Abstract
A significant fraction of disease-causing mutations affects pre-mRNA splicing. These mutations can generate both aberrant and correct transcripts, the level of which varies among different patients. An inverse correlation was found between this level and disease severity, suggesting a role for splicing regulation as a genetic modifier. Overexpression of splicing factors increased the level of correctly spliced RNA, transcribed from minigenes carrying disease-causing splicing mutations. However, whether this increase could restore the protein function was unknown. Here, we demonstrate that overexpression of Htra2-β1 and SC35 increases the level of normal cystic fibrosis transmembrane conductance regulator (CFTR) transcripts in cystic-fibrosis-derived epithelial cells carrying the 3849+10 kb C → T splicing mutation. This led to activation of the CFTR channel and restoration of its function. Restoration was also obtained by sodium butyrate, a histone deacetylase inhibitor, known to upregulate the expression of splicing factors. These results highlight the therapeutic potential of splicing modulation for genetic diseases caused by splicing mutations.
Keywords: cystic fibrosis, cystic fibrosis transmembrane conductance regulator functional restoration, sodium butyrate, splicing factors, splicing pattern modulation
Introduction
Cystic fibrosis (CF) is a common, severe autosomal recessive disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (Kerem et al, 1989). A significant fraction (13%) of CFTR mutations affects the pre-mRNA splicing of the gene (http://www.genet.sickkids.on.ca/cftr), by disrupting or generating intronic splicing motifs, required for exon recognition. In addition, there are mutations and polymorphisms that disrupt exonic splicing motifs, which also affect the splicing pattern. All these mutations can be divided into two groups on the basis of their effect on the splicing pattern. One group includes mutations that completely abolish exon recognition (such as 621+1 G → T, 711+1 G → T and 1525−1 G → A), resulting in a relatively severe disease due to a complete absence of correctly spliced transcripts (Zielenski et al, 1993; Ramalho et al, 2003). The second group includes mutations that generate both aberrantly and correctly spliced transcripts (such as 3849+10 kb C → T, 3272−26 A → G, IVS8-5T, D565G and G576A), the level of which varies among patients and among organs of the same patient (Ramalho et al, 2002; Pagani et al, 2003; reviewed in Nissim-Rafinia & Kerem, 2002). These patients often have a relatively mild phenotype, but with variable disease expression from minimal lung disease, pancreatic sufficiency and male fertility to relatively severe disease in all the involved organs (Augarten et al, 1993; Kerem et al, 1997). This variable disease expression is inversely correlated with the level of correctly spliced transcripts, such that lower levels are associated with severe disease and higher levels with a milder phenotype. These results suggested that splicing regulation has a role as a genetic modifier of disease severity in patients carrying splicing mutations that generate normal transcripts (Nissim-Rafinia & Kerem, 2002).
Splicing is regulated through the interaction of a complex repertoire of splicing factors with various splicing motifs (reviewed in Black, 2003). Differences in the levels of functional splicing factors were found among different tissues, which has been suggested to regulate the tissuespecific level of alternatively spliced transcripts. As the first step in understanding the role of splicing regulation as a modifier of CF, the effect of overexpression of splicing factors on the level of correctly spliced CFTR transcripts was studied in minigenes carrying CFTR splicing mutations (Nissim-Rafinia et al, 2000; Pagani et al, 2000, 2003; Aznarez et al, 2003). Among these were mutations that lead to partial skipping of exons 9 and 12, the 5′ end of exon 13 and the 3849+10 kb C → T mutation, which leads to partial inclusion of 84 base pairs (bp) from intron 19. Most (10 out of 11) of the minigenes were modulated by splicing factors. Higher levels of correctly spliced transcripts were generated by several of these factors (Htra-α and E4-ORF3, which promoted exons 13 and 9 inclusion, respectively, and hnRNP A1 and E4-ORF6, which promoted skipping over the cryptic 84 bp exon). However, whether the increased levels of correctly spliced transcripts can restore the forskolinstimulated CFTR function remains unknown.
Here, we demonstrate that an increase in the level of correctly spliced RNA, transcribed from an endogenous CFTR allele carrying the 3849+10 kb C → T mutation, activated the CFTR channel and restored its function. This was obtained by overexpression of SR (SC35) and SR-like (Htra2-β1) splicing factors, which promoted skipping of the 84 bp exon and increased the level of full-length CFTR transcripts. The same effect was obtained by administration of sodium butyrate (NaBu), a histone deacetylase inhibitor, previously shown to upregulate the expression of splicing factors (Chang et al, 2001; Brichta et al, 2003). These results explain the correlation between the level of correctly spliced CFTR transcripts and disease severity, and provide the required basis for the development of therapeutic approaches for patients carrying splicing mutations in CF as in other human diseases.
Results And Discussion
CFTR splicing modulation by splicing factors
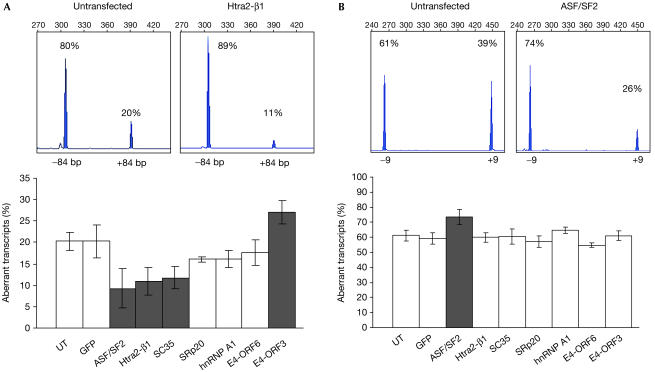
To study the potential of splicing modulation to restore the CFTR function, cells comprising the entire CFTR coding region, carrying a splicing mutation, were required. We established an epithelial cell line (CFP15a) from a nasal polyp of a CF patient carrying the 3849+10 kb C → T splicing mutation. This mutation generates a donor site in intron 19, which can lead to inclusion of 84 bp containing an in-frame stop codon (Highsmith et al, 1994). Reverse transcription–PCR (RT–PCR) showed that 20±2% of the transcripts include the cryptic exon (Fig 1A). These aberrant transcripts were transcribed only from the 3849+10 kb C → T allele. The correctly spliced transcripts (80%) were transcribed from both alleles, and as the relative contribution of each allele was unknown, we analysed throughout the study the relative level of aberrantly spliced transcripts.
Figure 1.
Effect of overexpression of splicing factors on the splicing pattern of CFTR exons in CFP15a cells carrying the 3849+10 kb C → T splicing mutation. Analysis of the splicing pattern of the cryptic 84 bp exon (A) and exon 9 (B). The top panels show examples of Genescan analysis of RT–PCR products from untransfected cells (left) and cells transfected (right) with Htra2-β1 (A) and ASF/SF2 (B). Percentages denote relative levels of correct and aberrant transcripts and numbers denote sizes of RT–PCR products (304 and 388 bp in (A), and 447 and 264 bp in (B)). The bottom panels show the effect of splicing factors on the level of aberrant transcripts. The grey columns represent a significant effect (P<0.01) using the Mann–Whitney U-test, adjusted with Bonferroni correction for eight comparisons. The white columns represent no effect. Error bars, standard deviation; UT, untransfected.
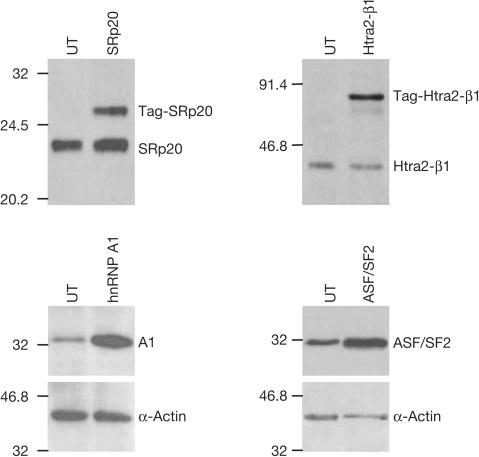
We analysed the effect of transient transfection of splicing factors on the level of aberrantly spliced CFTR transcripts in CFP15a cells, with cellular and viral factors. RT–PCR showed that Htra2-β1, SC35 and ASF/SF2 promoted skipping of the 84 bp exon and led to a significant decrease in the relative level of aberrantly spliced transcripts, whereas SRp20, hnRNP A1 and E4-ORF6 had no significant effect (Fig 1A). E4-ORF3 promoted inclusion and led to a significant increase in the relative level of aberrant transcripts. As a control, cells were transfected with green fluorescent protein. No effect was observed, indicating that the change in the splicing pattern was specific to the splicing factors. We further analysed the level of Htra2-β1, ASF/SF2, SRp20 and hnRNP A1 by immunoblotting (Fig 2). The results showed an increase in the expression of all factors, including those that had no effect (hnRNP A1 and SRp20). Furthermore, Htra2-β1 and hnRNP A1 are highly overexpressed; however, the latter did not affect the splicing pattern. This could result from different binding efficiencies of the splicing factors to splicing motifs and/or changes in relative concentrations of other cellular factors. In summary, our results show that overexpression of splicing factors can modulate the splicing pattern of the cryptic 84 bp exon in RNA transcribed from endogenous CFTR alleles carrying the 3849+10 kb C → T mutation.
Figure 2.
Increase of the protein level of splicing factors transfected into CFP15a cells. The top panels show tagged plasmids (endogenous and tagged of different sizes) and the bottom panels show untagged plasmids (endogenous and exogenous of same size). UT, untransfected.
Restoration of the CFTR function was expected to result from increasing the level of normal and full-length CFTR transcripts. The CFTR gene comprises 27 constitutively spliced exons; however, several exons (3, 4, 9, 12, 14a, 16, 17b and 22) undergo, in some individuals, partial aberrant splicing that generates nonfunctional CFTR proteins (Bienvenu et al, 1996). These exons might be modulated, leading to a decrease in the level of full-length CFTR transcripts. RT–PCR of exon 9 showed that the relative level of aberrantly spliced transcripts lacking the exon is 61±4% (Fig 1B). Overexpression of ASF/SF2 promoted skipping of exon 9 and significantly increased the level of aberrantly spliced transcripts, whereas overexpression of all other factors had no effect. For other exons, see supplementary information online. Taken together, Htra2-β1, SC35 and ASF/SF2 promoted skipping of the cryptic 84 bp exon. Whereas Htra2-β1 and SC35 had no effect on the splicing pattern of other exons and thus were expected to increase the level of normal and full-length CFTR transcripts, ASF/SF2 promoted exon 9 skipping, and therefore its effect on the level of normal transcripts could not be predicted.
CFTR activity restoration by Htra2-β1 and SC35
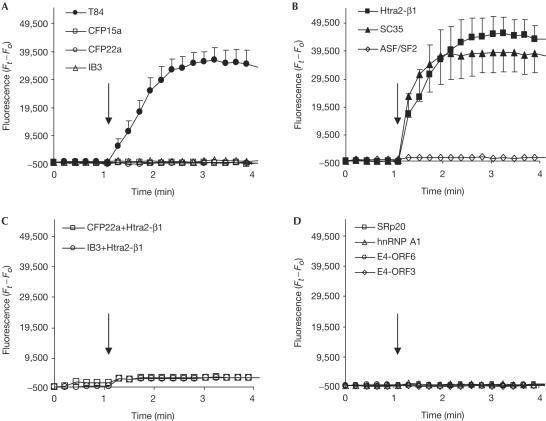
The CFTR protein is a cyclic AMP-stimulated Cl− channel, which also regulates other transport proteins. To study the effect of splicing modulation on the CFTR function, we measured Cl− efflux (Fig 3A). The analysis showed no forskolinstimulated chloride efflux, indicating that the CFTR channels in these cells are inactive, as found in CFP22a cells (ΔF508/ΔF508) and IB3 cells (W1282X/ΔF508).
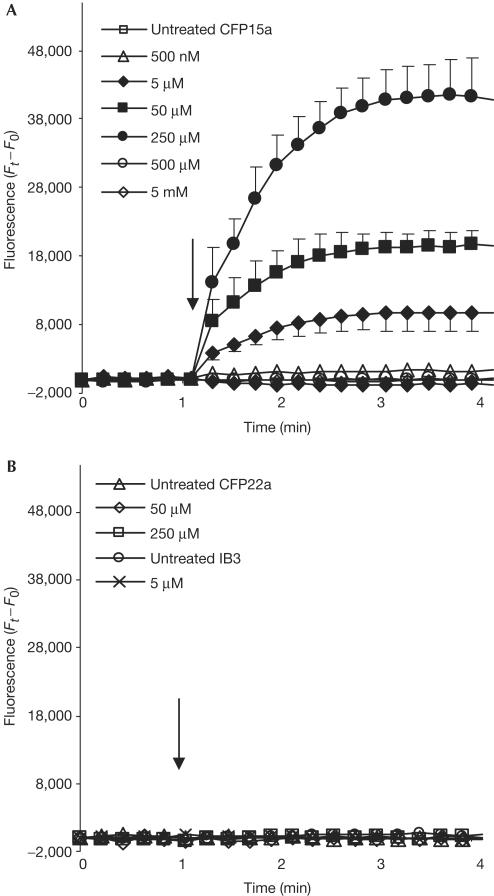
Figure 3.
Activation of CFTR Cl− efflux by overexpression of splicing factors. CFTR activity in (A) T84, CFP15a, CFP22a and IB3 cells, (B) CFP15a cells following transfection with Htra2-β1, SC35 and ASF/SF2, (C) CFP22a and IB3 cells following transfection with Htra2-β1 and (D) CFP15a cells following transfection with SRp20, hnRNP A1, E4-ORF6 and E4-ORF3. The filled symbols indicate activation of the CFTR Cl− efflux and the open symbols indicate no effect. The arrows indicate the time of forskolin administration.
Next, we analysed the effect of overexpression of splicing factors on the function of the CFTR protein. Overexpression of Htra2-β1 and SC35 activated the CFTR channel and restored its function, indicating that the increase in the level of normal transcripts was sufficient for this activation (Fig 3B). We studied the effect of Htra2-β1 on the CFTR function in CFP22a and IB3 cells, in which the CFTR RNA level is similar to that of CFP15a cells (data not shown). No restoration of the CFTR channel activity was found (Fig 3C), indicating that the Htra2-β1 restoration in CFP15a cells was obtained through modulation of the CFTR splicing pattern. Overexpression of ASF/SF2 did not activate the CFTR channel (Fig 3B), indicating that the level of normal and full-length CFTR transcripts was insufficient for CFTR activation. Overexpression of the other factors did not restore the CFTR function (Fig 3D). Altogether, these results show that only factors that significantly increased the relative level of normal CFTR transcripts led to restoration of the CFTR Cl− efflux. These results support our hypothesis that the splicing machinery is a modifier of disease severity in patients carrying splicing mutations (Nissim-Rafinia & Kerem, 2002). A recent study in mice identified a putative splicing factor (sodium channel modifier 1, SCNM1) that modifies disease severity in mice homozygous for a splicing mutation in the Scn8a gene. Mice carrying normal alleles of SCNM1 show chronic movement disorder; however, mice carrying a stop mutation in this gene show reduced levels of correctly spliced Scn8a transcripts and a lethal neurological disease (Buchner et al, 2003). These results provide the first evidence for the role of splicing regulation as a genetic modifier.
CFTR splicing modulation and activity restoration by NaBu
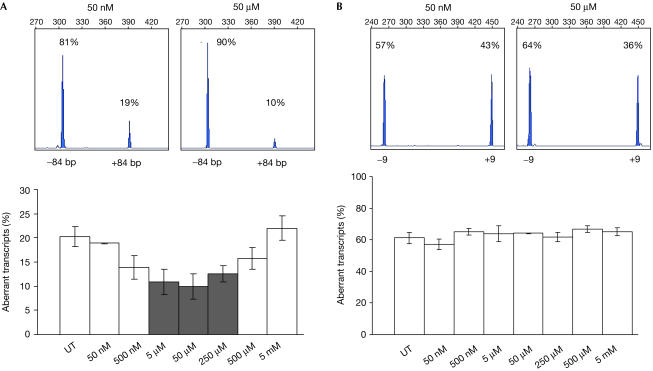
We further analysed the effect of NaBu on CFTR function, on the basis of previous studies showing that NaBu upregulated the expression of Htra2-β1 and SR proteins in spinal muscular atrophy (SMA) cells (Chang et al, 2001; Brichta et al, 2003). In CFP15a cells, NaBu affected the splicing pattern in an inverse ‘bellshape' manner, such that 5–250 μM NaBu significantly decreased the level of aberrantly spliced transcripts, whereas lower and higher concentrations had no effect (Fig 4A). NaBu had no effect on the splicing pattern of exon 9 (Fig 4B) and other CFTR exons studied (data not shown). We then analysed the effect of NaBu on the CFTR function. Activation of the CFTR channel was achieved only by 5–250 μM NaBu, which significantly increased the level of normal CFTR transcripts, whereas other concentrations had no effect on the CFTR Cl− efflux (Fig 5A). NaBu was also administered to CFP22a and IB3 cells and no activation was detected (Fig 5B), indicating that the NaBu restoration in CFP15a cells was obtained through modulation of the CFTR splicing pattern.
Figure 4.
Effect of NaBu on the splicing pattern of CFTR exons in CFP15a cells. Analysis of the splicing pattern of the cryptic 84 bp exon (A) and exon 9 (B). The top panels show examples of Genescan analysis of RT–PCR products from cells treated with 50 nM (left) and 50 μM (right) NaBu. Percentages and numbers in the top panels are as in Fig 1. The bottom panels show the effect of different NaBu concentrations on the level of aberrant transcripts. The grey bars show significant effect (P<0.01, Mann–Whitney U-test, adjusted with Bonferroni correction for seven comparisons) and the white bars show no effect. Error bars, standard deviation; UT, untreated.
Figure 5.
Activation of CFTR Cl− efflux by NaBu. Concentration-dependent activation in CFP15a cells (A) and CFP22a and IB3 cells (B). The arrows, filled and open symbols are as in Fig 3.
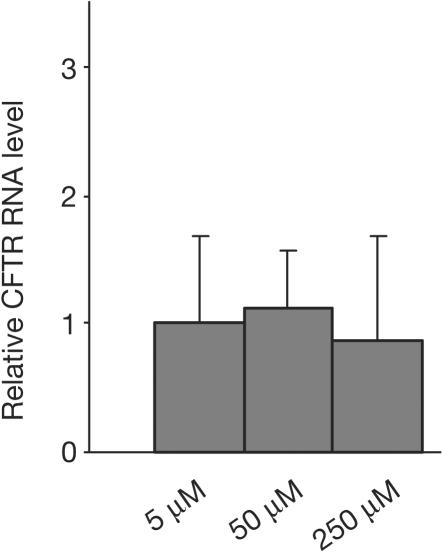
As NaBu has the potential to upregulate gene expression, we evaluated the possibility that NaBu, in addition to its effect on the splicing pattern, upregulated the expression of the CFTR gene in CFP15a cells. For this, we analysed the level of CFTR RNA before and after treatment with 5–250 μM NaBu. The results showed no change in the level of CFTR RNA (Fig 6), indicating no effect of NaBu on the expression of the CFTR gene. Thus, the restoration of the CFTR function by NaBu resulted from splicing modulation only. We then analysed the effect of NaBu on the endogenous level of several splicing factors by using anti-Htra2-β1 and a specific antibody for SR proteins, mAb104. Our results showed no significant increase in the expression of these factors (data not shown). However, as splicing is regulated by >100 factors, it is possible that other factors were affected by NaBu in the CF epithelial cell line used in this study.
Figure 6.
Effect of NaBu on the CFTR RNA level in CFP15a cells. The concentrations used are those that restored the CFTR function. Relative levels of CFTR RNA were quantified by real-time PCR, normalized to RPS9 for RNA loading, relative to untreated cells.
In many human genetic diseases, a substantial fraction of patients carry mutations that affect the correct splicing. A total of 10–15% of disease-causing mutations disrupt intronic splicing motifs and an additional ∼35% disrupt exonic motifs, which lead to aberrant splicing (reviewed in Cartegni et al, 2002). As found for CF, patients who carry splicing mutations causing SMA show extensive variability in disease severity, inversely correlated with the level of correctly spliced RNA containing exon 7 (Gavrilov et al, 1998). Overexpression of splicing factors (including Htra2-β1) promoted exon 7 inclusion and increased the level of correctly spliced SMN2 RNA (Hofmann et al, 2000). It is interesting to note that Htra2-β1 in our study also increased the level of correctly spliced transcripts, but by exon skipping.
Splicing modulation of SMN exon 7 was also demonstrated by treating SMA-derived cells with NaBu. This led to an increase in the level of correctly spliced RNA and SMN protein (Chang et al, 2001; Brichta et al, 2003). However, the effect of the splicing modulation on protein function was not studied. As found for overexpression of Htra2-β1, NaBu promoted skipping of the CFTR 84 bp exon, whereas it promoted inclusion of the SMN2 exon 7. NaBu treatment of SMA-like mice resulted in an increased level of full-length SMN protein, which resulted in a limited improvement of clinical symptoms (Chang et al, 2001).
Several small molecules, such as aclarubicin, sodium vanadate and valproic acid, also increased the level of correctly spliced SMN2 transcripts and proteins (Andreassi et al, 2001; Zhang et al, 2001; Brichta et al, 2003). Valproic acid also upregulated SMN2 gene expression and thus further contributed to this increase. Additional molecules such as EGCG and kinetin increased the level of correct IKAP spliced RNA and protein by splicing modulation in familial dysautonomia-derived cells carrying the IVS20+6 T → C splicing mutation (Anderson et al, 2003; Slaugenhaupt et al, 2004). Interestingly, in the case of EGCG, downregulation of hnRNP A2/B1 expression was found.
The small molecules used to modulate the splicing pattern are expected to affect the pattern of exons in the same or other genes, emphasizing the need to develop therapeutic approaches with specificity to the desired exon. One such approach is using antisense oligonucleotides designed to inhibit cryptic splicing. This resulted in a decrease in the level of aberrant CFTR transcripts containing the 84 bp exon (Friedman et al, 1999). In another approach, chimeric antisense oligonucleotides comprising two parts are designed: one is complementary to the aberrantly spliced exon and provides exon specificity, and the other contains binding motifs for recruitment of splicing factors to the mutation site (Cartegni & Krainer, 2003; Skordis et al, 2003). An increase in the binding of splicing factors by such oligonucleotides resulted in an increased level of SMN2 exon 7 and BRCA1 exon 18 inclusion. Another possible approach is identification of small molecules with specific effect.
In summary, the results presented in this study provide direct evidence that increasing the level of normal transcripts can restore protein function. This indicates that in cells carrying splicing mutations, the level of correctly spliced transcripts determines CFTR function and explains the previously found correlation between the level of correctly spliced CFTR transcripts and disease severity among patients and among organs of the same patient.
Methods
Transfection of splicing factors and NaBu administration. A total of 105 cells were plated for RNA analysis into six-well plates 24 h before transfection. A total of 104 cells were plated for functional analysis into 96-well plates 72 h before transfection. Transfection efficiency was optimized using variations in the amount of plasmid DNA and MAX-2 reagent (Eurogentec). A 1 and 0.15 μg portion of each plasmid was transfected into six- and 96-well plates, respectively. Cells were further grown for 48 h. Only experiments in which 50–60% of the cells were transfected were included in the analysis. NaBu was added for 4 h, 6 days after plating of cells. Experiments were repeated 3–7 times.
RNA extraction and RT–PCR analysis. Total RNA was purified using the ULTRASPEC RNA Kit (BIOTECX). Complementary DNA synthesis and splicing pattern analysis were performed as described previously (Rave-Harel et al, 1997; Nissim-Rafinia et al, 2000). Primers and PCR conditions are listed in supplementary Table 1 online.
CFTR functional analysis. CFTR function was measured by Cl− efflux using the Cl−-sensitive fluorescent indicator MQAE (Mansoura et al, 1999). For evaluation of CFTR activation, the difference between fluorescence intensity at the reading time (Ft) and at the beginning of the experiment (F0) was calculated.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor).
Supplementary Material
Supplementary Information
Acknowledgments
We thank A.R. Krainer, G. Akusjarvi, S. Stamm, B. Wirth, M. Brandeis, G. Dreyfuss and S.M. Berget for kindly providing expression vectors and antibodies, I. Cabantchik for the T84 and IB3 cells, J. Surowitz and D. Walstad for technical assistance and J. Olsen for producing the retroviral vectors. This research was supported by NACFF, ISF (grant number 490/03), and the Israeli Ministry of Health to B.K.
References
- Anderson SL, Qiu J, Rubin BY (2003) EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem Biophys Res Commun 310: 627–633 [DOI] [PubMed] [Google Scholar]
- Andreassi C et al. (2001) Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 10: 2841–2849 [DOI] [PubMed] [Google Scholar]
- Augarten A et al. (1993) Mild cystic fibrosis and normal or borderline sweat test in patients with the 3849+10 kb C → T mutation. Lancet 342: 25–26 [DOI] [PubMed] [Google Scholar]
- Aznarez I, Chan EM, Zielenski J, Blencowe BJ, Tsui LC (2003) Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum Mol Genet 12: 2031–2040 [DOI] [PubMed] [Google Scholar]
- Bienvenu T et al. (1996) Analysis of alternative splicing patterns in the cystic fibrosis transmembrane conductance regulator gene using mRNA derived from lymphoblastoid cells of cystic fibrosis patients. Eur J Hum Genet 4: 127–134 [DOI] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Brichta L et al. (2003) Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet 12: 2481–2489 [DOI] [PubMed] [Google Scholar]
- Buchner DA, Trudeau M, Meisler MH (2003) SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301: 967–969 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR (2003) Correction of disease-associated exon skipping by synthetic exonspecific activators. Nat Struct Biol 10: 120–125 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298 [DOI] [PubMed] [Google Scholar]
- Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H (2001) Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci USA 98: 9808–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K, Kole J, Cohn JA, Knowles MR, Silverman LM, Kole R (1999) Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J Biol Chem 274: 36193–36199 [DOI] [PubMed] [Google Scholar]
- Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH (1998) Differential SMN2 expression associated with SMA severity. Nat Genet 20: 230–231 [DOI] [PubMed] [Google Scholar]
- Highsmith WE et al. (1994) A novel mutation in the cystic fibrosis gene in patients with pulmonary disease but normal sweat chloride concentrations. N Engl J Med 331: 974–980 [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B (2000) Htra2-β1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc Natl Acad Sci USA 97: 9618–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem B et al. (1989) Identification of the cystic fibrosis gene: genetic analysis. Science 245: 1073–1080 [DOI] [PubMed] [Google Scholar]
- Kerem E et al. (1997) A cystic fibrosis transmembrane conductance regulator splice variant with partial penetrance associated with variable cystic fibrosis presentations. Am J Respir Crit Care Med 155: 1914–1920 [DOI] [PubMed] [Google Scholar]
- Mansoura MK, Biwersi J, Ashlock MA, Verkman AS (1999) Fluorescent chloride indicators to assess the efficacy of CFTR cDNA delivery. Hum Gene Ther 10: 861–875 [DOI] [PubMed] [Google Scholar]
- Nissim-Rafinia M, Kerem B (2002) Splicing regulation as a potential genetic modifier. Trends Genet 18: 123–127 [DOI] [PubMed] [Google Scholar]
- Nissim-Rafinia M, Chiba-Falek O, Sharon G, Boss A, Kerem B (2000) Cellular and viral splicing factors can modify the splicing pattern of CFTR transcripts carrying splicing mutations. Hum Mol Genet 9: 1771–1778 [DOI] [PubMed] [Google Scholar]
- Pagani F et al. (2000) Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J Biol Chem 275: 21041–21047 [DOI] [PubMed] [Google Scholar]
- Pagani F et al. (2003) New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum Mol Genet 12: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Ramalho AS, Beck S, Meyer M, Penque D, Cutting GR, Amaral MD (2002) Five percent of normal cystic fibrosis transmembrane conductance regulator mRNA ameliorates the severity of pulmonary disease in cystic fibrosis. Am J Respir Cell Mol Biol 27: 619–627 [DOI] [PubMed] [Google Scholar]
- Ramalho AS et al. (2003) Transcript analysis of the cystic fibrosis splicing mutation 1525−1G>A shows use of multiple alternative splicing sites and suggests a putative role of exonic splicing enhancers. J Med Genet 40: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rave-Harel N et al. (1997) The molecular basis of partial penetrance of splicing mutations in cystic fibrosis. Am J Hum Genet 60: 87–94 [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F (2003) Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci USA 100: 4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA et al. (2004) Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum Mol Genet 13: 429–436 [DOI] [PubMed] [Google Scholar]
- Zhang ML, Lorson CL, Androphy EJ, Zhou J (2001) An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: potential therapy of SMA. Gene Ther 8: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Zielenski J et al. (1993) Analysis of CFTR transcripts in nasal epithelial cells and lymphoblasts of a cystic fibrosis patient with 621+1G → T and 711+1G → T mutations. Hum Mol Genet 2: 683–687 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information