Abstract
Vacuolar H+-ATPase (V-ATPase) has a crucial role in the vacuolar system of eukaryotic cells. It provides most of the energy required for transport systems that utilize the proton-motive force that is generated by ATP hydrolysis. Some, but not all, of the V-ATPase subunits are homologous to those of F-ATPase and the nonhomologous subunits determine the unique features of V-ATPase. We determined the crystal structure of V-ATPase subunit C (Vma5p), which does not show any homology with F-ATPase subunits, at 1.75 Å resolution. The structural features suggest that subunit C functions as a flexible stator that holds together the catalytic and membrane sectors of the enzyme. A second crystal form that was solved at 2.9 Å resolution supports the flexible nature of subunit C. These structures provide a framework for exploring the unique mechanistic features of V-ATPases.
Keywords: V-ATPase, subunit C, 3D structure, mechanism, conformation
Introduction
Vacuolar H+-ATPase (V-ATPase) is functionally and structurally related to the mitochondrial and chloroplast F-ATPase. The most fundamental difference between F- and V-ATPases is a consequence of their unique function and origin. The main function of F-ATPases is to synthesize ATP at the expense of a proton-motive force generated by electron transport chains; the main function of V-ATPases in eukaryotic cells is to generate a proton-motive force at the expense of ATP and to cause limited acidification in the internal space (lumen) of several organelles of the vacuolar system (Stevens & Forgac, 1997; Nelson & Harvey, 1999; Nishi & Forgac, 2002). The origin of the eukaryotic F-ATPase is rooted in the eubacteria that evolved into chloroplasts and mitochondria, and because its assembly requires organellar gene products, it is confined to these organelles and shows no contact with the cytoplasm (Nelson, 1992). The origin of V-ATPase is related to the F-ATPase of archaebacteria, and evolved in eukaryotes concomitantly with the secretory pathway. In contrast to the F-ATPase, V-ATPase is vital to almost every eukaryotic cell, and except for yeast, its destruction causes lethality (Nelson & Harvey, 1999).
Both F- and V-ATPases are multisubunit protein complexes made up of distinct catalytic and membrane sectors. F-ATPase of Escherichia coli is composed of only eight subunits—five in the catalytic sector and three in the membrane sector—and the stoichiometry of its various subunits is 3α, 3β, 1γ, 1δ, 1ɛ, 1a, 2b and 10c (Jiang et al, 2001). The structural information that was obtained in the last decade on F-ATPase and its subunits shed light on one of the most intricate catalytic activities in nature (Abrahams et al, 1994; Stock et al, 1999; Gibbons et al, 2000). The crystal structure of bovine mitochondrial F1-ATPase showed an alternating arrangement of three αsubunits and three β-subunits around the central helical coiled-coil γ-subunit (Abrahams et al, 1994). The structure strongly supported the binding change mechanism, proposed by Boyer (1993), involving alterations at the nucleotide-binding sites of the catalytic βsubunits.
The intriguing question is how much of the structure, and consequently the mechanism of action, has been preserved in V-ATPases. In yeast, subunits A (Vma1p) and B (Vma2p) of the catalytic sector are assumed to function in similar if not identical fashion to F-ATPase subunits β and α, respectively. Subunits D (Vma8p) and F (Vma7p) and the proteolipids (Vma3p, Vma11p and Vma16p) are the best candidates for acting as the rotor of the enzyme. Subunits E and G (Vma4p and Vma10p) were postulated to function as stators holding the static parts of the enzyme that include subunits A, B and a (Nelson et al, 2002; Nishi & Forgac, 2002). Additional subunits are necessary for the assembly and/or function of V-ATPase, and in their absence yeast cells show V-ATPase null phenotype (Nelson & Harvey, 1999). Subunit d (Vma6p) is peripherally attached to the cytoplasmic face of the membrane sector (Stevens & Forgac, 1997). Recently, the crystal structure of subunit C of Thermus thermophilus V-ATPase was determined (Iwata et al, 2004). This subunit is not homologous to subunit C of eukaryotic V-ATPases and it shows low homology to V-ATPase subunit d (Nishi & Forgac, 2002). Subunit H (Vma13p) is required for the activity of the enzyme, but in its absence all the other subunits could be assembled. This subunit is the only one for which a high-resolution structure is available (Sagermann et al, 2001). Subunit C (Vma5p) is required for the proper assembly of V-ATPase, and may not only function as part of the stator but also show actin-binding properties (Beltrán et al, 1992; Curtis et al, 2002; Vitavska et al, 2003). Vma5p is the only subunit that reversibly leaves the enzyme in glucose deprivation, causing the catalytic subcomplex to detach from the membrane sector (Curtis et al, 2002). We sought to understand the interaction between Vma5p and other V-ATPase subunits and its possible role in directing the enzyme to its target membranes by interaction with the cytoskeleton. To this end, we determined the X-ray crystal structure of V-ATPase subunit C from yeast cells, and describe here a model at 1.75 Å resolution.
Structure of Vma5p at 1.75 Å resolution
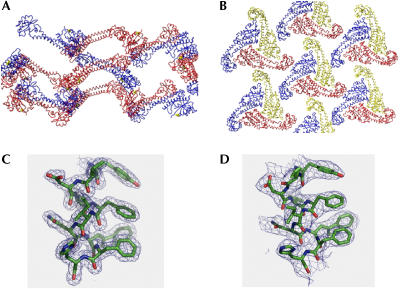
Data collection and phasing statistics, the refinement statistics and the model stereochemistry of the hanging-drop crystals have been reported (Drory et al, 2004; see supplementary information online). The two different crystal packings and the quality of the electron density maps of the two forms at the ‘foot' region are shown in Fig 1. In the hanging-drop crystals, the expressed protein crystallized as a head-to-tail dimer in the crystal lattice (Fig 1A). In sitting drops, Vma5p was crystallized as a monomer at a distinct conformation (Fig 1B). Fig 1C shows the experimental electron density map of a representative region of the hanging-drop crystal. The structure is resolved at 1.75 Å (Rcryst=0.21 and Rfree=0.23) and the fit to the electron densities is very good along the resolved polypeptide chains. Fig 1D shows the experimental electron density map of the same region of the sitting-drop crystal, with a lesser quality of the electron density map at 2.9 Å resolution. Despite the relatively low quality of this solution (Rcryst=0.29 and Rfree=0.35), the distinct conformation of the Vma5p in the sitting-drop crystal is quite apparent (Fig 1C,D).
Figure 1.
Crystal packing and electron density maps of Vma5p. (A) Crystal packing of the hanging-drop crystal, which was solved at 1.75 Å resolution. The protein monomers are in red and blue. A tartrate molecule is in yellow. (B) Crystal packing of the sitting-drop crystal, which was solved at 2.9 Å resolution. The protein monomers are in red, blue and yellow. (C) 2∣Fo∣−∣Fc∣ electron density map of the hanging-drop structure contoured at 1.5σ of a helical domain Tyr 296 to His 305. (D) 2∣Fo∣−∣Fc∣ electron density map of the sitting-drop structure contoured at 1.0σ of the same domain.
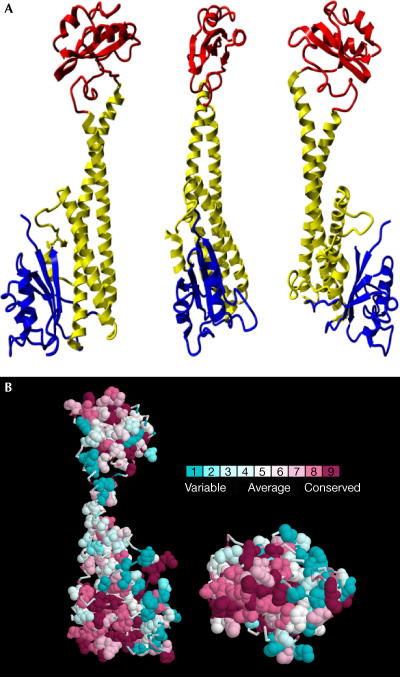
The expressed Vma5p is composed of 412 amino acids, 392 of which are of the native protein and 20 of which originated from the plasmid and the 6-histidine tag. In the structure depicted in Fig 2A (at 1.75 Å resolution), we could determine the coordinates of 364 amino acids and one tartrate molecule. The first 24 N-terminal amino acids and 24 amino acids in the solvent-exposed loop spanning amino acids 357–382 were disordered and did not appear in the structure. Thus, only the first four amino acids at the N-terminus of the reading frame are invisible. The protein is elongated and spans 103 Å from the upper amino acid (Pro 219) to the lowest amino acid (Pro 322; Fig 2A).
Figure 2.
Structure of Vma5p at 1.75 Å resolution. (A) Ribbon diagram of the Vma5p structure. The ‘head' domain is in red, the ‘neck' domain in yellow and the ‘foot' domain in blue. (B) Hydrophobic conserved patch in the base of the ‘neck' and ‘foot' domains. Aromatic and hydrophobic side chains (Ala, Leu, Val, Ile, Met, Pro, Phe, Trp and His) are shown in space-filling and coloured by conservation (Glaser et al, 2003). At the left, the orientation of the protein is as in the right representation in (A) and the representation at the right is tilted backward by 90°.
The structure of Vma5p consists of three distinct domains. (1) An upper globular domain (‘head') assembled by a polypeptide spans from amino acids Lys 166 to Ser 263. It is composed of four antiparallel β-sheets and two α-helices. (2) An elongated ‘neck' consisting of a helix bundle is held by conserved amino acids that are connected by salt bridges—Lys 165 with Glu 273, Asp 72 with Arg 310—as well as prolines and glycines at the end of the helices and hydrophobic interactions mainly at the base of the three-helix bundle (Fig 2B). (3) A lower globular ‘foot' composed of the N- and C-termini of the protein. This domain shows similar structural features to the ‘head', consisting of four anti-parallel βsheets with two intertwining α-helices. Even though there is no sequence similarity between the ‘head' and ‘foot' domains, they could be superimposed with an r.m.s. difference of 1.67 Å (61 Cα atoms). The overall structure is similar to the low-resolution small-angle X-ray scattering solution structure, which was recently published (Armbrüster et al, 2004). The assertion that subunits C and H share similar structure does not seem to hold up at high resolution.
Fitting into an EM structure of V-ATPase
The published low-resolution structures that are available from electron microscopy and single-particle averaging of mammalian and yeast V-ATPases showed a similar general structure to that of F-ATPase (Wilkens et al, 1999; Harrison et al, 2003; Zhang et al, 2003). It seems that, like F-ATPase, V-ATPase is composed of catalytic and membrane sectors that are held by a rotating shaft in the middle and a stator at the side of the enzyme. The structure of subunit C (Vma5p) that was solved in this work fits well with the apparent structure of the stator in the electron microscopy structure of yeast V-ATPase (Boekema et al, 1997; Harrison et al, 2003; Lolkema et al, 2003). Superimposing the Vma5p structure on the published electron microscopy structure (Wilkens et al, 1999, 2004; Harrison et al, 2003; Zhang et al, 2003) showed that the general shape and the structural properties of Vma5p fulfil the requirements of the stator.
We propose that the ‘head' domain is bound to the catalytic part of the enzyme, whereas the base of the ‘neck' and ‘foot' domains is bound to one of the membrane subunits. This notion is supported by the observation that V-ATPases are cold-sensitive enzymes (Moriyama & Nelson, 1989) and that, following cold treatment of the purified enzyme or glucose deprivation of yeast cells, the catalytic sector detaches from the membrane together with an Vma5p (Beltrán et al, 1992; Curtis et al, 2002). Fig 2B shows a molecular surface map of the lower face of the ‘foot' and ‘neck' domains, which presumably interacts with the membrane sector. An apparent hydrophobic surface composed of conserved amino acids is situated in a strategic position. Hydrophobic interactions are weakened at a low temperature, and this property may contribute to the cold-induced detachment of the membrane sector. In a recent publication, Wilkens et al (2004) suggested the location of subunit C together with an unidentified moiety as a second stator. Our structure of Vma5p suggests that subunit C itself made up these two identities.
Most importantly, the flexible structure of the ‘neck' domain fits the bill of a movable ratchet that functions in the conversion of the mechanical torque generated by the ATPase activity into proton pumping by the c-ring turbine (Junge et al, 2001).
Mechanistic implications of Vma5p structure
Models for rotation of the stalk have been proposed where the ring of c subunits rotates past a fixed subunit a in F-ATPases (Junge, 1999; Junge et al, 2001), and a similar mechanism was proposed to operate in V-ATPases (Perzov et al, 2001; Nelson et al, 2002; Hirata et al, 2003; Imamura et al, 2003). The structure of F-ATPase that was assembled from the crystal structure of partial complexes of mitochondrial F-ATPase (Abrahams et al, 1994; Stock et al, 1999; Gibbons et al, 2000; Kagawa et al, 2004) strongly supports the proposed mechanism. Thus, a complete cycle of the rotor (subunits c, γ and ɛ) yields three ATP molecules. The structure of the central stalk of an intact F1 domain has recently been determined at a high resolution (Gibbons et al, 2000; Kagawa et al, 2004). The structure of yeast F1 attached to a ring of ten c subunits is also known at 3.9 Å resolution (Stock et al, 1999). The close contact between subunits γ and ɛ and the c ring supports the idea that this central stalk and the c ring form the rotary ensemble of the ATPase motor. The reverse reaction of ATP-dependent proton uptake involves the hydrolysis of the three ATP molecules that cause a full cycle of the rotor.
Recent structural data from yeast (Stock et al, 1999) and chloroplast (Seelert et al, 2000) indicate that there are 10 and 14 c subunits per complex, respectively. Recently, the structure of the membrane domain of the Na+-motive V-ATPase from Enterococcus hirae was solved and was shown to be composed of heptameric subunit K that is equivalent to the c subunit of eukaryotic V-ATPases (Murata et al, 2003). Therefore, hydrolysis of a single ATP molecule will result in the pumping of 3.3 protons across the mitochondrial membrane, 4.7 protons across the chloroplast membrane and 2.3 Na+ across the E. hirae membrane. It was proposed that the coupling between the unmatched quantal values is mediated by the torque generated at the flexible stator of the respective enzymes (Junge et al, 2001). Numerous biochemical and molecular biology experiments showed that the stator of F-ATPases is composed of a dimer of the b subunit of Fo (Grabar & Cain, 2003). The structure of the stator could be detected by electron microscopy (Rubinstein et al, 2003). The two parallel helices, formed by the subunit b dimer, provide the flexible structure that may function in the translation of gradual torque into a 120° quantal jump, and vice versa. V-ATPase contains subunit G (Vma10p), which is homologous to subunit b of F-ATPases (Supekova et al, 1995). However, in V-ATPases, it lacks the membranespanning region, and it was assumed that it may fulfil a similar function to the b subunit of F-ATPase by binding to a membrane subunit at one end and to a catalytic domain at the other. We propose that the unique properties of V-ATPases require a new structure to fulfil the stator function, and that subunit C replaces the b subunit for this function.
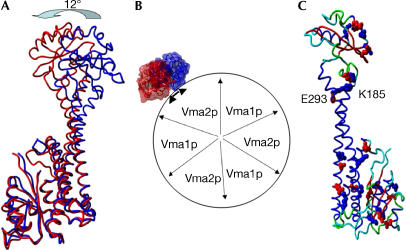
The structure of Vma5p reveals two slightly twined, long α-helices that provide flexibility and enable movement of the ‘head' in relation to the ‘foot' domain (Fig 2A). Indeed, the structure that was solved from the sitting-drop crystals shows a remarkable movement of the ‘head' domain in relation to the ‘foot' domain (Fig 3A,B). Whereas minor structural changes were detected in the ‘foot' domain, most of the structural changes took place in the middle of the ‘neck' and in the ‘head' domains. The free movement of about 12° to both sides may provide the flexibility required for the smooth operation of the ATP-dependent proton transport by V-ATPase.
Figure 3.
Elasticity of Vma5p. (A) Superposition of the two conformations of Vma5p. The movement of the ‘head' is indicated. The 1.75 Å structure is in red and the 2.9 Å structure in blue. (B) Top view of possible interaction between the catalytic sector (modelled as F-ATPase) with subunit C in the two conformations. (C) Salt bridges in the high-resolution structure of Vma5p. The protein is coloured according to the secondary structures. Blue, α-helix; red, βsheet; green, turns; cyan, loops. Negatively charged amino acids are in red, and positively charged in blue. The only salt bridge that connects the two long α-helices is indicated.
Evolutionary forces and structural constraints
The most recently published low-resolution structures of V-ATPases suggested the presence of two stators. One of them contains E and G (and probably parts of subunits a and H), and the other one contains subunit C together with an unidentified moiety (Wilkens et al, 2004). The fabric of the first stator contained a conserved subunit G that is homologous to the b subunit of F-ATPase and novel subunits that were added after the V-ATPases diverged from the F-ATPases. We propose that a second flexible stator was added during the evolution of eukaryotic cells to fulfil the requirements of the highly adjustable proton pumping system that functions differentially in numerous organelles and various membranes.
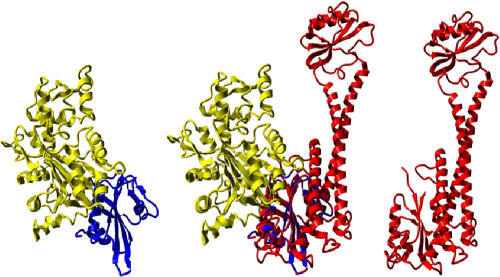
The cellular location of V-ATPase that faces the cytoplasm dictated several of its unique properties. As the organization of internal membranes in eukaryotic cells is facilitated by interactions with the cytoskeleton, it was not surprising that V-ATPase, which constitutes a major structure in specific organelles, directly interacts with the cytoskeleton. Both subunit B and subunit C were shown to have the property of actin binding in mammalian and insect V-ATPases, respectively (Holliday et al, 2000; Vitavska et al, 2003). A structural search for proteins that bind actin showed that gelsolin contains similar structural features of the ‘head' and ‘foot' of Vma5p. Fig 4 shows the published structure of gelsolin–actin complex (Vorobiev et al, 2003), and the fit of gelsolin and its complex with actin into the structure of Vma5p. It is apparent that gelsolin fits very well with the ‘foot' and/or the structurally similar ‘head' domains. The mode of precise interaction between Vma5p and F-actin may be determined in the future by their co-crystallization.
Figure 4.
The proposed actin-binding site on the foot of Vma5p. The complex between yeast actin (yellow) and human gelsolin (blue) is fitted onto the ‘foot' domain of Vma5p (red). The structure of actin–gelsolin complex is illustrated according to PDB 1YAG.
Methods
The reading frame of VMA5 gene from Saccharomyces cerevisiae was amplified by PCR and cloned into the Nde1 and Xho1 restriction sites of a pET28a+ (Novagen Inc., Madison, WI, USA) expression vector to give a 6-His tag at its C-terminus. The protein was expressed in E. coli C43 or C41 cells (a generous gift from John E. Walker). The protein was purified in a four-step procedure and crystallized as previously described (Drory et al, 2004). Two different crystal forms were obtained by the hanging- and sitting-drop methods after 1 week, and diffracted to 1.75 and 2.9 Å, respectively. The crystals that were obtained by hanging drops belong to space group P43212 (unit-cell parameters were a=b=62.54, c=337.37 Å, α=β=γ=90°) with one molecule per asymmetric unit. The phases were solved by SIRAS with Lu(O2C2H3), which gave one site (Drory et al, 2004). The crystals that were obtained by the sitting-drop method belong to space group H3 (unit-cell parameters were a=b=132.23, c=34.18 Å, α=β=90°, γ=120°) with one molecule per asymmetric unit. The structure was solved by molecular replacement, using the ‘foot' domain of the P43212 crystals as a search model and manual rebuilding with O of the upper part of the ‘neck' and the ‘head' domains (Drory et al, 2004).
Atomic coordinates are deposited in the Protein Data Bank under accession number 1U7L.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v5/n12/extref/7400294s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
This project has been funded by the BMBF (German Federal Ministry of Education and Research) and supported by BMBF's international bureau at the DLR (German Aerospace Center).
References
- Abrahams JP, Leslie AGW, Lutter R, Walker JE (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370: 621–628 [DOI] [PubMed] [Google Scholar]
- Armbrüster A, Svergun DI, Coskun U, Juliano S, Bailer SM, Grüber G (2004) Structural analysis of the stalk subunit Vma5p of the yeast V-ATPase in solution. FEBS Lett 570: 119–125 [DOI] [PubMed] [Google Scholar]
- Beltrán C, Kopecky J, Pan Y-CE, Nelson H, Nelson N (1992) Cloning and mutational analysis of the gene encoding subunit C of yeast V-ATPase. J Biol Chem 267: 774–779 [PubMed] [Google Scholar]
- Boekema EJ, Ubbink-Kok T, Lolkema JS, Brisson A, Konings WN (1997) Visualization of a peripheral stalk in V-type ATPase: evidence for the stator structure essential to rotational catalysis. Proc Natl Acad Sci USA 94: 14291–14293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer PD (1993) The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim Biophys Acta 1140: 215–250 [DOI] [PubMed] [Google Scholar]
- Curtis KK, Francis SA, Oluwatosin Y, Kane PM (2002) Mutational analysis of the subunit C (Vma5p) of the yeast vacuolar H+-ATPase. J Biol Chem 277: 8979–8988 [DOI] [PubMed] [Google Scholar]
- Drory O, Mor A, Frolow F, Nelson N (2004) Expression, crystallization and phasing of vacuolar H+-ATPase subunit C (Vma5p) of Saccharomyces cerevisiae. Acta Crystallogr D 60: 1906–1909 [DOI] [PubMed] [Google Scholar]
- Gibbons C, Montgomery MG, Leslie AGW, Walker JE (2000) The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat Struct Biol 7: 1055–1061 [DOI] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bell RE, Bechorshental D, Martz E, Ben-Tal N (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19: 163–164 [DOI] [PubMed] [Google Scholar]
- Grabar TB, Cain BD (2003) Integration of b subunits of unequal lengths into F1F0-ATP synthase. J Biol Chem 278: 34751–34756 [DOI] [PubMed] [Google Scholar]
- Harrison M, Durose L, Song CF, Barratt E, Trinick J, Jones R, Findlay JB (2003) Structure and function of the vacuolar H+-ATPase: moving from low-resolution models to high-resolution structures. J Bioenerg Biomembr 35: 337–345 [DOI] [PubMed] [Google Scholar]
- Hirata T, Iwamoto-Kihara A, Sun-Wada GH, Okajima T, Wada Y, Futai M (2003) Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J Biol Chem 278: 23714–23719 [DOI] [PubMed] [Google Scholar]
- Holliday LS, Lu M, Lee BS, Nelson RD, Solivan S, Zhang L, Gluck SL (2000) The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J Biol Chem 275: 32331–32337 [DOI] [PubMed] [Google Scholar]
- Imamura H, Nakano M, Noji H, Muneyuki E, Ohkuma S, Yoshida M, Yokoyama K (2003) Evidence for rotation of V1-ATPase. Proc Natl Acad Sci USA 100: 2312–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M et al. (2004) Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc Natl Acad Sci USA 101: 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hermolin J, Fillingame RH (2001) The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc Natl Acad Sci USA 98: 4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge W (1999) ATP synthase and other motor proteins. Proc Natl Acad Sci USA 96: 4735–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge W, Panke O, Cherepanov DA, Gumbiowski K, Muller M, Engelbrecht S (2001) Intersubunit rotation and elastic power transmission in F0F1-ATPase. FEBS Lett 504: 152–160 [DOI] [PubMed] [Google Scholar]
- Kagawa R, Montgomery MG, Braig K, Leslie AG, Walker JE (2004) The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J 23: 2734–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolkema JS, Chaban Y, Boekema EJ (2003) Subunit composition, structure, and distribution of bacterial V-type ATPases. J Bioenerg Biomembr 35: 323–335 [DOI] [PubMed] [Google Scholar]
- Moriyama Y, Nelson N (1989) Cold inactivation of vacuolar H+-ATPases. J Biol Chem 264: 3577–3582 [PubMed] [Google Scholar]
- Murata T, Arechaga I, Fearnley IM, Kakinuma Y, Yamato I, Walker JE (2003) The membrane domain of the Na+-motive V-ATPase from Enterococcus hirae contains a heptameric rotor. J Biol Chem 278: 21162–21167 [DOI] [PubMed] [Google Scholar]
- Nelson N (1992) Evolution of organellar proton-ATPases. Biochim Biophys Acta 1100: 109–124 [DOI] [PubMed] [Google Scholar]
- Nelson N, Harvey WR (1999) Vacuolar and plasma membrane V-ATPases. Phys Rev 79: 361–385 [DOI] [PubMed] [Google Scholar]
- Nelson N, Sacher A, Nelson H (2002) The significance of molecular slips in transport systems. Nat Rev Mol Cell Biol 3: 876–881 [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M (2002) The vacuolar (H+)-ATPases—nature's most versatile proton pumps. Nat Rev Mol Cell Biol 3: 94–103 [DOI] [PubMed] [Google Scholar]
- Perzov N, Padler-Karavani V, Nelson H, Nelson N (2001) Features of V-ATPases that distinguish them from F-ATPases. FEBS Lett 504: 223–228 [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Walker JE, Henderson R (2003) Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J 22: 6182–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagermann M, Stevens TH, Matthews BW (2001) Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98: 7134–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelert H, Poetsch A, Dencher NA, Engel A, Stahlberg H, Muller DJ (2000) Structural biology: proton-powered turbine of a plant motor. Nature 405: 418–419 [DOI] [PubMed] [Google Scholar]
- Stevens TH, Forgac M (1997) Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Dev Biol 13: 779–808 [DOI] [PubMed] [Google Scholar]
- Stock D, Leslie AGW, Walker JE (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286: 36–40 [DOI] [PubMed] [Google Scholar]
- Supekova L, Supek F, Nelson N (1995) The Saccharomyces cerevisiae VMA10 is an intron-containing gene encoding a novel 13-kDa subunit of vacuolar H+-ATPase. J Biol Chem 270: 13726–13732 [DOI] [PubMed] [Google Scholar]
- Vitavska O, Wieczorek H, Merzendorfer H (2003) A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J Biol Chem 278: 18499–18505 [DOI] [PubMed] [Google Scholar]
- Vorobiev S, Strokopytov B, Drubin DG, Frieden C, Ono S, Condeelis J, Rubenstein PA, Almo SC (2003) The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA 100: 5760–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens S, Vasilyeva E, Forgac M (1999) Structure of the vacuolar ATPase by electron microscopy. J Biol Chem 274: 31804–31810 [DOI] [PubMed] [Google Scholar]
- Wilkens S, Takao I, Forgac M (2004) Three-dimensional structure of the vacuolar ATPase—localization of subunit H by difference imaging and chemical cross-linking. J Biol Chem 279: 41942–41949 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Charsky C, Kane PM, Wilkens S (2003) Yeast V1-ATPase: affinity purification and structural features by electron microscopy. J Biol Chem 278: 47299–47306 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information