Abstract
Chromosome dimers, which frequently form in Escherichia coli, are resolved by the combined action of two tyrosine recombinases, XerC and XerD, acting at a specific site on the chromosome, dif, together with the cell division protein FtsK. The C-terminal domain of FtsK (FtsKC) is a DNA translocase implicated in helping synapsis of the dif sites and in locally promoting XerD strand exchanges after synapse formation. Here we show that FtsKC ATPase activity is directly involved in the local activation of Xer recombination at dif, by using an intermolecular recombination assay that prevents significant DNA translocation, and we confirm that FtsK acts before Holliday junction formation. We show that activation only occurs with a DNA segment adjacent to the XerD-binding site of dif. Only one such DNA extension is required. Taken together, our data suggest that FtsK needs to contact the XerD recombinase to switch its activity on using ATP hydrolysis.
Keywords: FtsK, site-specific recombination, XerCD
Introduction
Repair of circular DNA molecules by homologous recombination at stalled or broken replication forks can lead to crossing over and hence dimer formation (Cox et al, 2000; Michel et al, 2001). Bacteria containing chromosome dimers cannot segregate their DNA, or divide correctly, and form filaments before dying. Chromosome dimers form every seven generations on average in laboratory-grown Escherichia coli, so mechanisms that ensure that the chromosome is monomeric are central to bacterial survival and growth (Steiner & Kuempel, 1998; Pérals et al, 2000). In E. coli, Xer sitespecific recombination converts chromosome dimers into monomers by the addition of a crossover at a 28 base-pair (bp) recombination site, dif, located in the replication termination region of the chromosome. Recombination is mediated by two related tyrosine recombinases, XerC and XerD, within a nucleoprotein complex, (XerCD–dif)2, which contains two molecules each of XerC and XerD and two synapsed dif sites. XerC and XerD catalyse two sequential pairs of DNA strand exchanges such that recombination proceeds via a Holliday junction (HJ) intermediate (reviewed in Barre & Sherratt, 2002).
Chromosome dimer resolution also requires FtsK, a large, multifunctional, integral membrane protein, which coordinates chromosome segregation and cell division (Liu et al, 1998; Capiaux et al, 2001). Such coordination is crucial in bacteria where DNA replication and segregation are not separated in time, and can occur as cells divide.
FtsK can be divided into three domains: a membrane-spanning N-terminal domain (FtsKN), which localizes to the division septum and is essential for cell division (Draper et al, 1998); a long linker (600 amino acids) of unknown function; and a C-terminal AAA+ ATPase domain (FtsKC; Yu et al, 1998; Barre et al, 2000; Aussel et al, 2002). Two roles have been assigned to FtsK in chromosome dimer resolution. First, FtsK has been implicated in positioning the terminus regions of chromosome dimers at mid-cell and synapsing their dif sites (Capiaux et al, 2002; Corre & Louarn, 2002). Consistent with such a role, FtsK50C, an active derivative of FtsKC, was shown to be an ATP-dependent DNA translocase in vitro (Aussel et al, 2002). Second, FtsKC is directly involved in Xer recombination. In vitro analyses of FtsK-dependent Xer recombination between plasmid-borne dif sites suggest that FtsK50C can switch the catalytic state of (XerCD–dif)2 so that recombination is initiated by XerD rather than XerC (Aussel et al, 2002). ‘Mix and match' experiments between Haemophilus influenzae and E. coli Xer–FtsK, and the exploitation of FtsK chimaeras containing different segments of the C-terminal domains of H. influenzae and E. coli FtsK have shown that the C-terminal 140 amino acids of FtsKC interact with (XerCD–dif)2 during recombination activation (Yates et al, 2003).
Is ATP hydrolysis by FtsKC necessary for local recombination activation, or is it only required for synapse formation between dif sites carried on long DNA molecules? We have analysed the local role of FtsKC in the activation of Xer recombination by using a new in vitro intermolecular Xer recombination assay that does not provide an opportunity for significant DNA translocation. We show that activation by FtsK50C still depends on ATP hydrolysis. Furthermore, the presence of nicks as close as 3 bp from the recombination site did not prevent recombination activation, supporting the idea that the motor domain of FtsK functions in the local activation of the recombinases independently of global DNA translocation. In addition, we show that Xer activation by FtsK50C requires only one duplex DNA arm protruding from the (XerCD–dif)2 complex, adjacent to an XerD-binding site. This observation is the first evidence for asymmetry in the recognition of (XerCD–dif)2 by FtsK, and suggests a specific interaction between the recombinase that carries out the first pair of strand exchanges, XerD, and the FtsK activator. This asymmetry implies that the orientation of dif sites with respect to the septum-localized FtsK must be nonrandom in vivo.
Results
FtsK50C activates intermolecular Xer recombination
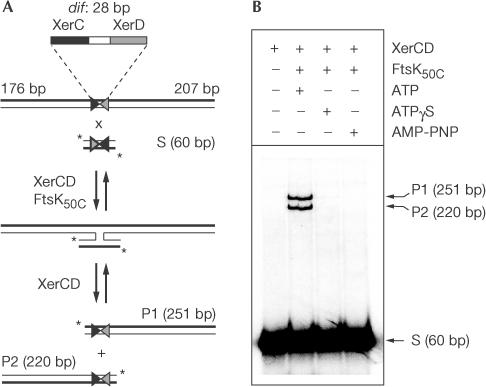
To analyse the local role of FtsKC in the activation of Xer recombination in the absence of significant DNA translocation, we developed an in vitro intermolecular recombination reaction between two short dif-containing DNAs. The lengths of the DNAs were chosen so as to preclude significant translocation by FtsK50C (Fig 1A). In initial experiments, one DNA molecule contained a dif site flanked by arms of 176 and 207 bp, whereas the second DNA molecule consisted of dif flanked by arms of 16 bp, and was 5′-end-labelled on both strands with 32P. Incubation of equimolar concentrations of the two DNAs with XerCD in the presence of FtsK50C and ATP led to a complete intermolecular sitespecific recombination reaction (Fig 1B). The reaction was absolutely dependent on ATP hydrolysis by FtsK50C, as nonhydrolysable AMP-PNP and Walker A and B box mutants of FtsK50C, which cannot hydrolyse ATP, did not support recombination (Fig 1B and data not shown). The poorly hydrolysable nucleotide ATP-γS supported a low but detectable level of recombination, correlating with its expected rate of hydrolysis.
Figure 1.
Minimal XerCD–FtsK recombination system. (A) Schematic illustrating an intermolecular Xer recombination reaction between a 411 bp unlabelled DNA fragment containing a centrally located dif and a short, dif-containing DNA (60 bp) 5′-end-labelled with 32P on both strands. (B) Intermolecular recombination by XerCD is absolutely dependent on FtsK50C and ATP. Reactions were incubated at 37°C for 60 min and analysed by 7% TBE–PAGE.
XerCD activation needs DNA on one side of dif only
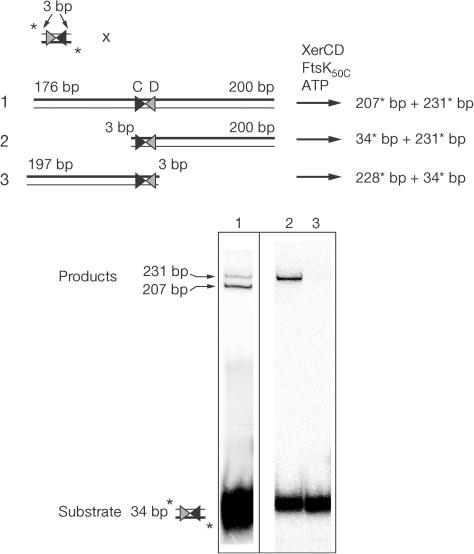
Short unlabelled DNAs with either one or two dif flanking arms were reacted with a minimal 32P-labelled dif site flanked on either side by just 3 bp of DNA (3CD3) to explore the requirements for FtsK50C-dependent activation of XerCD recombination. Remarkably, an absolute asymmetry was observed: only unlabelled DNA fragments with a flanking arm adjoining the XerD-binding site of dif (D-side DNA) could be recombined with the minimal labelled fragment. No reaction was observed when the flanking DNA was adjoining the XerC-binding site (C-side DNA; Fig 2). This asymmetric requirement for flanking DNA indicates either that FtsK50C needs to activate the (XerCD–dif)2 nucleoprotein complex from a position on DNA adjacent to bound XerD or that FtsK50C has a DNA sequence or structure dependence, with only a specific Dside DNA sequence permitting efficacious FtsK50C loading.
Figure 2.
FtsK-dependent Xer recombination requires DNA adjacent to the XerD-binding site only. The schematic illustrates reactions between short, labelled dif-containing DNAs and unlabelled DNAs with either (1) two arms flanking dif or one arm adjacent to dif on either the XerD side (2) or the XerC side (3). Recombination reactions were incubated at 37°C for 60 min and analysed by 7% TBE–PAGE.
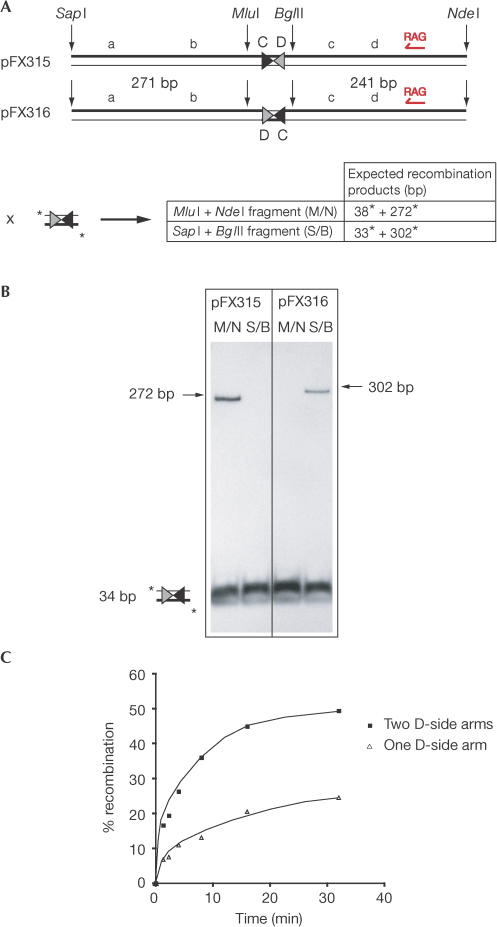
To distinguish these two possibilities, unlabelled DNAs with identical DNA sequences adjoining either the XerC- or XerD-binding sites of dif were compared for their ability to support FtsK50C-dependent stimulation of XerCD–dif recombination. These DNA fragments were derived from two dif-containing plasmids that were identical apart from an inverted dif orientation (pFX315 and pFX316; Fig 3A). Only those DNA molecules with an arm adjoining the XerD-binding site of dif recombined with 3CD3 in an FtsK50C-dependent reaction (Fig 3B). In total, three different Dside DNA sequences could support recombination. These data demonstrate that the presence rather than the sequence of D-side DNA is required for FtsK50C activation of Xer recombination.
Figure 3.
Asymmetric DNA requirement of FtsK50C is independent of DNA sequence. (A) Orientation of dif sites in pFX315 and pFX316, and the recombination products expected from reactions between dif-containing restriction fragments and a short, labelled dif-containing DNA (3CD3). Lowercase letters indicate DNA orientation. The single RAG sequence element is annotated. (B) Electrophoretic analysis of 60 min reactions detailed in (A). (C) Recombination over time for reactions between short, dif-containing DNAs where either one or both short fragments have a Dside extension. The reactions with just one D-side arm are between 3CD3 and 315 M/N.
A second asymmetry arises from the observation that only one D-side DNA segment is required for activation of Xer recombination in the intermolecular assay (Figs 2, 3). Adding a second Dside arm into the reaction (on the labelled 3CD3 fragment) doubles the proportion of recombination without affecting the reaction profile, as expected if FtsK50C has only to approach the (XerCD–dif)2 synaptic complex on one D-side arm to activate recombination (Fig 3C).
D-side DNA length affects recombination efficiency
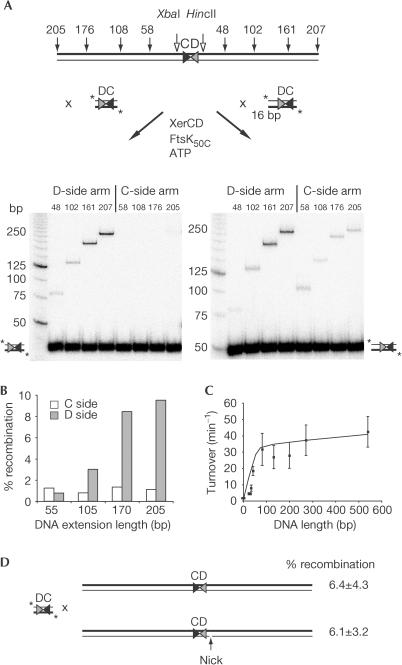
Having shown that D-side DNA is essential for activation of Xer recombination by FtsK50C, we investigated whether the length of this DNA arm affected the efficiency of the reaction. Unlabelled short fragments with various lengths of C- or D-side DNA extension were reacted with 32P-labelled 3CD3 DNA or DNA with 3 bp on the C side and 16 bp on the D side (‘3CD16'; Fig 4A). Increasing Dside DNA length from 48 to 207 bp increased the level of labelled recombinant product to a maximum of approximately 10% (Fig 4A,B). Further increases in DNA length did not increase the levels of recombination observed (data not shown). As expected, none of the DNAs with extensions only on the C side of dif reacted. However, adding a short 16-bp arm on the D side of the labelled fragment enabled recombination to occur at a low level independently of unlabelled Cside arm length, presumably due to inefficient FtsK50C loading on the 16-bp D-side arm (Fig 4A, right panel).
Figure 4.
Effects of DNA arm length and backbone continuity on FtsK50C activities. (A) Unlabelled DNAs with different lengths of arm adjoining either the XerC side or the XerD side of dif were reacted with short, labelled dif-containing DNAs with either 3-bp arms (left panel) or 3 bp on the XerC side and 16 bp on the XerD side (3CD16; right panel), in the presence of XerCD, FtsK50C and ATP for 60 min. (B) Histogram comparing the levels of recombination observed when unlabelled DNAs with XerC side (white bars) or XerD side (grey bars) extensions were reacted with labelled 3CD16. The results are the means of three experiments. (C) Graph of DNA length against FtsK50C ATPase activity (mean turnover±s.d. from three independent reactions, in the absence of XerCD). (D) A singlestrand nick does not affect FtsK50C activation of recombination. Unlabelled DNA without (top reaction) or with (bottom reaction) a single-stranded nick 3 bp along the XerD DNA arm was reacted with labelled 3CD3 in the presence of XerCD, FtsK50C and ATP for 60 min. The results are the mean±s.d. of three independent experiments.
Why does FtsK50C activation of Xer recombination exhibit such a DNA length dependence? Examination of FtsK50C ATPase activity as a function of DNA length (Fig 4C) demonstrates a biphasic relationship between ATPase level and DNA length. The first phase shows a linear relationship between ATPase activity and DNA length, as DNA length increases from 0 to ∼80 bp. The second phase, of shallower gradient, shows that further increases in DNA length lead to only small increases in ATPase turnover. The DNA length threshold of 80 bp could reflect either the amount of DNA required to support stable and productive FtsK50C–DNA interaction, or the length of DNA translocated in a single catalytic cycle (if ATP hydrolysis is correlated with translocation). We cannot distinguish between these two possibilities, although the observation that a circular plasmid stimulates ATP hydrolysis by FtsK50C up to four times as strongly as when it is linear (data not shown) suggests that FtsK50C translocates processively in steps of more than 80 bp. Also, related AAA+ ring proteins, such as RuvB and MCM, cover between 20 and 70 bp DNA per active oligomer (Chen et al, 2002; Fletcher et al, 2003), so the 80-bp ATPase threshold for FtsK50C could reflect the FtsK50C ‘footprint' of a single FtsK50C oligomer. The DNA length threshold of 150–200 bp for recombination could indicate that more than a single FtsK50C oligomer is required for recombination activation.
D-side DNA nicks do not prevent Xer activation
As FtsK50C can generate superhelical torsion in DNA as it translocates, we reasoned that even the local activation of Xer recombination by FtsK50C might require the continuity of the phosphodiester backbone on the D-side DNA. Addition of a single-strand nick at various positions on the DNA segment adjacent to the XerD-binding site did not compromise the activation step. Even a nick just three nucleotides away from the XerD-binding site of dif did not compromise activation of Xer recombination by FtsK50C (Fig 4D), thereby showing that activation is not dependent on torsional changes that need to be transmitted through an uninterrupted phosphodiester backbone.
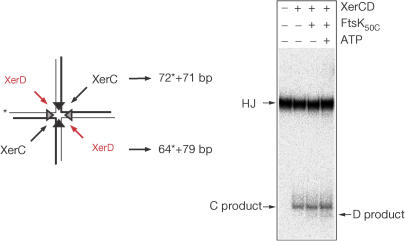
FtsK50C Functions before HJ formation
Previous work showed that FtsK50C activates Xer recombination by mediating a switch in the catalytic state of the (XerCD–dif)2 complex, so that recombination is initiated by XerD rather than by XerC (Aussel et al, 2002). Nevertheless, it remained unclear as to whether FtsK50C was only stimulating the firststrand exchange or whether it could also direct HJ resolution. We constructed a dif-containing HJ and analysed the effect of FtsK50C on its resolution by XerCD (Fig 5). In the absence of FtsK50C, the HJ is resolved primarily by XerC-mediated cleavage into two duplex products. The presence of FtsK50C, with or without ATP, has little effect on the HJ resolution reaction. The small increase in the level of XerD cleavage product in the presence of FtsK50C and ATP reflects intermolecular recombination between the two duplex products of XerC-mediated HJ cleavage. Therefore, FtsK50C appears not to modulate Xer recombination on an HJ substrate. Instead it acts only to promote the initial strand exchanges by XerD, generating an HJ intermediate that can spontaneously undergo XerC strand exchanges to form products in a reaction independent of FtsK50C. This confirms the view obtained from studies of plasmid resolution (Aussel et al, 2002).
Figure 5.
FtsK50C does not affect the direction or efficiency of resolution of a dif-containing HJ. The HJ schematized was prepared by annealing four oligonucleotides and then reacted with XerCD, in the presence or absence of FtsK50C and ATP, for 60 min at 37°C. Reactions were deproteinized before 6% TBE–PAGE.
Discussion
We have shown that FtsK50C can activate intermolecular Xer recombination between two short, linear dif-containing DNA fragments. In this ATP hydrolysis-dependent reaction, there is no room on the DNA arms for significant translocation by FtsK50C, suggesting that the enzyme can use ATP hydrolysis to activate XerCD recombination directly. The observation that nicks on the DNA substrate, even as close as 3 bp from the dif site, do not prevent recombination activation by FtsK50C implies that changes in DNA torsion induced by FtsK50C are not essential to the activation mechanism. Furthermore, the fact that recombination activation by FtsK50C requires only one DNA arm adjoining an XerD-binding site in the (XerCD–dif)2 synaptic complex suggests that FtsKC must interact with (XerCD–dif)2 in a precisely oriented manner, possibly by directly contacting one XerD molecule. Such a model fits with the observation that XerD initiates strand exchange at dif, in contrast to FtsK-independent Xer reactions where XerC initiates recombination (Blakely et al, 2000; Aussel et al, 2002; Bregu et al, 2002). If the (XerCD–dif)2 complex adopts a structure similar to that of the well-characterized tyrosine recombinase–DNA complex Cre-loxP (Guo et al, 1997, 1999), then switching on XerD catalysis and switching off XerC catalysis within (XerCD–dif)2 must involve substantial reorganization of the macromolecular complex by FtsKC, a process presumably driven by ATP hydrolysis. However, we cannot yet pinpoint the exact role of ATP hydrolysis by FtsK in the reaction mechanism: it may even be involved at a late stage to release the motor from the recombinase complex.
It is intriguing to note that the length of chromosomal DNA from the origin to the XerD half-site of dif is 29 kb (approximately 30 s replication time) shorter than the length from the origin to the XerC half-site of dif. If both replication forks moved at the same rate, the replicated dif sites would tend to be oriented such that the XerD half-sites are more distal to the moving fork than the XerC half-sites. This orientation might make the XerD arms more accessible to a translocase such as FtsK and could explain the evolution of an activatory FtsK–XerD interaction.
FtsK has been implicated in sensing chromosome polarity to set up productive translocation and dif synapsis (Pérals et al, 2000; Corre and Louarn, 2002). It has been hypothesized that chromosome polarity is defined by RAG elements, eight nucleotide DNA sequences with a highly skewed chromosomal distribution inverting at dif (Capiaux et al, 2001; Lobry & Louarn, 2003). In the experiments reported here, FtsK50C action was not influenced by the presence or absence of RAGs in either orientation on either arm of the unlabelled DNA (Fig 3A and data not shown). Although our data do not preclude a role for RAGs in global translocation, they clearly show that RAGs do not affect the local action of FtsK, and that although the FtsKC motor may contribute to dif synapsis in vivo, it must also be able to activate nucleoprotein complexes that have formed by random collision.
The assay developed here will facilitate further dissection of the molecular interactions involved in the chromosome dimer resolution reaction and should lead to a detailed view of how a radially symmetrical ring-shaped motor protein can lock onto and then actively remodel a protein–DNA complex to make that complex competent for recombination.
Methods
Intermolecular recombination assays. Plasmids pFX315 and pFX316 were constructed by inserting wild-type dif into a derivative of pNB0 (Morgan et al, 2000). Short unlabelled dif-containing DNA fragments of variable length were produced by PCR on pMIN33 or by restriction digestion of pFX315 or pFX316. N.BstNBI (NEB) was used to create a single nick 3 bp away from dif adjacent to either the XerC-binding site (pFX316) or the XerD-binding site (pFX315). Short, 32P-labelled, dif-containing DNA fragments were produced by annealing 5′-end-labelled oligonucleotides and purifying duplex DNA by 6% Tris–borate–EDTA polyacrylamide gel electrophoresis (TBE–PAGE). The short DNAs were eluted, ethanol-precipitated and resuspended in 50–200 μl TE (10 mM Tris–Cl, pH 8.0, 1 mM EDTA). XerCD and FtsK50C proteins were purified as described previously (Aussel et al, 2002).
Intermolecular recombinations were carried out in 10 μl of reaction buffer (10 mM Tris–Cl, pH 7.5, 1 mM DTT, 10 mM MgCl2, 1 mg/ml BSA, 40 mM NaCl) and contained approximately 2 nM labelled DNA, 2 nM unlabelled DNA, 150 nM XerC, 75 nM XerD, 300 nM FtsK50C (monomer concentrations) and 2.5 mM ATP or ATP-γS. FtsK50C was added last, to start the reaction. Reactions were incubated at 37°C for up to 60 min before being stopped, deproteinized and analysed by 7% native TBE–PAGE. Gels were imaged and quantified using a Fuji PhosphorImager and ImageQuant software.
ATPase assays. ATP hydrolysis assays were carried out by thin-layer chromatography (TLC). Reaction mixtures (20 μl) containing 20 mM Tris–Cl, pH 7.5, 1 mM DTT, 40 mM NaCl, 10 mM MgCl2, 50 ng DNA, 2 mM ATP and 1 nM [α32-P] ATP were pre-incubated at 37°C for 5 min before FtsK50C addition (150 nM). Reactions were incubated for a further 30 min before being stopped with EDTA (0.5 M, 2.5 μl), kept at 20°C for 10 min and analysed by TLC on PEI–cellulose plates (Merck). ATP hydrolysis was directly proportional to time for at least 30 min, enabling enzyme turnover to be calculated in the presence of different DNAs.
HJ resolution. dif-containing HJs were prepared by annealing oligonucleotides (used to prepare untethered HJ in Arciszewska et al, 1997). HJs were purified twice by TBE–PAGE and ethanol precipitation before being used in resolution reactions at 2 nM. Reactions were performed using the same buffer and reactant concentrations as the intermolecular recombination reactions. Reactants were mixed on ice, and then FtsK50C was added immediately to some reactions. Reactions were incubated at 37°C for 60 min before being stopped and analysed in the same way as the intermolecular recombination reactions.
Acknowledgments
This research was supported by the Wellcome Trust and the Royal Society. T.H.M. received an MRC postgraduate training award.
References
- Arciszewska LK, Grainge I, Sherratt DJ (1997) Action of sitespecific recombinases XerC and XerD on tethered Holliday junctions. EMBO J 16: 3731–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aussel L, Barre F-X, Aroyo M, Stasiak A, Stasiak AZ, Sherratt DJ (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108: 195–205 [DOI] [PubMed] [Google Scholar]
- Barre F-X, Sherratt DJ (2002) Xer sitespecific recombination: promoting chromosome segregation. In Mobile DNA II, Craig NL, Craigie R, Gellert M, Lambowitz A (eds) Vol. 1, pp 149–161. Washington, DC: ASM Press [Google Scholar]
- Barre F-X, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ (2000) FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev 14: 2976–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely GW, Davidson AO, Sherratt DJ (2000) Sequential strand exchange by XerC and XerD during sitespecific recombination at dif. J Biol Chem 275: 9930–9936 [DOI] [PubMed] [Google Scholar]
- Bregu M, Sherratt DJ, Colloms SD (2002) Accessory factors determine the order of strand exchange in Xer recombination at psi. EMBO J 21: 3888–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capiaux H, Cornet F, Corre J, Guijo M-I, Pérals K, Rebollo JE, Louarn J-M (2001) Polarization of the Escherichia coli chromosome. A view from the terminus. Biochimie 83: 161–170 [DOI] [PubMed] [Google Scholar]
- Capiaux H, Lesterlin C, Pérals K, Louarn J-M, Cornet F (2002) A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep 3: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-J, Yu X, Egelman EH (2002) The hexameric ring structure of the Escherichia coli RuvB branch migration protein. J Mol Biol 319: 587–591 [DOI] [PubMed] [Google Scholar]
- Corre J, Louarn J-M (2002) Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli. J Bacteriol 184: 3801–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ (2000) The importance of repairing stalled replication forks. Nature 404: 37–41 [DOI] [PubMed] [Google Scholar]
- Draper GC, McLennan N, Begg K, Masters M, Donachie WD (1998) Only the N-terminal domain of FtsK functions in cell division. J Bacteriol 180: 4621–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS (2003) The structure and function of MCM from archaeal M. thermoautotrophicum. Nat Struct Biol 10: 160–167 [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul DN, Van Duyne GD (1997) Structure of Cre recombinase complexed with DNA in a sitespecific recombination synapse. Nature 389: 40–46 [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul DN, Van Duyne GD (1999) Asymmetric DNA bending in the Cre-loxP sitespecific recombination synapse. Proc Natl Acad Sci USA 96: 7143–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Draper GC, Donachie WD (1998) FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol 29: 893–903 [DOI] [PubMed] [Google Scholar]
- Lobry JR, Louarn J-M (2003) Polarisation of prokaryotic chromosomes. Curr Opin Microbiol 6: 101–108 [DOI] [PubMed] [Google Scholar]
- Michel B, Flores MJ, Viguera E, Grompone G, Seigneur M, Bidnenko V (2001) Rescue of arrested replication forks by homologous recombination. Proc Natl Acad Sci USA 98: 8181–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RD, Calvet C, Demeter M, Agra R, Kong H (2000) Characterization of the specific DNA nicking activity of restriction endonuclease N.BstNBI. Biol Chem 381: 1123–1125 [DOI] [PubMed] [Google Scholar]
- Pérals K, Cornet F, Merlet Y, Delon I, Louarn JM (2000) Functional polarization of the Escherichia coli chromosome terminus: the dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol Microbiol 36: 33–43 [DOI] [PubMed] [Google Scholar]
- Steiner WW, Kuempel PL (1998) Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol Microbiol 27: 257–268 [DOI] [PubMed] [Google Scholar]
- Yates J, Aroyo M, Sherratt DJ, Barre F-X (2003) Species specificity in the activation of Xer recombination at dif by FtsK. Mol Microbiol 49: 241–249 [DOI] [PubMed] [Google Scholar]
- Yu X-C, Weihe EK, Margolin W (1998) Role of the C-terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol 180: 6424–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]