Abstract
The cleavage of proteins within their transmembrane domain by Presenilin (PS) has an important role in different signalling pathways and in Alzheimer's disease. Nevertheless, not much is known about the regulation of PS activity. It has been suggested that substrate recognition by the PS complex depends only on the size of the extracellular domain independent of the amino-acid sequence and that PS activity is constitutive in all cells that express the minimal components of the complex. We report here the development of an in vivo reporter system that allowed us to analyse the processing of human amyloid precursor protein (APP) and the Notch receptor tissue specifically during Drosophila development in the living organism. Using this system, we demonstrate differences between APP and Notch processing and show that PS-mediated cleavage of APP can be regulated in different cell types independent of the size of the extracellular domain.
Keywords: APP, Drosophila, Presenilin, Notch, RIP
Introduction
During the pathogenesis of Alzheimer's disease (AD), the amyloid β-peptide (Aβ) accumulates in plaques. The Aβ peptide is produced by the sequential cleavage of amyloid precursor protein (APP) by two proteases termed β- and γ-secretase (see Annaert & De Strooper, 2002). The first cleavage occurs in the extracellular domain (EC) near the transmembrane region of APP by the βsecretase BACE. The remaining C-terminal fragment (CTF) serves as a substrate for the γ-secretase, which mediates proteolysis inside the membrane region, releasing the cytoplasmic domain (AICD) and the Aβ peptide. The production of Aβ is blocked by different α-secretases, which cleave within the Aβ sequence. γ-Secretase activity is carried out by a complex consisting of four proteins that form the minimal active complex (see Aguzzi & Haass, 2003). PS contains two aspartate residues, which are considered to form the active centre of the protease, whereas Nicastrin (Nct), Aph-1 and Pen-2 stabilize the complex.
The PS-complex cleaving APP comprises the same core components necessary for the cleavage of the Notch (N) receptor, but it is unclear whether both complexes are molecularly identical. Following ligand binding, Notch is cleaved near the transmembrane domain, rendering the remaining CTF a substrate for the PS complex, which results in the release of NICD. Recently, this coordinated processing of APP and Notch has been termed regulated intramembrane proteolysis (RIP; Brown et al, 2000). Although the proteases involved in RIP of Notch and APP have been identified, little is known about their regulation. It has been suggested that substrate recognition by PS depends only on the size of the EC and is sequence independent. This indicates that cleavage of the EC of any type-I transmembrane protein could transform it into a substrate for PS (Struhl & Adachi, 2000) and that the cleavage within the EC is the regulatory step. First evidence that such a model might be imperfect was provided by Lieber et al (2002). In their transgenic Drosophila assay, two proteases were able to cleave the EC of Notch at a similar position. Nevertheless, the generated products were processed with a different efficiency by PS, suggesting a regulatory step after the release of the EC.
To address the question of how the proteases involved in RIP are regulated and behave in the context of a living animal, we developed an in vivo reporter system that allows us to analyse the processing of APP and Notch during the development of Drosophila directly and tissue specifically. Using this system, we demonstrate differences between APP and Notch processing and show that PS-dependent cleavage of APP is modulated in different cell types.
Results And Discussion
Drosophila as a model system to study rip
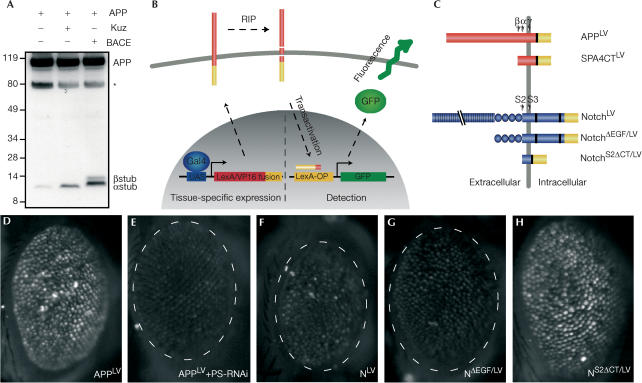
Previous studies showed that in embryonic Drosophila S2 cells, human APP is cleaved by α-secretase and PS (Fossgreen et al, 1998; Takasugi et al, 2003). To determine whether there is αsecretase activity also in the adult fly, we performed western blot analysis with APP-transgenic lines (Fig 1A). In extracts from heads, a CTF could be detected corresponding in size to APP cleaved at the αsite. One of the best candidates responsible for this α-secretase activity in Drosophila is the protease Kuzbanian (Kuz). When we overexpressed Kuz and APP together, the amount of the α-CTF of APP strongly increased, showing that Kuz can indeed cleave APP. As there is no homologue for BACE in Drosophila, we established transgenic flies coexpressing human BACE and APP, which led to the accumulation of a CTF similar in size to APP cleaved at the β-site. The nature of the CTFs was further confirmed with an antibody against amino acids (aa) 1–17 of the Aβ sequence that recognized the β-CTF but not the α-CTF (data not shown).
Figure 1.
Drosophila as a model system to study the processing of APP and Notch. (A) Western blot of head extracts from flies expressing either APP alone or together with Kuz and BACE under the control of GMR-Gal4. Antibody CT13 against the AICD was used for detection. The asterisk marks an unspecific crossreaction. The positions of molecular-mass markers are shown (in kDa) at the left. (B,C) Diagram of the reporter system and the LexA–VP16 (LV) fusion constructs. LV is shown in yellow, and nuclear localization sequences are shown as black bars. The LNR repeats of Notch are represented by circles, and the EGF domains by narrow boxes. (D–H) GFP signals generated by the reporter system in the compound eye of the adult fly. The expression of APPLV leads to a robust GFP signal (D). RNAi against PS results in a loss of the signal (E). No GFP is detected in flies expressing either the full-length Notch receptor (F) or the truncated form (G). (H) Constitutive activation of the reporter system by NS2dCT/LV. For genotypes, see Methods.
To examine the processing of APP in vivo, we created a reporter system, which is based on the fusion of the LexA DNA-binding domain and the VP16 activator domain (LV) to APP (Fig 1C). The construct is expressed tissue specifically under the control of Gal4 (Brand & Perrimon, 1993). Upon subsequent processing by αsecretase and PS, AICD/LV is released, translocates to the nucleus and binds to LexA operators upstream of a green fluorescent protein (GFP) reporter. The processing event is then visualized by the expression of GFP (Fig 1B). Owing to the close similarities between the cleavage of Notch and APP, and to investigate whether particular features of the two processes can be distinguished in living tissues, we included a fusion construct of Drosophila Notch (Fig 1C). A truncated form of Notch lacking the EGF repeats (NΔEGF/LV), which abolishes the binding to the ligand (Kidd et al, 1998), served as negative control.
As we have already revealed the presence of α-secretase activity in the adult eye, we first analysed the processing of our constructs by expression in the eye cells with the GMR-Gal4 line (Freeman, 1996). A strong GFP signal was detected with APPLV, indicating that αsecretase and PS are indeed active (Fig 1D). In contrast, NLV was not processed (Fig 1F), suggesting that there is no significant activation of the Notch receptor in the adult eye. Likewise, we observed no GFP signal after expression of NΔEGF/LV (Fig 1G). These results are consistent with earlier genetic studies, which did not reveal any function for Notch in the adult eye (Shellenbarger & Mohler, 1978). A strong activation of the GFP reporter was observed only with a precleaved form of Notch. This construct mimics the ligand-induced cleavage of the EC and is a direct substrate for PS (Fig 1H). To confirm the specificity for PS-mediated cleavage of APP, we knocked down the components of the PS complex by RNA interference (RNAi). When we coexpressed hairpin RNAs complementary to PS, Nct, Aph-1 or Pen-2 (Fig 1E, and not shown), the GFP signal generated by APPLV was abolished. Analogous observations in Drosophila have recently been published (Guo et al, 2003). Taken together, these results demonstrate that our reporter system specifically visualizes RIP of APP and Notch.
Restriction of efficient APP processing to neurons
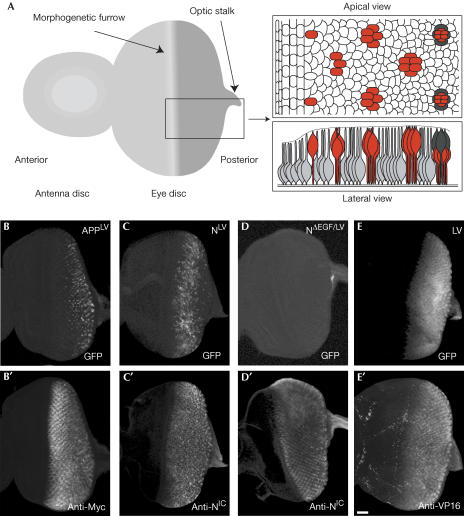
We next addressed the question of whether there are differences between APP and Notch processing in the developing Drosophila eye. At the end of the larval stages, the patterning of the future retina starts with the selection of the first photoreceptor (R8) of each ommatidium in the morphogenetic furrow of the eye imaginal disc (Fig 2A; Wolff & Ready, 1993). The morphogenetic furrow moves from the posterior end through the eye disc, patterning each row of founder cells. The R8 photoreceptor then recruits neighbouring cells to form additional cell types (Fig 2A, apical view). These fate decisions and the specification of R8 involve the Notch pathway (Frankfort & Mardon, 2002). Using GMR–Gal4, we expressed our fusion constructs in all cells posterior to the furrow. For APPLV, we detected a strong GFP signal (Fig 2B). Surprisingly, this was restricted to cells at the posterior end of the disc, although the construct was expressed homogeneously (Fig 2B′). In contrast, Notch processing could be visualized in a broad stripe of cells in the vicinity of the morphogenetic furrow, and the GFP signal vanished at the posterior end of the disc (Fig 2C). No GFP signal was observed for the negative control NΔEGF/LV (Fig 2D).
Figure 2.
The reporter system reveals spatial and temporal differences in the processing of APP and Notch in the developing eye. (A) Schematic representation of an eye disc from a third-instar larva. The expression domain of GMR–Gal4 starting after the morphogenetic furrow is shown in dark grey. The magnification shows the progressive patterning of the disc. The neurons of the photoreceptor clusters are shown in red, cone cells in dark grey and undetermined cells in light grey. (B–E) GFP signals generated by the reporter system in the developing eye disc. APPLV (B), NotchLV (C), NotchΔEGF/LV (D) and LexA–VP16 (E) were expressed under the control of GMR–Gal4. To confirm that the fusion proteins are expressed homogeneously in all cells behind the morphogenetic furrow, the discs were stained with anti-Myc (B′), anti-NICD (C′,D′) and anti-VP16 (E′). All pictures are projections of optical zsections covering the complete epithelium. Scale bar, 20 μm.
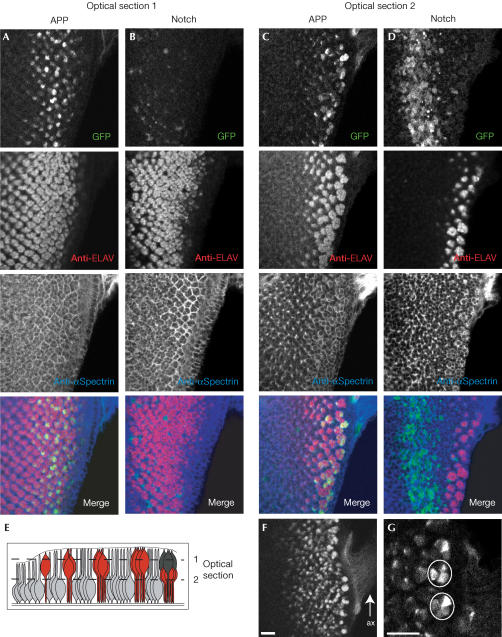
To study the pattern of APP and Notch processing in more detail, we stained the discs for the neuronal marker ELAV and αSpectrin, which outlines the cell boundaries. Figure 3 shows two optical sections through an eye disc expressing APPLV (A+C) or NLV (B+D). The majority of the GFP signal representing APP processing is detected in the nuclei and soma of differentiating neurons, overlapping with ELAV. There is no detectable GFP in undetermined cells (Fig 3C). Further evidence for the elevated processing in neuronal cells is provided by the GFP signal in the axons projecting from the differentiating neurons to the optic lobe (Fig 3F). At the beginning of differentiation, not all of the ELAV-positive cells show a GFP signal, whereas at the posterior end of the disc more clusters of photoreceptors reveal an activated reporter in all cells (Fig 3G).
Figure 3.
Elevated processing of APP in differentiating neurons. Two optical sections through the epithelium of the eye disc are shown. Optical section 1 (A, B) comprises mainly nuclei and cell bodies of the neurons, whereas optical section 2 (C, D) contains undetermined cells (see E). The GFP signal representing APP (A, C) or Notch processing (B, D) is shown together with antibody stainings for ELAV and αSpectrin. (E) Schematic drawing of a lateral view of the eye disc showing the location of neuronal cells (red), cone cells (dark grey) and unpatterned cells (light grey). (F) The axons (ax) of the photoreceptors projecting to the optical lobes show a strong GFP signal in flies expressing APPLV. (G) As development continues, all neuronal cells of the photoreceptor cluster show APP processing, although there are variations in the intensity of the GFP signal. Scale bar, 10 μm.
Processing of Notch follows a different pattern: GFP is expressed only in undetermined cells at the basal side of the epithelium, showing no overlap with ELAV (Fig 3D). The pattern may reflect the role of Notch in spacing the ommatidia in the morphogenetic furrow through lateral inhibition (Frankfort & Mardon, 2002). This process is required for the restriction of neuronal competence to a group of cells that is subsequently resolved into a single R8 founder cell for every ommatidium.
In summary, these results show differences in APP and Notch processing in the eye imaginal disc. The processing of Notch depends on ligand binding and the activity of the protease Kuz, and is restricted to undetermined cells posterior to the morphogenetic furrow. Although these cells are competent to release NICD, there is no RIP of APP detectable. APP only becomes a good substrate for the PS complex in differentiating neurons. However, as we used a full-length form of APP, we could not at this point distinguish whether the change in APP processing in the different cell types was due to altered α-secretase or PS activity.
Regulated PS activity restricts efficient APP processing
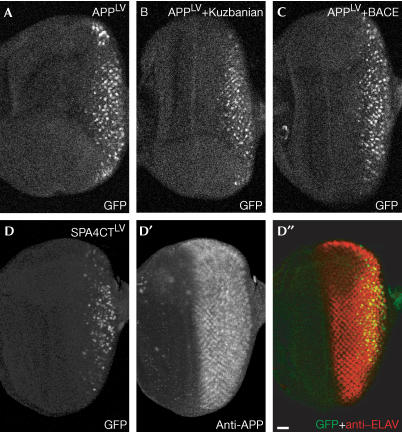
To address this question, we overexpressed Kuz and BACE to increase the cleavage of APP in all cells. As shown previously (Fig 1A), both proteases induce a massive production of CTFs. Surprisingly, there was no change in the pattern of GFP reporter expression (Fig 4A–C), indicating that the production of CTFs from APP is not the limiting step in cells with no detectable RIP of APP. Instead, the activity of the PS–protease complex seems to be regulated, resulting in an increased release of AICD in differentiating neurons. To abolish completely the need for α- or βsecretase cleavage, we designed a fusion construct that mimics the APP fragment after release of the EC by BACE (SPA4CTLV; Fig 1C). It has previously been shown that this construct is a direct substrate for PS, leading to the generation of Aβ and the release of AICD in cell lines and in Drosophila (Fossgreen et al, 1998; Lichtenthaler et al, 1999; Struhl & Adachi, 2000). Again, our reporter system revealed an identical pattern in reporter gene activation between this direct PS substrate and full-length APP (Fig 4D).
Figure 4.
APP processing is regulated at the PS level. (A) The restricted pattern of GFP expression representing APP processing does not change upon coexpression of Kuz (B) or BACE (C). (D) GFP expression induced by the processing of a truncated form of APP, SPA4CTLV, which mimics precleavage by BACE. (D′) SPA4CTLV is expressed in every cell posterior to the morphogenetic furrow. (D″) Overlay of the GFP signal with antibody staining for ELAV. (D–D″) Projections of optical sections covering the complete epithelium. Scale bar, 20 μm.
Thus, using two independent approaches to generate direct substrates for PS from APP, we could not change the pattern of APP processing visualized by the reporter system. These results clearly suggest that the activity of the PS complex can be modulated, leading to the restriction of efficient APP cleavage to neuronal cells in the eye disc. This finding is surprising, as previous studies in Drosophila embryos suggested that any integral membrane protein is constitutively cleaved by PS when its EC is shorter than 200 aa (Struhl & Adachi, 2000). Further decrease in the size of the EC resulted in a progressive increase in cleavage efficiency with a threshold level at 50 aa. Below this size, the rate of cleavage did not change further. With an EC of 30 aa, SPA4CTLV is under this threshold level and should be recognized by the PS complex with the same efficiency in all cells. However, we observed a strong increase in PS-mediated cleavage of APP in differentiating neurons compared with the undetermined cells.
Notch and APP cleavage in the wing disc
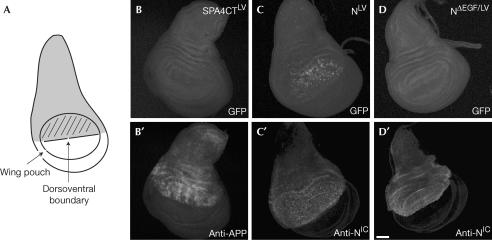
To confirm the results obtained in the eye disc, we expressed our constructs with the driver line ap-Gal4 in the wing imaginal disc (Fig 5A; Calleja et al, 1996). Using full-length APPLV (not shown) or the precleaved SPA4CTLV (Fig 5B), we were not able to detect any efficient release of AICD, although the constructs were expressed at high levels (Fig 5B′). The potential αsecretase and PS activity of the cells could be visualized by the expression of NLV, which leads to GFP-positive cells in the wing pouch (Fig 5C). In these cells, activation of the Notch pathway promotes proliferation and outgrowth of the future wing blade (Baonza & Garcia-Bellido, 2000). As expected, the nonprocessable NΔEGF/LV showed no signal (Fig 5D). These experiments underline the different requirements for efficient PS-mediated release of AICD and NICD and the cell-type-dependent variation in the level of APP cleavage by PS.
Figure 5.
Notch and APP cleavage in the wing disc. (A) Schematic representation of a wing disc from a third-instar larva. The ap-Gal4 line allows expression in the dorsal part of the disc (grey). The cells of the wing pouch proliferate and give rise to the adult wing. The GFP signals generated by the processing of SPA4CTLV (B), NotchLV (C) and NotchΔEGF/LV (D) are shown. (C′,D′) Expression of the fusion constructs detected with antibodies for APP or NICD. Scale bar, 40 μm.
Conclusions
We have developed an in vivo reporter system in Drosophila, which demonstrates that the release of AICD, and thus of the amyloidogenic Aβ peptide, is regulated in different cell types by modulated PS activity. Interestingly, the processing seems to be upregulated in neurons, which are most affected by the toxic effects of Aβ in AD patients. Although this study does not provide a mechanistic understanding of why processing might vary across cell types, it provides evidence that challenges the model that PS activity is not regulated and depends only on the size of the EC. How can the differences between the results of our reporter system and previous studies be explained? One possible answer is that most previous studies were carried out in cell culture using either fibroblasts or transformed cell lines. At the moment, we are not aware of any study that directly compares the processing efficiency of APP in various cell types. Other studies performed in vivo used very sensitive enzymatic methods to detect cleavage activities. However, the high sensitivity and signal amplification makes quantitative analysis by in situ staining very difficult. In contrast, our GFP-based assay is less sensitive, so that we do not expect to see the constitutive cleavage that is probably taking place at a low level in all cells. For example, when we analysed embryonic stages where previous studies using a lacZ reporter revealed cleavage of a similar SPA4CTLV construct (Struhl & Adachi, 2000), we could only detect faint GFP signals (data not shown). The versatility of our system, however, allowed us to examine different tissues in the developing organism, enabling us to compare directly different cell types and to identify those cells with elevated cleavage activity.
The ICD of APP, in complex with additional cofactors, shows transcriptional activity after translocation to the nucleus (Cao & Südhof, 2001). Therefore, it is possible that the binding of a repressor to AICD in the undetermined cells could give rise to the cellspecific GFP signal observed. To circumvent such a possibility, we chose VP16 for our reporter system, which is one of the strongest known transcriptional activators. It has been shown, for instance, that the fusion of VP16 to the Notch pathway component Suppressor of Hairless counteracts repression in Drosophila (Furriols & Bray, 2000). Thus, although we cannot formally rule out modulated transcription of the reporter gene, we reason that the results obtained with our reporter system reliably reflect the processing of APP.
The PS-dependent cleavage of APP could be regulated in different ways. First, three different splice variants of Drosophila PS have been reported, which are differentially expressed during development (Nowotny et al, 2000). Alternative splicing is also predicted for Nct (Krause et al, 2002), leading to a variety of possible PS complexes consisting of the minimal protein components. Furthermore, studies in mammalian cells described complexes varying in size, suggesting that there might be additional subunits, which would offer further possibilities to modulate PS activity (see De Strooper, 2003). Yet another possibility would be a change in trafficking of APP, which would prevent the physical contact between the substrate and the protease in the undetermined cells. However, we have not been able to detect any alterations in the localization of APP in our studies. One could also envisage that the cleavage within the EC has to be accompanied by additional modifications within the remaining CTF and that these modifications are cell-type specific, an argument that could also explain the observation of Lieber et al (2002) that Notch CTFs generated by TACE are less efficiently cleaved by PS than those generated by Kuz.
The differences we observe between Notch and APP processing by PS in Drosophila could provide new hope for the use of inhibitors as therapeutics for AD. Currently, the benefit of lowering Aβ production by interfering with PS activity would be accompanied by the deleterious effects of simultaneously blocking the Notch signalling pathway. If it were possible to design drugs against the distinct types of the PS complex or secondary factors responsible for effective APP cleavage, the two processes could be separated. In addition, our results clearly show the need for similar in vivo studies in a vertebrate AD model to determine the exact mechanisms underlying the normal and aberrant processing of APP in a real in vivo situation in a complex neuronal network.
Methods
Plasmid construction. The fusion constructs were made by homologous recombination in Escherichia coli. The nucleotides 1281–1781 of the PS cDNA were cloned into the pWIZ vector (Lee & Carthew, 2003; see also supplementary information online). The aa sequences of the fusion parts of the LV constructs are: NLV …QANKGSEAIYI-MKALTARQQ… NΔEGF/LV …QANKGSEAIYI-MKALTARQQ… NS2ΔCT/LV …FFGMVLSTQRKR-PPKKKRKVMKALTARQQ… APPLV …NPTYKFFEQMQN-PPKKKRKVMKALTARQQ… SPA4CTLV …NPTYKFFEQMQN-PPKKKRKVMKALTARQQ…
Fly stocks. Transgenic fly lines were established by standard procedures. Two insertions of the LexA–GFP reporter on the second chromosome were recombined. The genotype of the flies is as follows: 2xLexA-hrGFP; UAS-LVfusion/GMR-Gal4∥2xLexA-hrGFP/PS-RNAi;UAS-APPLV GMR-Gal4/+∥2xhrGFP/ap-Gal4; UAS-LVfusion/+. The RNAi experiment was carried out at 28°C, and all other crosses were carried out at 25°C.
Immunocytochemistry. Tissues were dissected in phosphate-buffered saline (PBS), fixed in PBS/3% paraformaldehyde for 30 min, blocked in PBS/5% bovine serum albumin (BSA)/0.3% Triton, incubated 14 h with primary antibody in PBS/1% BSA/0.1% Triton, washed, incubated with antibodies coupled to Alexa546 or Alexa647 (Molecular Probes), washed and embedded in Mowiol. The antibodies used were as follows: anti-Myc, anti-VP16 (Santa Cruz); anti-NICD, anti-ELAV, anti-αSpectrin (Developmental Studies Hybridoma Bank, University of Iowa).
Western blotting. Ten fly heads per sample were homogenized in SDS sample buffer.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v5/n4/extref/74000122s1.pdf).
Supplementary Material
supplementary information
Acknowledgments
We thank D. Kuttenkeuler, M.W. Young, T. Lieber, A.F. Stewart, D. Pan, S. Cohen, R.W. Carthew and M. Bienz for material, M.V. Bilic and L. Ringrose for comments on the manuscript and M. Mueller for excellent technical assistance. This work was supported by a grant from the DFG (SPP ‘Cellular Mechanisms of AD') to G.M.
References
- Aguzzi A, Haass C (2003) Games played by rogue proteins in prion disorders and Alzheimer's disease. Science 302: 814–818 [DOI] [PubMed] [Google Scholar]
- Annaert W, De Strooper B (2002) A cell biological perspective on Alzheimer's disease. Annu Rev Cell Dev Biol 18: 25–51 [DOI] [PubMed] [Google Scholar]
- Baonza A, Garcia-Bellido A (2000) Notch signaling directly controls cell proliferation in the Drosophila wing disc. Proc Natl Acad Sci USA 97: 2609–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100: 391–398 [DOI] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G (1996) Visualization of gene expression in living adult Drosophila. Science 274: 252–255 [DOI] [PubMed] [Google Scholar]
- Cao X, Südhof TC (2001) A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293: 115–120 [DOI] [PubMed] [Google Scholar]
- De Strooper B (2003) Aph-1, Pen-2, and Nicastrin with Presenilin generate an active γsecretase complex. Neuron 38: 9–12 [DOI] [PubMed] [Google Scholar]
- Fossgreen A, Bruckner B, Czech C, Masters CL, Beyreuther K, Paro R (1998) Transgenic Drosophila expressing human amyloid precursor protein show γsecretase activity and a blistered-wing phenotype. Proc Natl Acad Sci USA 95: 13703–13708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G (2002) R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development 129: 1295–1306 [DOI] [PubMed] [Google Scholar]
- Freeman M (1996) Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87: 651–660 [DOI] [PubMed] [Google Scholar]
- Furriols M, Bray S (2000) Dissecting the mechanisms of Suppressor of Hairless function. Dev Biol 227: 520–532 [DOI] [PubMed] [Google Scholar]
- Guo M, Hong EJ, Fernandes J, Zipursky SL, Hay BA (2003) A reporter for amyloid precursor protein γsecretase activity in Drosophila. Hum Mol Genet 12: 2669–2678 [DOI] [PubMed] [Google Scholar]
- Kidd S, Lieber T, Young MW (1998) Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev 12: 3728–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause A, Haas SA, Coward E, Vingron M (2002) SYSTERS, GeneNest, SpliceNest: exploring sequence space from genome to protein. Nucleic Acids Res 30: 299–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW (2003) Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30: 322–329 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler SF, Multhaup G, Masters CL, Beyreuther K (1999) A novel substrate for analyzing Alzheimer's disease γsecretase. FEBS Lett 453: 288–292 [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Young MW (2002) kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev 16: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny P et al. (2000) Posttranslational modification and plasma membrane localization of the Drosophila melanogaster Presenilin. Mol Cell Neurosci 15: 88–98 [DOI] [PubMed] [Google Scholar]
- Shellenbarger D, Mohler J (1978) Temperaturesensitive periods and autonomy of pleiotropic effects of l(1)Nts1, a conditional Notch lethal in Drosophila. Dev Biol 62: 432–446 [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A (2000) Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell 6: 625–636 [DOI] [PubMed] [Google Scholar]
- Takasugi N et al. (2003) The role of presenilin cofactors in the γsecretase complex. Nature 422: 438–441 [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF (1993) Pattern formation in the Drosophila retina. In The Development of Drosophila melanogaster, Bate M, Martinez Arias A (eds) pp 1277–1326. Plainview, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information