specific primers are shown in (C); for their sequence, see Material and methods. (E) Anti-SelB/eEFsec antibody staining of western blots loaded with protein extracts of wild-type flies (OreR), homozygous knockout mutant flies (KO#24 and KO#46) or flies overexpressing SelB/eEFsec from a cDNA-derived transgene (see text). Note that SelB/eEFsec is left undetected in the KO#24 and KO#46 mutant individuals even in the presence of higher amounts of loaded protein (see loading control provided by anti-α-tublin staining).
SelB/eEFsec Mutants Lack Differential Decoding Activity
Mouse mutants that are unable to synthesize selenoproteins do not survive (Bosl et al, 1997; Kumaraswamy et al, 2003). In contrast, SelB/eEFsec mutants of the genotypes KO#24/KO#24, KO#46/KO#46 or KO#46/Df(2R)PK1 develop into fertile flies, indicating that the gene has no vital function.
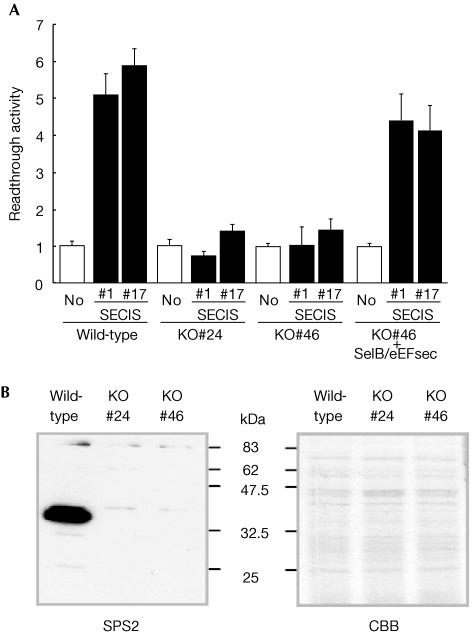
We next examined whether SelB/eEFsec mutants are able to decode differentially the UGA codon. We generated transgenic flies carrying the previously described LacZ/UGA/luciferase reporter genes either with or without a SECIS element (Hirosawa-Takamori et al, 2000). Wild-type transformants containing the SECIS-bearing reporter gene contained luciferase activity, whereas homozygous SelB/eEFsec mutants contained only background levels both in the absence and presence of the SECIS element (Fig 3A). The ability to produce SECIS-dependent readthrough activity was rescued by the expression of one copy of SelB/eEFsec transgene under control of the actin 5C promotor (see Hirosawa-Takamori et al, 2000) in the mutant flies (Fig 3A). Furthermore, antibodies directed against Drosophila selenoprotein SPS2 (Hirosawa-Takamori et al, 2000; Castellano et al, 2001; Martin-Romero et al, 2001) detect the selenoprotein on western blots containing wild-type protein extracts, whereas no SPS2 was found in SelB/eEFsec mutants (Fig 3B). Collectively, these results establish that flies lacking SelB/eEFsec activity are unable to decode UGA as selenocysteine.
Figure 3.
Lack of readthrough activity in SelB/eEFsec knockout mutants carrying a LacZ/UGA/luciferase reporter gene. (A) LacZ/UGA/luciferase reporter gene activity of transgenes lacking (No) or containing the SECIS element (#1 and #17 represent different transgenic lines) in wild type, in two different SelB/eEFsec mutants (KO#24 and KO#46) and SelB/eEFsec mutants that contain a cDNA-based SelB/eEFsec-expressing transgene (KO#46+SelB/eEFsec). For details of the assay system, see Hirosawa-Takamori et al (2000). Bars represent the mean values of relative luciferase activities from six independent experiments; standard deviation is indicated. Note SECIS-dependent readthrough activity in wild type, lack of readthrough activity in SelB/eEFsec mutants and the SelB/eEFsec transgene-dependent rescue of the readthrough activity. (B) Anti-dSPS2 antibody staining of western blots containing protein extracts of wild-type (white) and homozygous knockout mutant (KO#24 and KO#46) flies. Note the absence of SPS2 protein in the SelB/eEFsec mutant individuals. CBB, Coomassie brilliant blue stained gel.
No Effect On Longevity And Oxidative Stress Defence
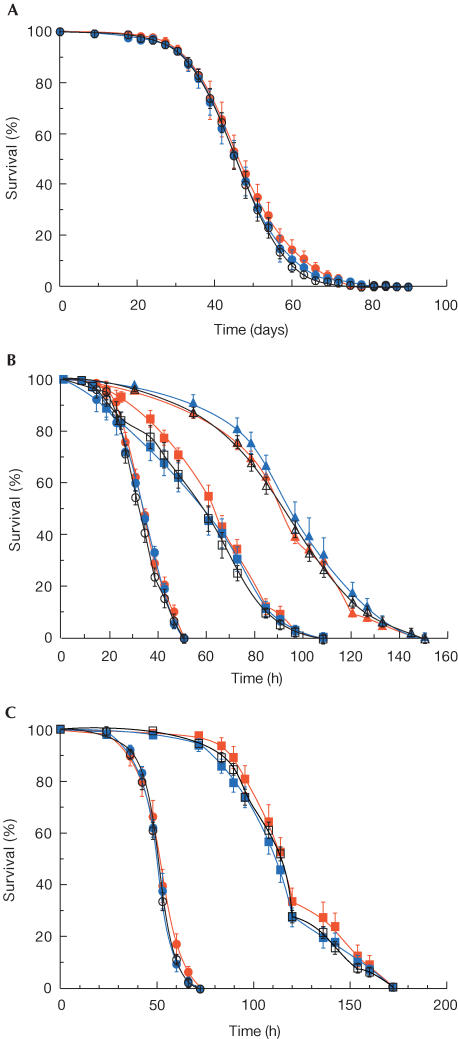
Homozygous SelB/eEFsec mutant individuals develop into fertile adults, which can be propagated in a wild-type-like manner. This finding shows that the mutation has no immediate effect on viability. Furthermore, the lifespan of the mutant individuals (Fig 4A) and their response to oxidative stress (Fig 4B,C) was indistinguishable from wild type.
Figure 4.
Determination of lifespan and sensitivity to oxidative stress of SelB/eEFsec mutant flies. (A) Comparison of the lifespan (in days) of wild-type (open circles) and SelB/eEFsec mutant (red: KO#24; blue: KO#46) individuals (n>150 males per genotype; mean value of three independent experiments). (B) Survival of wild-type and mutant flies (for coding, see (A)) at different paraquat concentrations (40 mM: circles; 10 mM: squares; 2.5 mM: triangles). (C) Survival of wild-type and mutant flies (for coding, see (A)) at different hydrogen peroxide concentrations (3.0%: circles; 0.3%: squares).
The finding that selenoproteins are not essential for redox homeostasis of Drosophila is consistent with the observation that Drosophila and Anopheles homologues (H.-R. Chung, unpublished observation) of vertebrate antioxidant enzymes such as glutathion peroxidase, thioredoxin reductase or SelR are non-selenocysteine (Kryukov et al, 2002; Missirlis et al, 2002, 2003). However, it seems to contradict recent results showing that suppression of Drosophila SelH (BthD) by RNAi expression reduces the viability of flies (Kwon et al, 2003). In this context, it is important to note that the Drosophila genes encoding SelH, SelD and SelK are duplicated and code for non-selenoproteins, which contain cysteine or arginine instead of the selenocysteine residue (Adams et al, 2000; Castellano et al, 2001; Martin-Romero et al, 2001). In the case of the two SelD homologues (SPS1 and SPS2), the non-selenoprotein SPS1 was shown to exert a vital function and participates in the oxidative stress defence (Morey et al, 2003a, 2003b). Thus, its activity may provide a backup function to support selenoprotein SPS2 activity in the SelB/eEFsec mutant individuals. By analogy, the non-selenoprotein homologue of SelH might compensate for the loss of selenoprotein SelH function in SelB/eEFsec mutant individuals, whereas in RNAi knockdown mutants the activities of both SelH proteins might be impaired on the basis of their similar mRNA sequences. This may therefore explain the different effects of RNAi knockdown experiments (Kwon et al, 2003) versus the selective loss of selenoproteins in SelB/eEFsec mutants described here.
Speculation
Our results present evidence that Drosophila selenoproteins are not essential for viability, longevity or oxidative stress defence as observed with vertebrates (Low & Berry, 1996; Driscoll & Copeland, 2003). Thus, selenoproteins may have undergone an insect-specific adoption of novel function(s) once the components of their oxidative stress defence system became independent of selenoprotein biosynthesis. This conclusion is consistent with the most recent finding of species-specific selenoproteins in lower vertebrates, which are not conserved in mammals (Castellano et al, 2004).
The fact that selenoprotein biosynthesis is maintained in flies suggests that following initial gene duplication events in ancestral organisms, selenoprotein-coding genes may have adopted new and possibly important, but nonvital, functions. Such functions may affect, for example, behaviour, learning and/or memory processes, which were not addressed by our present study, and may account for the continued requirement for selenoprotein synthesis once the redox homeostasis system became independent of selenocysteine-bearing enzymes during the course of insect evolution.
Material and Methods
Fly stocks and genetics. Flies were kept as described (Missirlis et al, 2002). To obtain the donor flies for homologous recombinations, the targeting DNA construct was transformed into flies by P-element transformation (Rubin & Spradling, 1982). Stocks for homologous recombination were yw (v); P{ry+,70FLP}4, P{v+,70I-SceI}2B, Sco/S2, CyO and yw; P{I-CreI}/TM3, Sb, Ser. Targeting was performed as described (Rong & Golic, 2000; Rong et al, 2002). Transgenic flies (No, #1, #17) and the assay used to examine the readthrough activity of the UGA codon are described by Hirosawa-Takamori et al (2000). The DNA fragment containing the SelB/eEFsec open reading frame was cloned into the pUAST vector DNA (pUAST-eEFsec) and used for fly transformation (see above). Crosses were carried out to obtain KO#24 or KO#46 mutant flies bearing the transgene to assay whether selenocysteine coding is restored in mutants. Protein extracts were prepared for each genotype (Fig 3B), and LacZ and luciferase activity were assayed (Hirosawa-Takamori et al, 2000).
Molecular characterization of the target event. Southern blot analysis was performed using the EcoRI/XhoI fragment of LP02881 DNA as probe (Fig 2B). Preparation of genomic DNA, total RNA and amplification by genomic PCR have been described (Missirlis et al, 2002). PCR involved wild-type primers (5′-ATGCCGATCAACTTTAATATTGGC-3′) and KO-specific primers (5′-ATGCCGATCAACTTTTAAACTCGAG-3′) and the common reverse primer (5′-GAGCATGAGATCAATGATCTGGGCACCTCC-3′). RT–PCR was carried out as outlined by the manufacturer (Promega, Madison, USA).
Targeting vector construct. Two genomic DNA fragments (fragments 1 and 2) were amplified by genomic PCR. Primers were P1F (5′-GCGGCCGCGCCTTCACCTGAGCATGTCGCATC-3′) and P2R (5′-GCTAGCCCGCAAGCGTAGCTTCTCCAGCTT-3′) for fragment 1; and P3F (5′-AAGCTTCTCGCAAAAACTTTGGAAGCCACC-3′) and P4R (5′-GGTACCACCAGGCGCTGTGCTGCCTTAACC-3′) for fragment 2. To introduce base-pair changes, fragments were cloned into pCRII-TOPO DNA (Invitrogen, Groningen, The Netherlands). Mutagenesis was performed with the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, USA). Primers were 5′-GTACAATGCCGATCAACTTTTAAACTCGAGTTGCTGG GACATGTTGAC-3′ and 5′-GTCAACATGTCCCAGCAACTCGAGTTTAAAAGTTGATC GGCATTGTAC-3′ to introduce TAA stop codons, a XhoI site and a frame shift in the wild-type sequence. The primers to generate a SceI site were 5′-CTGGCTAGGGATAACAGGGTAATA-3′ and 5′-AGCTTATTACCCTGTTATCCCTAG-3′. Mutated fragment 1 DNA was generated by NotI/NheI treatment, and mutated fragment 2 DNA by HindIII/KpnI digestion. The fragments were ligated into the NotI/KpnI sites of pTV2 vector DNA (Rong et al, 2002) using the linker SceI oligos.
Developmental expression analysis and western blot analysis. Developmental northern blot analysis was performed as described (Grönke et al, 2003). Radioactively labelled SelB/eEFsec antisense RNA probe was prepared by in vitro transcription on an EcoRI-linearized LP02881 cDNA template using SP6 polymerase (Strip-EZ RNA kit; Ambion, Austin, TX, USA). As loading control, the blot was reprobed with RpL9 antisense RNA (Grönke et al, 2003). Anti-SelB/eEFsec-specific rabbit antiserum (Eurogentec, Seraing, Belgium) was produced against the amino-acid stretch GEKGRIERTFGQTSK (positions 458–472). Western blots prepared from protein extracts of females were stained with anti-SelB/eEFsec rabbit serum (1:2,000 dilution) or anti-SPS2 rabbit serum (1:1,000 dilution). As secondary antibodies, horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:10,000 dilution; Sigma-Aldrich, Steinheim, Germany) was used and visualized by SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, USA). As loading control, the blot was reprobed with antitubulin antibodies (E7; 1:5,000 dilution; DSHB, Iowa City, IA, USA) and HRP-conjugated anti-mouse IgG (secondary antibodies; 1:10,000 dilution; Sigma-Aldrich, Steinheim, Germany).
Paraquat and hydrogen peroxide assays and lifespan measurements. Up to 15 adult males (3- to 4-day-old) were kept in vials with 1.5 ml medium (1% sucrose, 1.3% low melting agarose) and specified concentrations of paraquat or hydrogen peroxide (Sigma-Aldrich, Steinheim, Germany). Survival and lifespan measurements were described earlier (Missirlis et al, 2002).
Acknowledgments
We thank K.G. Golic who provided strains for homologous recombination and D.L. Hatfield for the anti-SPS2 antibodies. We are grateful to G. Vorbrüggen, R.P. Kühnlein, F. Missirlis and S. Takamori for various important contributions. This work was supported by the DFG (SP selenoprotein). H.-R.C. is a Boehringer Ingelheim Fonds fellow.
References
- Adams MD et al. ( 2000) The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Atkins JF, Gesteland RF ( 2000) The twenty-first amino acid. Nature 407: 463–465 [DOI] [PubMed] [Google Scholar]
- Böck A ( 2000) Synthesis of selenoproteins—an overview. Biofactors 11: 77–78 [DOI] [PubMed] [Google Scholar]
- Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM ( 1997) Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc Natl Acad Sci USA 94: 5531–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigo R ( 2001) In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep 2: 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S, Novoselov SV, Kryukov GV, Lescure A, Blanco E, Krol A, Gladyshev VN, Guigó R ( 2004) Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep 5: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM ( 2000) A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J 19: 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR ( 2003) Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr 23: 17–40 [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A ( 2000) Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J 19: 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S, Beller M, Fellert S, Ramakrishnan H, Jäckle H, Kühnlein RP ( 2003) Control of fat storage by a Drosophila PAT domain protein. Curr Biol 13: 603–606 [DOI] [PubMed] [Google Scholar]
- Hirosawa-Takamori M, Jäckle H, Vorbrüggen G ( 2000) The class 2 selenophosphate synthetase gene of Drosophila contains a functional mammalian-type SECIS. EMBO Rep 1: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN ( 2002) Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci USA 99: 4245–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN ( 2003) Characterization of mammalian selenoproteomes. Science 300: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Kumaraswamy E et al. ( 2003) Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol Cell Biol 23: 1477–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SY, Badenhorst P, Martin-Romero J, Carlson BA, Paterson BM, Gladyshev VN, Lee BJ, Hatfield DL ( 2003) The Drosophila selenoprotein BthD is required for survival and has a role in salivary gland development. Mol Cell Biol 23: 8495–8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SC, Berry MJ ( 1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci 21: 203–208 [PubMed] [Google Scholar]
- Martin-Romero FJ, Kryukov GV, Lobanov AV, Carlson BA, Lee BJ, Gladyshev VN, Hatfield DL ( 2001) Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J Biol Chem 276: 29798–29804 [DOI] [PubMed] [Google Scholar]
- Melov S ( 2002) Animal models of oxidative stress, aging, and therapeutic antioxidant interventions. Int J Biochem Cell Biol 34: 1395–1400 [DOI] [PubMed] [Google Scholar]
- Missirlis F, Ulschmid JK, Hirosawa-Takamori M, Grönke S, Schafer U, Becker K, Phillips JP, Jäckle H ( 2002) Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability. J Biol Chem 277: 11521–11526 [DOI] [PubMed] [Google Scholar]
- Missirlis F, Rahlfs S, Dimopoulos N, Bauer H, Becker K, Hilliker A, Phillips JP, Jäckle H ( 2003) A putative glutathione peroxidase of Drosophila encodes a thioredoxin peroxidase that provides resistance against oxidative stress but fails to complement a lack of catalase activity. Biol Chem 384: 463–472 [DOI] [PubMed] [Google Scholar]
- Morey M, Serras F, Corominas M ( 2003a) Halving the selenophosphate synthetase gene dose confers hypersensitivity to oxidative stress in Drosophila melanogaster. FEBS Lett 534: 111–114 [DOI] [PubMed] [Google Scholar]
- Morey M, Corominas M, Serras F ( 2003b) DIAP1 suppresses ROS-induced apoptosis caused by impairment of the selD/sps1 homolog in Drosophila. J Cell Sci 116: 4597–4604 [DOI] [PubMed] [Google Scholar]
- Moustafa ME, Kumaraswamy E, Zhong N, Rao M, Carlson BA, Hatfield DL ( 2003) Models for assessing the role of selenoproteins in health. J Nutr 133: 2494S–2496S [DOI] [PubMed] [Google Scholar]
- Rong YS, Golic KG ( 2000) Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG ( 2002) Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev 16: 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC ( 1982) Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353 [DOI] [PubMed] [Google Scholar]
- Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ ( 2000) Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep 1: 158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]