Abstract
The importance of coupling the process of endocytosis to factors regulating actin dynamics has been clearly demonstrated in yeast, and many proteins involved in these mechanisms have been identified and characterized. Here we demonstrate the importance of two additional cortical components, Ysc84p and Lsb5p, which together are essential for the organization of the actin cytoskeleton and for fluid phase endocytosis. Both Ysc84p and Lsb5p were identified through two-hybrid screens with different domains of the adaptor protein Sla1p. Ysc84p colocalizes with cortical actin and requires the presence of an intact actin cytoskeleton for its cortical localization. Ycl034w/Lsb5p localizes to the cell cortex but does not colocalize with actin. The Lsb5 protein contains putative VHS and GAT domains as well as an NPF motif, which are all domains characteristic of proteins involved in membrane trafficking. Deletion of either gene alone does not confer any dramatic phenotype on cells. However, deletion of both genes is lethal at elevated temperatures. Furthermore, at all temperatures this double mutant has depolarized actin and an almost undetectable level of fluid phase endocytosis. Our data demonstrate that Ysc84p and Lsb5p are important components of complexes involved in overlapping pathways coupling endocytosis with the actin cytoskeleton in yeast.
INTRODUCTION
There is an increasing body of evidence that suggests a requirement for a dynamic actin cytoskeleton to facilitate the process of endocytosis. Initial evidence arising from studies in yeast demonstrated that strains mutated in actin-binding proteins caused concomitant defects in cortical actin organization and in endocytosis (Raths et al., 1993; Benedetti et al., 1994 and reviewed in Geli and Riezman, 1998). However, it has not always been clear from such studies which proteins are playing a direct role in coupling actin to the endocytic machinery and which proteins have simply disrupted actin, thereby having an indirect effect on endocytosis. Studies on mammalian cells have generally been less conclusive for a role for the actin cytoskeleton in endocytosis, possibly because of further levels of complexity in the processes (Qualmann et al., 2000; Schafer, 2002). Here too, many studies are now indicating the importance of cortical actin in facilitating the endocytic process. Evidence includes an increase in the number of clathrin-coated pits at the cell surface in response to disruption of actin using cytochalasin-D (Shurety et al., 1996) and in the demonstration of a number of actin-binding proteins localizing to clathrin-coated pits (Engqvist-Goldstein et al., 2001; Kessels et al., 2001). Furthermore, reports of association of dynamic actin with phagosomes (May et al., 2000) and of dynamin association with actin tails on moving vesicles (Lee and De Camilli, 2002) have suggested a mechanism for driving movement of vesicles from the plasma membrane into the cell.
A potential reason for the lack of a clearly defined link between actin and endocytosis is that the coupling of the processes is highly dynamic and may involve interactions between distinct protein complexes, depending on the nature of the cargo being internalized. In addition, studies from this laboratory and others have demonstrated that in yeast there is considerable redundancy among the proteins involved in organizing the cortical actin cytoskeleton and linking it to the endocytic machinery (Wesp et al., 1997; Wendland and Emr, 1998; Ayscough et al., 1999; Tang et al., reviewed in Goode and Rodal, 2001). It seems likely that the situation in higher eukaryotes will prove to be even more complex. However, based on research in both yeast and higher organisms, a number of models have been suggested for the role of actin in the endocytic process. Possibilities include cortical actin being involved in localizing and stabilizing an endocytic complex, to a more active role in driving invagination, scission, and possibly movement of the vesicle from the plasma membrane (reviewed in Qualmann et al., 2000). To date evidence does not strongly point to any of these individual models, and it is possible that in some cell types there is more than one requirement for actin that will add further to the complexity.
One protein that we have shown to be important both in regulating actin dynamics as well as being required for wild-type levels of endocytosis is Sla1p. Sla1p is a multifunctional protein required for cortical actin patch structure, dynamics, and organization in budding yeast (Holtzman et al., 1993; Ayscough et al., 1999; Warren et al., 2002). Sla1p has three SH3 domains in its N-terminal third and a C-terminal domain comprised of multiple repeats that interact with the N-terminal EH-domain of End3p and the LR1 domain of Pan1p (Tang et al., 2000). The interaction with End3p is required for Sla1p localization to the plasma membrane (Warren et al., 2002). End3p and Pan1p are EH-domain containing proteins shown to be required for endocytosis and normal actin organization in yeast (Raths et al., 1993; Benedetti et al., 1994; Tang et al., 1997; Wendland and Emr, 1998; Tang et al., 2000). EH-domain–containing proteins in mammalian cells have also been shown to localize to clathrin-coated pits and be involved in endocytic events (Tebar et al., 1996). Despite localizing with the endocytic machinery, the most notable phenotypes in sla1 null cells are on the cortical actin cytoskeleton. In this regard we have demonstrated interactions of Sla1p with a subpopulation of actin-binding proteins, Abp1p and Las17p/Bee1p, both of which are activators of Arp2/3 in yeast (Warren et al., 2002). Previously, we proposed that Sla1p is localized to the endocytic machinery in order to restrict its effect on actin dynamics to the appropriate regions of the cell (Warren et al., 2002). In the studies we report here, we have identified two proteins that interact with Sla1p. One of these localizes to the cortical actin cytoskeleton and the second localizes to the cell cortex in an actin-independent manner. We demonstrate that these proteins, Ysc84p and Lsb5p, are important components of complexes involved in functionally overlapping pathways coupling endocytosis with the actin cytoskeleton in yeast.
MATERIALS AND METHODS
Materials
Chemicals used were obtained from BDH/Merck (Poole, Dorset, United Kingdom) unless otherwise stated. Media was from Melford Laboratories (yeast extract, peptone, agar; Ipswich, United Kingdom) or Sigma (minimal synthetic medium and amino acids; Poole, Dorset, United Kingdom). Latrunculin-A was a gift from Phil Crews (UC Santa Cruz, Santa Cruz, CA).
Yeast Strains and Cell Growth
The yeast strains used in this study are listed in Table 1. Plasmids and the oligonucleotides used to generate PCR products for direct deletions and epitope tagging of the genomic copies of genes are given in Tables 2 and 3, respectively. All tagging and deletions were generated using the plasmids described by Longtine and colleagues (1998).Unless otherwise stated yeast cells were grown with rotary shaking at 29°C in liquid YPD medium (1% yeast extract, 2% Bacto-peptone, 2% glucose supplemented with 40 μg/ml adenine). Tetrads were dissected using a Singer Instruments MSM Manual micromanipulator (Watchet, Somerset, UK). Transformations were performed using lithium acetate as previously described (Kaiser et al., 1994). Cell growth and generation times were assessed using a CASY Model TT Schärfe (Reutlingen, Germany) cell counter according to the manufacturer's instructions.
Table 1.
Yeast strains used in this study
| Strain name | Genotype | Origin/Reference |
|---|---|---|
| PJ269-2A | MATa trp1-901, leu2-3,112, ura3-52, his3Δ200, gal4Δ, gal80Δ, LYS2∷GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2 | James et al. (1996) |
| KAY302 | MATα trp1-1, leu 2-3,112, lys2-801, his3-Δ200, ura 3-52 | Warren et al. (2002) |
| KAY355 | KAY302 + integrated SLA1-3xHA∷TRP1 | This study |
| KAY564 | KAY302 +YSC84-7xalanine-13xmyc∷TRP1 | This study |
| KAY525 | KAY302 + integrated GFP-Ysc84∷TRP1 | This study |
| KAY537 | KAY302 + integrated GFP-Lsb3∷HIS3 | This study |
| KAY397 | KAY302 + integrated Sla1-GFP∷HIS3 | Warren et al. (2002) |
| KAY300 | MATa trp1-1, leu 2-3,112, his3-Δ200, ura 3-52, Δsla1∷URA3 | Warren et al. (2002) |
| KAY527 | KAY300 + integrated GFP-Ysc84 | This study |
| KAY126 | MATa leu 2-3,112, his3-Δ200, ura 3-52, Δabp1∷LEU2 | Drubin (Berkeley) |
| KAY544 | KAY126 + integrated GFP-Ysc84∷HIS3 | This study |
| KAY473 | MATα, his3Δ1, leu2Δ1, ura3Δ, lys2Δ, Δlas17∷KanMx | Research Genetics |
| KAY547 | KAY473 + integrated GFP-Ysc84∷HIS3 | This study |
| KAY514 | KAY302 + integrated LSB5-13xmyc∷TRP1 | This study |
| KAY510 | MATa, his3Δ200, leu2-3,112, ura3-52, trp1-1, lys2Δ, Δysc84∷HIS3 | This study |
| KAY515 | MATα, his3Δ1, leu2Δ1, ura3Δ, lys2Δ, Δlsb5∷KanMx | Research Genetics |
| KAY516 | MATα, his3Δ, leu2Δ, ura3Δ, lys2Δ, Δlsb5∷KanMx, Δysc84∷HIS3 | This study |
| KAY447 | MATα, his3Δ1, leu2Δ1, ura3Δ, lys2Δ | Research Genetics |
| KAY601 | KAY300 + integrated Lsb5-13xmyc∷HIS3 | This study |
| KAY604 | KAY447 + plasmid p416MET25 | This study |
| KAY605 | KAY447 + plasmid p416MET25-SH3yl-1 (pKA269) | This study |
Table 2.
Plasmids used in this study
| Plasmid | Description | Origin/reference |
|---|---|---|
| pGBDU-C1 | Yeast 2 hybrid binding domain plasmid | James et al. (1996) |
| pGAD-C1 | Yeast 2 hybrid activation domain plasmid | James et al. (1996) |
| pKA51 | pRS313 +SLA1 | Ayscough et al. (1999) |
| pKA116 | pRS313 +SLA1ΔSH3#3 | Ayscough et al. (1999) |
| pKA237 | pGBDU-C1 + SLA1(118-511). Sequence 118-511 cut using EcoR1 from pKA51 |
This study |
| pKA238 | pGBDU-C1 + SLA1(118-511) with sequence corresponding to SH3#3 excised. Sequence inserted was cut using EcoR1 from pKA116 |
This study |
| pKA251 | pGAD-C1 + Ysc84ΔSH3. Isolated activation plasmid carrying Ysc84 211-468 was mutated at residue 416 to create a stop codon prior to the C-terminal SH3 domain. | This study |
| pKA38 | pGEX1-Sla1(118-511). Sequence 118-511 cut using EcoR1 from pKA51 and cloned into pGEX1 | This study |
| pKA272 | pGAD-C1 + Lsb3(86-451). Lsb3 from nucleotide 258 was cloned following PCR from genomic DNA. | This study |
| pKA260 | Nucleotides 1461-2192 of SLA1 corresponding to regions HD1+HD2 were amplified by PCR using pKA51 as the template. Primers were designed with BamH1/Pst1 restriction sites flanking the region of interest. The PCR product was then cloned into pGBDU-C1. | This study |
| pKA267 | pGEX2T + Lsb5. LSB5 was cloned from genomic yeast DNA using oligos oKA242 and 244 with flanking BamH1 and BglII restriction sites. | This study |
| pME18S-FL | Plasmid carrying cDNA of SH3yl-1 (human gene) | Hata (Tokyo) |
| p416MET25 | MET promoter plasmid | Mumberg et al. (1994) |
| pKA269 | SH3yl-1 was excised from pME18S-FL using Xho1 and cloned into the appropriate site in p416MET25 | This study |
Table 3.
Oligonucleotides used in this study
| Oligo | Sequence | Description |
|---|---|---|
| oKA165 | ACTTCACAGTAAAATTTTCAAGAGTAGTGGTAATATCAAAT CCAAAATCCAAAAGAATTCGAGCTCGTTTAAAC |
Ysc84 tagging N-term. (5′) |
| oKA166 | AACATACTTGGTCTCGCTTTTCAAGCTTCGAGGAATTGGAT TATTGCTACCCATTTTGTATAGTTCATCCATGC |
Ysc84 tagging N-term. (3′) |
| oKA216 | GGACTAACGGAAAAGAAGGAATATTCCCTGCAAACTACGTT AGAGTTTCTGCTGCAGCCGCTGCAGCTGCACGGATCCCCGGGTTAATTAA |
Ysc84 tagging C-term. (5′) + 7xala linker |
| oKA198 | GGCTTTGGATCACGGGTGGCAAAGAAATCAAAAACAAAAGC TGTATCACACGCAGAATCCGAGCTCGTTTAAAC |
Lsb3 N-term tagging (5′) |
| oKA199 | AACATACTTTGTCTCACTCTTTAAACTCCTTGGAATAGGAT TGTTAATACCCATTTTGTATAGTTCATCCATGC |
Lsb3 N-term tagging (3′) |
| oKA132 | CTCAACAATCAAGGCAAGCCAACATATTCAATGCTACTGCA TCAAATCCGTTTGGATTCGCTGCAGCCGCTGCAGCTGCACG GATCCCCGGGTTAATTAA |
Sla1 tagging C-term. +7xala linker (5′) |
| oKA133 | ACGAAACTATTTCATATATCGTGTTTTAGTTATTATCCTAT AAAATCTTAAAATACATTAATCGAATTCGAGCTCGTTTAAAC |
Sla1 tagging C-term (3′) |
| oKA224 | ACGATCTCAAACCCTTTCGGTGATCATAACAAAGCTGCAGC CGCTGCAGCTGCACGGATCCCCGGGTTAATTAA |
Lsb5 tagging C-term (5′) |
| oKA225 | GGATTAAATGTACTATATATATATATATATATATGTGTGTA TGCATACGTACATACTGAATTCGAGCTCGTTTAAAC |
Lsb5 tagging C-term (3′) |
| oKA242 | CGCGGATCCATGGGGTTTCTTTCGGATC | LSB5 cloning 5′ |
| oKA244 | GGCAGATCTGGGTGGAGATATGTTGTGC | LSB5 cloning 3′ |
| oKA136 | CACAGTAAAATTTTCAAGAGTAGTGGTAATATCAAATCCAA AATCCAAAACGGATCCCCGGGTTAATTAA |
Δysc84 (5′) |
| oKA137 | TCTAAAAAAAAATTCTATATATAGGAACGAGACATATGGAG GACGATATTGAATTCGAGCTCGTTTAAAC |
Δysc84 (3′) and C-term tagging |
Two-hybrid Screens
The yeast two-hybrid screens used bait and activation plasmids, and a yeast strain pJ69–2A was designed and constructed by Philip James (James et al., 1996). The regions of Sla1p tested in the two-hybrid screens were generated as described in Table 2. The bait plasmids were checked for self-activation before the library screen was carried out. For the screen, 2 μg of bait plasmid was transformed into the two-hybrid yeast strain pJ69–2A. Bait-transformed cells, 2 × 109, were grown up in liquid media lacking uracil and were transformed with 20 μg genomic Saccharomyces cerevisiae library DNA from each reading frame (library was a gift from Francis Barr, Munich, Germany). Colonies that grew within 7 d were picked from plates lacking histidine, uracil, and leucine. After restreaking and selection for the inability to grow on media lacking adenine and containing 5-fluororotic-acid (5-FOA), plasmids were extracted from remaining strains. The plasmids were retransformed into pJ69–2A with the bait plasmid to ensure that activation of reporter genes still occurred. The plasmids were sequenced to identify the region responsible for the two-hybrid interaction, and the resulting sequence was compared with S. cerevisiae DNA sequences using the BLAST alignment program found at the Saccharomyces Genome Database Website (http://genome-www.stanford.edu/Saccharomyces/).
GST Pull-down Assays
GST fusions proteins and GST alone were induced from 100 ml bacterial cultures for 2 h by addition of 1 mM IPTG. Purification of the proteins onto glutathione-sepharose 4B beads was performed according to the Pharmacia protocol (Piscataway, NJ). To detect binding of yeast proteins, cell extracts were prepared as described previously (Warren et al., 2002), except that the final spin was performed at 45,000 rpm in a TLA100 ultracentrifuge (Beckman Instruments, Fullerton, CA). Extract, 100 μl, was incubated with 20 μl beads for 3 h at 4°C with rotation. The beads were pelleted at 500 × g for 3 min and washed three times. Bound proteins were eluted in sample buffer and separated on a 10% SDS polyacrylamide gel, before transfer to PVDF for analysis. Blotting for anti-myc was performed using the antibody A14 and for anti-HA using the antibody Y11. Both were used at 1/1000 dilution and are rabbit polyclonal antibodies from Santa Cruz Biotechnology (Santa Cruz, CA).
Fluorescence Microscopy Procedures
Endocytosis of the fluid phase marker lucifer yellow (Fluka, Buchs, Switzerland) was performed according to the method of Dulic and colleagues (Dulic et al., 1991). Quantitation of fluorescence intensity for staining vacuoles was performed using Scanalytics IP laboratory software (Billerica, MA). FM4–64 staining was performed essentially as described (Vida and Emr, 1995), except that cells were visualized at time points immediately after addition of the dye.
Rhodamine-phalloidin (Molecular Probes, Eugene, OR) staining for visualizing F-actin was performed as previously described (Hagan and Ayscough, 2000). Cells were processed for immunofluorescence essentially as described (Ayscough and Drubin, 1998). After fixation with formaldehyde, cells adhered to slides with poly-l-lysine were treated for 1 min with 0.1% SDS in PBS before incubating with antibodies. Primary antibodies used in this study were A14 anti-myc at a dilution of 1:100 (Santa Cruz Biotechnology). Secondary antibodies used were fluorescein-isothiocyanate (FITC)-conjugated goat anti-guinea pig (Cappell/Organon Technika, Malvern, PA) at a dilution of 1:1000. Cells were viewed with a Olympus BX-60 fluorescence microscope with a 100 W mercury lamp and an Olympus100X Plan-NeoFluar oil-immersion objective (Tokyo, Japan). Images were captured using a Roper Scientific MicroMax 1401E (Tucson, AZ) cooled CCD camera using Scanalytics IP lab software on an Apple Macintosh 7300 computer (Cupertino, CA).
Strains expressing fluorescently tagged proteins were made by integration of PCR-generated DNA fragments onto the genomic sequences of the appropriate genes as described by Longtine et al. (1998). Cells expressing GFP-tagged Sla1p, GFP-tagged Ysc84p, or GFP-tagged Lsb3p were visualized after growing to log phase in suspension in YPD media (Sla1-GFP) or after induction of expression after growth for 4 h in YP + 2% galactose (Ysc84-GFP or Lsb3-GFP) supplemented with adenine or after being taken from a freshly growing colony on a plate. For imaging 3 μl of cells was put on a slide, covered with a coverslip, and sealed with nail polish. For single images, GFP-expressing cells were viewed, and images were recorded as described above. For double labeling with rhodamine-phalloidin, cells were fixed using ethanol as follows: Log phase cells were pelleted then gently resuspended in ice-cold 70% ethanol and left on ice for 10 min. Cells were pelleted for 20 s and resuspended in rhodamine-phalloidin (5 mg/ml) in PBS containing 1 mg/ml BSA. After cells were left on ice for 5 min, they were washed three times with PBS. Cells were resuspended in PBS and placed on poly-lysine–treated slides.
For time-lapse live cell imaging, exponentially growing cells were harvested and resuspended in a smaller volume of synthetic complete medium containing glucose (SCD) and then applied to a slide to which a thin pad of 25% gelatin (Porcine 300 bloom; Sigma) in SCD had been applied. After sealing with a coverslip and rubber cement, cells were imaged for GFP fusion proteins expression using a DeltaVision Restoration Microscope System (Applied Precision Inc., Issaquah, WA), equipped with a Nikon Plan Apo 100× (1.4 NA; Garden City, NY) objective and a Roper Scientific Interline Cooled CCD camera (5 MHz MicroMax1300YHS; Tucson, AZ). Optical sectioning was performed at 0.4-μm intervals to encompass the entire cell-limiting exposure times to 0.09 s. Data were collected in fast acquisition mode. 3D image data sets were deconvolved using the SoftWoRx application (Applied Precision Inc.) running on a Silicon Graphics Octane workstation (Silicon Graphics Inc., Mountain View, CA). 2D maximum-intensity projections were generated from the 3D datasets using SoftWoRx and images captured in TIFF format using MediaRecorder (Silicon Graphics Inc.) and assembled using Adobe PhotoShop (Adobe Inc., San Jose, CA).
Electron Microscopy
Log phase cells were fixed using freshly prepared 2% potassium permanganate for 45 min at room temperature. After washing, the pellets were processed by dehydration through a series of ethanol from 50 to 100% and then in propylene oxide. Samples were then incubated overnight in a resin (Durcapan, Sigma): propylene oxide 1:1 mix before embedding in resin and curing. Sections were cut and stained with uranyl acetate and lead citrate before viewing on a Zeiss 902 transmission electron microscope (Thornwood, NY).
RESULTS
Sla1p Interacts with Ysc84p in a Two-hybrid Screen
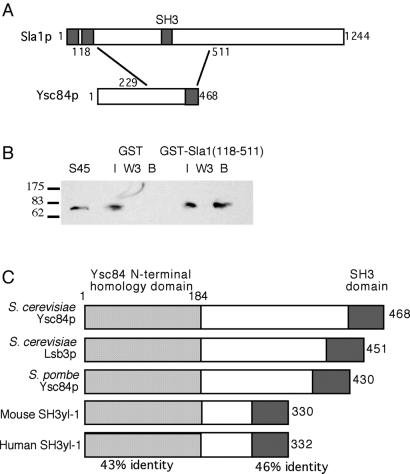
Analysis of Sla1p has previously identified a region which, when deleted, confers an aberrant actin phenotype on cells expressing the mutant protein. Deletion of other regions of the protein has little or no detectable phenotype on actin organization (Ayscough et al., 1999). The responsible region lies between amino acids 118 and 511 and included the sequence between the second and third SH3 domains (Gap1) as well as the third SH3 domain (SH3#3; Figure 1). Deletion of either Gap1 or SH3#3 domain alone does not confer an actin phenotype (Ayscough et al., 1999). To identify proteins that interact with this domain, a two-hybrid screen was performed with Sla1(118–511) as the bait. The screen was performed as outlined in MATERIALS AND METHODS. Of 31 positive interactions, 17 were subsequently sequenced and shown to encode a partial YSC84 gene. All of the clones isolated contained sequence corresponding to the C-terminal half of Ysc84p, although three different start sites were identified at the 5′ end of the clones. All 3′ ends of the clones were in the noncoding sequence beyond the YSC84 gene. The minimum interacting sequence that was identified in the screen started at sequence corresponding to residue 229 of Ysc84p and encompassed the remaining C-terminal sequence of YSC84. Previous large-scale two-hybrid studies had shown an interaction between full-length Sla1p and Ysc84p when each was expressed as a fusion with the bait or activation domain (Uetz et al., 2000; Drees et al., 2001). The screen described here allows the interaction domains to be further defined to the C-terminal 239-amino acid region of Ysc84p and a region of Sla1p encompassing its third SH3 domain.
Figure 1.
Ysc84p interacts with Sla1p. (A) Sla1p and Ysc84p were demonstrated to interact by two-hybrid approaches. The minimum interacting sequence was Sla1(118–511) and Ysc84(229–468). The interaction required the SH3 domain of Ysc84p but not the third SH3 domain of Sla1p. The Ysc84p related protein in S. cerevisiae, Lsb3p, was also shown to interact by two-hybrid approaches with the same region of Sla1p, and the sequence between residues 86–451 of Lsb3p. (B) GST-Sla1(118–511) and GST (as a control) were expressed in bacteria and purified. Whole cell extracts were made from a strain KAY564 expressing Ysc84-myc, where the myc tag was integrated following an seven-residue alanine linker. Ysc84-myc was clearly present in the 45K supernatant made from the extracts. The protein is seen only to bind to the beads carrying the GST-Sla1(118–511) fusion. I is input material, W3 is the third wash of the beads, and B is the material bound to the beads. (C) A cartoon showing the domain structure of the Ysc84p family members identified to date and the regions of highest homology between these proteins.
To determine whether the third SH3 domain of Sla1p is required for this interaction, a plasmid was constructed that contained the Gap1 region alone. When this was coexpressed in cells with a plasmid expressing the smallest YSC84 fragment, activation was still detected, indicating that the Sla1p third SH3 domain is not required for the interaction. A construct was then generated by in vitro mutagenesis in which a stop codon was introduced before the sequence encoding the SH3 domain of Ysc84p. When this Ysc84ΔSH3 construct was coexpressed with the Sla1(118–511) bait, activation was no longer detected. Therefore, the Ysc84p:Sla1p interaction occurs via the SH3 domain of Ysc84p. As described in MATERIALS AND METHODS, interactions in the two-hybrid screen were detected as growth on synthetic growth media lacking specific nutrients. In all cases the level of growth for the positive interactions appeared equivalent, with no combination of bait and prey giving rise to significantly faster or slower growing strains.
To verify the two-hybrid interaction between Ysc84p and Sla1p, a biochemical approach was used. The sequence encoding the amino acid 118–511 fragment of Sla1p was ligated to sequence encoding GST, and a fusion protein was produced (described in MATERIALS AND METHODS). A cell strain was generated expressing an myc-tagged form of Ysc84p. The myc tag was integrated into the genome at the 3′ end of YSC84 after addition of sequence for an seven-residue alanine linker. A linker was used to avoid disruption of interactions of the SH3 domain, which is at the extreme C terminus of the protein. A C-terminal tag was used so that the protein was expressed at endogenous levels. Extracts were made from the Ysc84-myc strain (KAY564) and were passed over beads carrying GST alone or GST-Sla1(118–511). The beads were washed extensively, and then bound proteins were separated using SDS-PAGE. The gels were blotted and probed with anti-myc antibody. As shown in Figure 1B, the GST-Sla1 beads, but not the GST beads alone, were able to bind Ysc84-myc from the cell extracts, thus providing complementary evidence for an Sla1p–Ysc84p interaction.
Ysc84p Is a Member of a Family of SH3 Domain-containing Proteins Conserved from Yeast to Humans
Sequence analysis using BLAST revealed a close homologue of Ysc84p in S. cerevisiae and also homologues in Schizosaccharomyces pombe, mouse, and humans. The S. cerevisiae homologue (Yfr024c-a) has been previously called Lsb3, so we have also adopted this nomenclature (Madania et al., 1999). Alignment of the two S. cerevisiae proteins reveals high homology across the entire length of the protein, although the N-terminal 210 amino acids and the C-terminal SH3 domain show the highest level of identity (91 and 86%, respectively). The high level of conservation at the termini was further accentuated when alignments of the homologues were generated from all organisms in which they have been identified (Figure 1C). Comparison of the N-terminal 184 amino acids from the five proteins revealed a 43% identity (65% similarity), and the C-terminal SH3 domain showed 46% identity (69% similarity).
A further observation from sequence analysis is that both YSC84 and LSB3 genes contain introns that are relatively infrequent in genes of the S. cerevisiae genome. However, the introns themselves are not in the same relative position in the genes. The intron in YSC84 is 169 nucleotides in length and lies between nucleotides 47 and 217 of the gene, and in LSB3 the intron is 118 nucleotides in length and lies between nucleotides 53 and 171 of the gene. It is not yet clear whether these introns have a functional role in expression of these two genes.
To assess whether the Ysc84-related protein Lsb3p also interacts with Sla1p, an expression plasmid was generated containing a region of Lsb3p equivalent to the domain of Ysc84p known to interact with Sla1p (amino acids 86–451). When assessed in a two-hybrid assay using the Sla1p(118–511) bait, an activation was detected (data not shown). Therefore, Lsb3p is also able to interact with Sla1p.
Ysc84p Localizes with the Cortical Actin Cytoskeleton
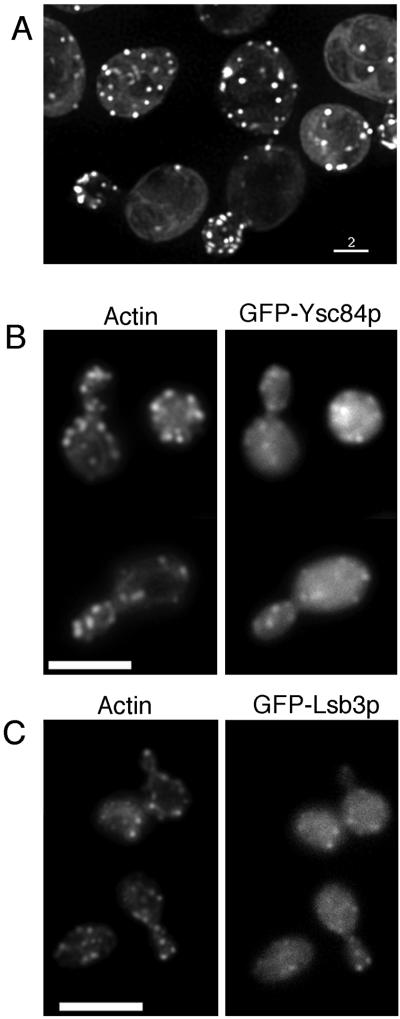
To investigate the possible role of the Ysc84p in cells, a sequence encoding a GFP tag was inserted onto the N-terminus of the gene. Attempts to insert a GFP sequence at the C terminus were also tried, but no transformants with a correct insertion were obtained. It is possible that such a large and structured tag would interfere with the C-terminal SH3 domain and create a dominantly negative mutant in the cell. The GFP tag did not affect the viability of the strain when inserted onto YSC84 in a wild-type or an Δlsb5 background, indicating that the tagged protein was functional (a combination of Δlsb5 and Δysc84 generates severe phenotypic defects; for fuller explanation see below). Furthermore, after 4 h in galactose medium neither the actin phenotype nor endocytosis was adversely affected. When cells carrying the GFP-YSC84 gene were grown in galactose-containing medium, the expression of the gene was induced, and GFP localization could be seen in cells. As shown in Figure 2 GFP-Ysc84p displayed a polarized cortical patch localization highly reminiscent of actin staining (Figure 2A). When the cells were fixed and their actin was stained, the GFP and rhodamine-phalloidin staining could be seen to colocalize (Figure 2B). It should be noted that to maintain the GFP fluorescence, fixation for this double staining was achieved using ethanol (see MATERIALS AND METHODS), which is less than optimal for preservation of cell structures, including the actin cytoskeleton, but both fluorescent signals were detectable. GFP-Lsb3p was also expressed and shown to colocalize to the cortical actin patches (Figure 2C).
Figure 2.
Ysc84p and Lsb3p colocalize with cortical actin patches. Cells expressing GFP-tagged Ysc84p (KAY525) or its homologue Lsb3 (KAY537) were grown to log phase. Expression of the tagged gene was induced by growth for 4 h in galactose-containing medium as outlined in MATERIAL AND METHODS. (A) GFP-Ysc84p live cells were visualized, and images were taken using the Deltavision deconvolution microscope as described. Bar, 2 μm. (B) GFP-Ysc84p–expressing cells were fixed using ethanol and costained with rhodamine-phalloidin to allow visualization of GFP and the actin cytoskeleton. (C) GFP-Lsb3p–expressing cells were fixed using ethanol and costained with rhodamine-phalloidin to allow visualization of GFP and the actin cytoskeleton. Bar, 10 μm.
The localization of Ysc84p was studied in live cells to determine whether it moved with similar kinetics to actin patches (∼0.06–0.3 μm/s; Doyle and Botstein, 1996; Waddle et al., 1996) or whether it behaved more similarly to Sla1p, which is fairly static at the cell membrane (Warren et al., 2002). Time lapse movies of the GFP-tagged Ysc84p indicate that patches containing Ysc84p move at rates of up to 0.25 μm/s. Thus, in respect to its behavior in cells, Ysc84p is more akin to actin patches. This also fits with its colocalization with actin patches compared with the partial colocalization of Sla1p and actin (Ayscough et al., 1999; Warren et al., 2002).
Localization of Ysc84p to the Cortex Requires F-actin and Abp1p but not Sla1p
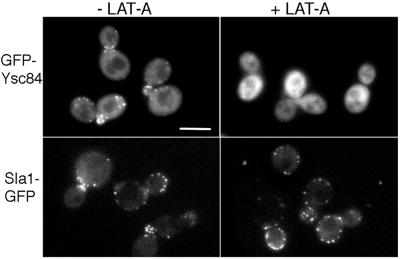
Further evidence for close association of Ysc84p with the actin cytoskeleton was investigated using the actin-disrupting drug latrunculin-A (LAT-A). In yeast, addition of LAT-A causes the rapid disassembly of actin patches and prevents the reincorporation of actin monomers into new actin filaments (Ayscough et al., 1999). Within 10 min of LAT-A addition, cortical actin structures can no longer be detected in cells. Sla1p-containing complexes, however, continue to localize to the cell cortex (Figure 3, lower panels). In cells expressing GFP-Ysc84p, addition of LAT-A caused a rapid loss in localization of the protein in patches at the cell cortex with kinetics similar to the loss of actin patches (Figure 3, upper panels). Therefore, localization of Ysc84p is primarily dependent on an intact actin cytoskeleton, whereas Sla1p complexes at the cell cortex localize independently of actin.
Figure 3.
Ysc84p requires F-actin for its cortical localization. Cells expressing GFP-Ysc84p (top panels) or Sla1p-GFP (bottom panels) were treated with the actin-disrupting drug latrunculin-A (200 μM for 15 min) and visualized by fluorescence microscopy as described. As shown in the right panels, in the presence of LAT-A Ysc84p loses its cortical punctate staining, whereas Sla1p remains at the cortex. Bar, 5 μm.
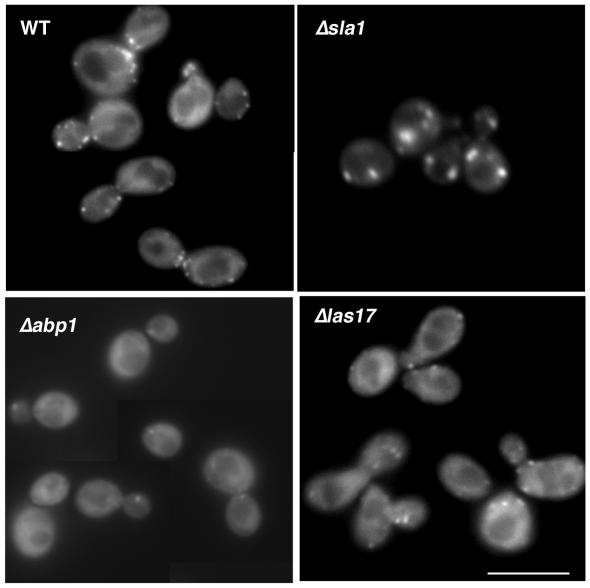
To investigate links between Ysc84p and Sla1p further, Sla1-GFP localization was observed in strains in which YSC84 was deleted. In this case, Sla1p continues to localize to the cell cortex with a distribution similar to that found in wild-type cells. In strains in which SLA1 is deleted, cortical actin is somewhat aberrant, in that the patches are larger and fewer than in wild-type cells. However, Ysc84p continues to localize to these actin structures in the absence of Sla1p (Figure 4 top right). These data demonstrate that although Ysc84p and Sla1p can interact in cells, their localization at the cell cortex is not dependent on one another. Rather Ysc84p interacts with cortical actin structures and requires the presence of F-actin to localize to the cell membrane, whereas Sla1p localizes to the cortex in an F-actin–independent manner and does not require the presence of Ysc84p to achieve this.
Figure 4.
Ysc84p requires Abp1p and Las17p but not Sla1p to associate with cortical actin patches. Ysc84p was tagged with GFP in wild-type cells or in cells in which sla1, abp1, or las17/bee1 had been deleted (strain KAY525, 527, 544, 547, respectively). Fluorescence microscopy was used to assess localization of GFP in each strain. In the absence of abp1 and las17/bee1, there was little or no localization of GFP-Ysc84p to punctate patches at the cell cortex, indicating that these proteins are important for Ysc84p localization. Bar, 5 μm.
Analysis of the Ysc84p sequence does not reveal any known actin-binding motifs so it is most likely that its localization to the actin patches is mediated by another protein. To investigate this, we integrated the Ysc84 GFP tag into strains in which either ABP1 or LAS17/BEE1 is deleted. Abp1p is a protein that binds directly to yeast actin in vivo and in vitro and has been demonstrated to activate Arp2/3 activity. However, deletion of ABP1 does not confer an obvious actin phenotype, and actin patches remain wild-type in appearance and localization. Las17/Bee1p is the yeast WASP homologue that also activates Arp2/3, and its deletion causes cells to have an aberrant actin phenotype. In the absence of LAS17/BEE1 expression Ysc84p still shows some localization to cortical complexes (Figure 4, bottom right). However, these do not resemble the large aggregates of F-actin structures seen in this deletion strain (Li, 1997; and our unpublished results), indicating that Las17p is important in mediating Ysc84p interaction with actin patches. Even more dramatic, however, is the consequence of deletion of abp1 on Ysc84p localization. In the absence of Abp1p, despite a normal distribution of actin patches (Drubin et al., 1988), Ysc84p is expressed but is no longer able to localize to the cell cortex (Figure 4, bottom left). These data indicate that both Las17p and Abp1p are necessary for localization of Ysc84p to the cell cortex.
YCL034w/Lsb5 Interacts with the Central Region of Sla1p in a Two-hybrid Screen
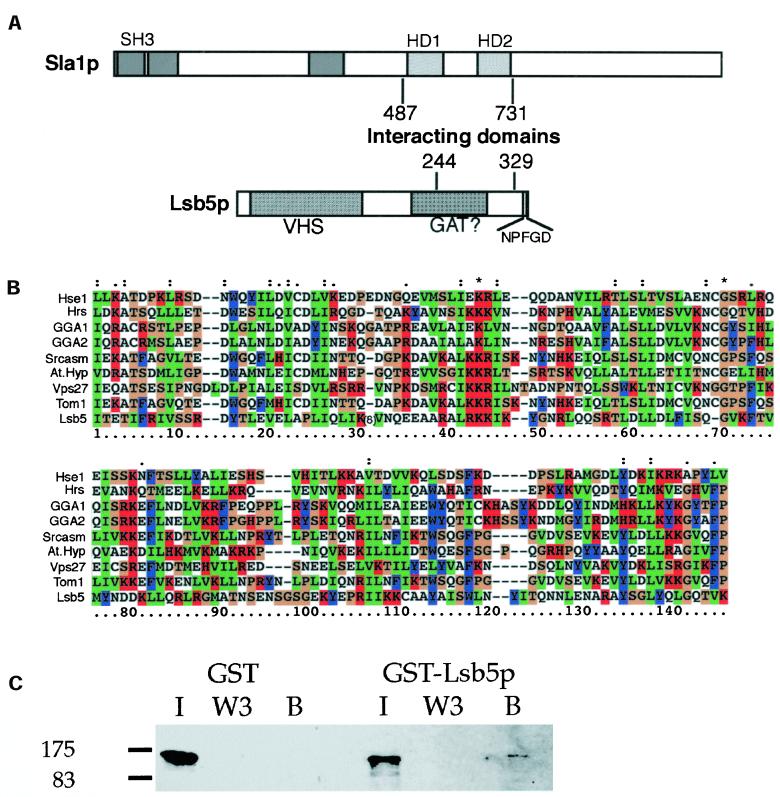
Our earlier studies on Sla1p had indicated that the most highly conserved region of the protein between homologues in other species (S. pombe, Neurospora crassa, Aspergillus nidulans, and Candida albicans; Ayscough et al., 1999; and our unpublished observations) were two regions, each of ∼60 amino acids in the central third of the protein. A two-hybrid screen was conducted with a bait containing these two domains (Sla1 residues 487–731). Of 30 positive interactions, 7 were subsequently sequenced and shown to encode a partial YCL034w gene. All of these clones contained sequence corresponding to the C-terminal region of Ycl034w, although each isolate contained distinct fragments of the gene. The minimum interacting sequence that was identified in the screen was amino acids 244 and 329, and this region was common to all isolates. This region is distinct from the region isolated by Madania and colleagues (1999), in which Las17p was shown to interact with a fragment of YCL034w between amino acids 40 and 213. Because YCL034w has been previously described as a Las17-binding protein (Madania et al., 1999) and has not been described elsewhere, we will refer to it henceforth by the given name of Lsb5p.
Database searches reveal homologues of Lsb5p in S. pombe and C. albicans but as yet, not in higher eukaryotes. Domain searches do, however, reveal a putative VHS domain at its N-terminus; there is also a possible GAT domain in the central region as well as an NPFXD motif at its extreme C terminus (Figure 5; http://smart.embl-heidelberg.de). Such a domain structure is reminiscent but not identical to the GGA proteins, which are proposed to act as adaptin-like proteins in vesicle trafficking (Boman et al., 2000; Hirst et al., 2001; Zhdankina et al., 2001). The putative VHS domain lies between residues 15 and 159, and the homology is given a significance (e-value) of 4.0e-08. A potential alignment of several VHS domains from a range of organisms and protein families is shown in Figure 5B.
Figure 5.
Sla1p interacts with Lsb5p. (A) Schematic outlining the regions of interaction between Sla1p and Lsb5p. Lsb5p was identified in a two-hybrid screen for proteins interacting with the central region of Sla1p. An 85-amino acid region of Lsb5p was found to interact with the 244-amino acid region of Sla1p. (B) Homology searches reveal a possible VHS domain at the N-terminus of Lsb5p. Shown here is a multiple sequence alignment generated using ClustalX software with recognized VHS domains from a range of other proteins. Hse1p, Gga1p, Gga2p, Vps27p, and Lsb5p are all S. cerevisiae sequences available through the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). Hrs (rat: AF036344); Tom1 (human: AJ010071); Srcasm (mouse: AF395837) and AtHyp (a hypothetical protein from Arabidopsis thaliana: AC012562). All the latter sequences are from EMBL databases retrieved through http://www.ebi.ac.uk/cgi-bin/emblfetch. To enable the alignment to proceed a sequence of eight amino acids was deleted from Lsb5 at the position indicated. This position corresponds to a region between alpha helices as determined by secondary structure prediction software. (C) To verify the two-hybrid interaction, GST-Lsb5 or GST alone were expressed and purified from bacteria using glutathione-sepharose beads. Extracts were made from cells expressing HA-tagged Sla1p (KAY355). After separation using SDS-PAGE and blotting, antibodies to HA detected Sla1p only on the GST-Lsb5p beads and not on those carrying GST alone. I is input material, W3 is the third wash of the beads, and B is the material bound to the beads.
Further evidence for the Sla1p, Lsb5p interaction was sought using a complementary approach. A GST-Lsb5 fusion protein was generated after cloning the LSB5 sequence into a pGEX vector (Pharmacia) and expressing the protein in bacteria. Cell extracts were prepared from cells expressing HA-tagged Sla1p (KAY355) and incubated with either GST-Lsb5 beads or with GST alone. After washing, samples were separated by SDS-PAGE and blotted onto PVDF membranes. Sla1-HA was detected using anti-HA antibody. As shown in Figure 5C, a small proportion of Sla1-HA binds to the GST-Lsb5p, but there is no binding to beads carrying GST alone.
Lsb5p Localizes to Cortical Complexes Independently of F-Actin
To investigate the role of Lsb5p, it was epitope tagged to allow localization to be observed using immunofluorescence microscopy. Attempts to GFP tag were not successful, possibly because the insertion of a relatively large and structured tag interfered with the correct functioning of the protein. A myc tag did not affect the viability of strains when inserted onto LSB5 in a wild-type or a Δysc84 background, indicating that the tagged protein was functional (a combination of Δlsb5 and Δysc84 generates severe phenotypic defects; for fuller explanation, see below).
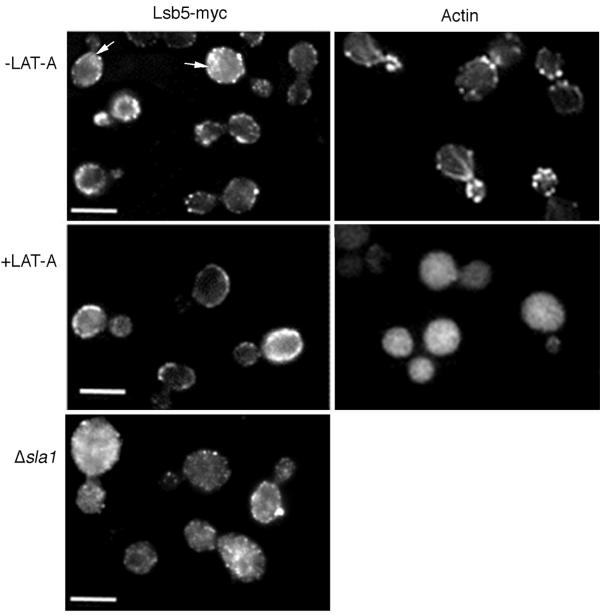
As shown in Figure 6, an Lsb5p-myc localizes to punctate spots at the cell cortex. Its distribution is not dramatically polarized, with similar levels of cortical staining in mother and daughter cells. This localization does not coincide with actin localization, suggesting Lsb5p is not tightly coupled to the actin cytoskeleton. Furthermore, addition of LAT-A disrupted the actin-staining pattern but not the cortical localization of Lsb5p, demonstrating that Lsb5p localizes independently of F-actin.
Figure 6.
Localization of Lsb5p to the cell cortex is independent of F-actin. (A) Actively growing Lsb5p-myc–expressing cells (KAY514) were fixed and processed for immunofluorescence microscopy as described in MATERIALS AND METHODS (left panels). A parallel population of cells were stained with rhodamine-phalloidin to assess the state of the actin cytoskeleton in the population (right panels). In actively growing cells Lsb5p localizes to the cell cortex in small punctate structures; there are also a significant number of cells that appear to show some staining of internal organelles, indicated with arrows. The localization does not appear to be polarized and does not closely resemble staining for actin, which is highly polarized, especially in small/medium budded cells. Half of each sample was then treated with 200 μM LAT-A for 5 min (lower panels). Despite a complete disruption of the actin cytoskeleton, Lsb5p continues to localize to the cell cortex in punctate structures. These however appear to be punctate than in the untreated population. (B) Using the same approach actively growing cells expressing Lsb5-myc, but in which sla1 is deleted (KAY601), were fixed and processed for immunofluorescence microscopy. In these cells Lsb5-myc appears to localize less well to the cell cortex, and there is increased cytoplasmic staining. Bar, 10 μm
Lsb5-myc was also localized in an Δsla1 deletion strain (KAY601). In these strains the actin is aberrant, with the cortical actin being found in fewer larger chunks compared with the small patches in wild-type cells (Holtzman et al., 1993; Ayscough et al., 1999). In the absence of Sla1p, Lsb5-myc did show some localization to the plasma membrane of cells, but this was significantly reduced compared with the wild-type situation (Figure 6B). In addition, the level of cytosolic staining was increased. This result indicates that Sla1p is a significant factor responsible for the cellular localization of Lsb5p.
Genetic Interactions of Δysc84 and Δlsb5 Strains
Deletion of eitherYSC84 or LSB5 from yeast did not produce a distinct phenotype and strains containing either single deletion were viable and able to grow at all temperatures tested (20–37°C). To investigate possible roles of the proteins further, crosses were made between strains deleted for a number of genes known to be important in actin organization. Crosses were made between Δysc84 and Δlsb3, Δsla1, Δlas17, and Δabp1. In all cases the double-deleted strains generated were viable and did not show a more serious growth phenotype than the parent strains. The similarity between Δysc84 and its homologue Δlsb3 suggested they might act redundantly with one another in an essential role. However, this was not the case, and the double mutant Δysc84 Δlsb3 was able to grow at a normal range of temperatures.
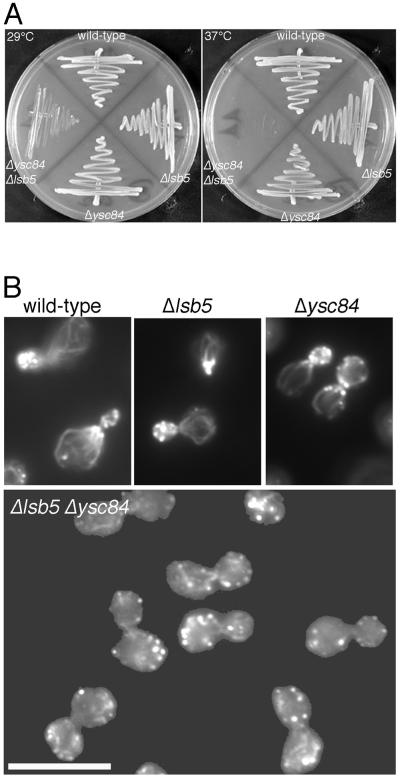
Δlsb5 was crossed with Δsla1, Δabp1, and also with Δysc84 and Δlsb3. Again, double mutants with Δsla1, Δabp1, and Δlsb3 were viable and able to grow at all temperatures tested. However, the cross between Δlsb5 and Δysc84 generated a strain that had a very significant growth defect (Figure 7A). The Δlsb5 Δysc84 mutant could not grow at elevated temperatures, and even at 29°C its generation time was 5.5 h (compared with 2.5 h for our wild-type strain). This synthetic interaction indicated that the two proteins might be normally operating in redundant pathways.
Figure 7.
Combination of Δysc84 and Δlsb5 deletions produces severe growth and actin phenotypes. (A) Strains carrying deletions of several genes encoding proteins associating with actin or the endocytic machinery were combined. Only the combination of Δysc84 and Δlsb5 gave a severe phenotype in growth. (B) Actively growing wild-type (KAY302), Δlsb5 (KAY515), Δysc84 (KAY510), and Δlsb5 Δysc84 (KAY516) cells were fixed and stained with rhodamine-phalloidin. Fluorescence microscopy was used to visualize the actin cytoskeleton. In the absence of both ysc84 and lsb5 but in neither single mutant, severe disorganization of the actin cytoskeleton was observed. Bar, 10 μm.
The Δysc84 Δlsb5 Strain Has Defects in the Actin Cytoskeleton
Because the double mutant had a severe growth defect, we investigated whether the actin cytoskeleton in these cells was disrupted. As shown in Figure 7B, although the single deletion mutant strains both contain an apparently wild-type actin cytoskeleton, the actin cytoskeleton in the double mutant strain was extremely depolarized. When small/medium budded cells were assessed for a polarized actin cytoskeleton, nearly 100% of wild-type, Δlsb5, and Δysc84 mutants showed polarity. However, fewer than 10% of the double mutant strain cells (Δlsb5 Δysc84) showed actin patches polarized to the bud. Interestingly, in the Δlsb5 Δysc84 strain, actin was still observed to localize to the cytokinetic ring, suggesting that a distinct set of actin-binding proteins mediate formation of this structure.
Double mutants of Δysc84 Δlsb3, Δysc84 Δabp1, Δlsb5 Δabp1, Δysc84 Δsla1, and Δlsb5 Δsla1 were also visualized using rhodamine-phalloidin. In all cases their actin was not changed by the presence of the Δysc84 or Δlsb5 deletion, such that Δabp1 strains were similar to wild-type cells (as are the single deletion Δabp1 cells), and the Δsla1 deletion strains had the same aberrant actin phenotype as the single Δsla1 mutants with fewer, larger actin patches.
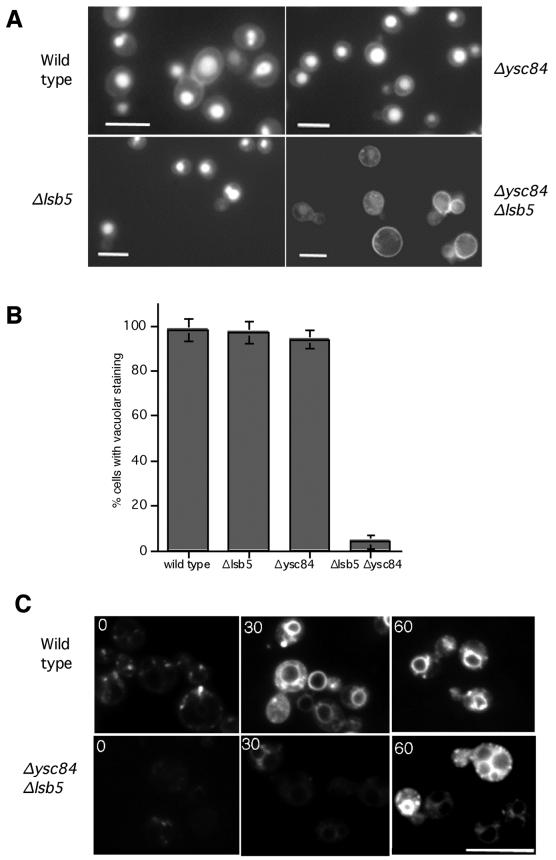
The Δysc84 Δlsb5 Strain Has a Severe Defect in Fluid Phase Endocytosis and Vesicle Trafficking
The recognized interactions between the actin cytoskeleton and the endocytic machinery indicated that the disruption of actin in the double mutant Δlsb5 Δysc84 may cause an effect on endocytosis. To assess fluid phase endocytosis uptake of the dye lucifer yellow was followed. In wild-type cells this dye can be seen to accumulate in the vacuoles after its internalization (Figure 8A). In cells in which lsb5 or ysc84 alone were deleted, no apparent effect was observed on lucifer yellow uptake. However, in the Δlsb5 Δysc84 double mutant we observed a dramatic decrease in lucifer yellow uptake (Figure 8, A and B). Quantitation revealed that <5% of cells in the Δlsb5 Δysc84 cell population appeared to endocytose the dye, compared with >95% in the wild-type or single mutant populations. The image shown for the Δlsb5 Δysc84 double mutant is overexposed to show that the cells are present and that any dye associated with the cells appears to be at the cell wall. Images taken with the same acquisition time as the wild-type cell images did not show any detectable cell staining.
Figure 8.
Disruption of fluid phase endocytosis and FM4–64 uptake in Δysc84 Δlsb5 mutant cells. (A) Lucifer yellow was used to assess fluid phase endocytosis in wild-type (KAY302), Δlsb5 (KAY515), Δysc84 (KAY510), and Δlsb5 Δysc84 (KAY516) cells. Actively growing cells were incubated at room temperature for 1 h in the presence of Lucifer yellow before visualization by fluorescence microscopy. Bar, 10 μm. (B) The percentage of cells containing vacuolar Lucifer yellow staining was also assessed and shown here graphically. (C) To monitor whether membrane trafficking was affected and the initial stages of endocytosis, uptake of FM4–64 was monitored. As shown, vacuolar staining could be seen in wild-type cells within 10 min of incubation, whereas a similar level of uptake was not seen in Δysc84 Δlsb5 cells until after 30 min. Even after 60 min the level of staining in the mutant cells was significantly lower than in wild-type cells. Bar, 10 μm.
To determine whether there were defects in membrane trafficking steps as well as in the initial endocytosis step, uptake of the lipophylic dye FM4–64 was followed. Wild-type and single Δysc84 or Δlsb5 mutants all showed a rate of uptake similar to one another, with vacuolar staining being clear after 10 min and with most cells having vacuolar membrane staining after 30 min (Figure 8C). However, movement of FM4–64 to vacuoles in the double mutant was significantly reduced, with no vacuolar membrane staining being observed until 30 min, and even after this period uptake appeared significantly lower than in the wild-type cells (Figure 8C). These data suggest that this strain is severely compromised in endocytosis but also has defects in later membrane trafficking steps.
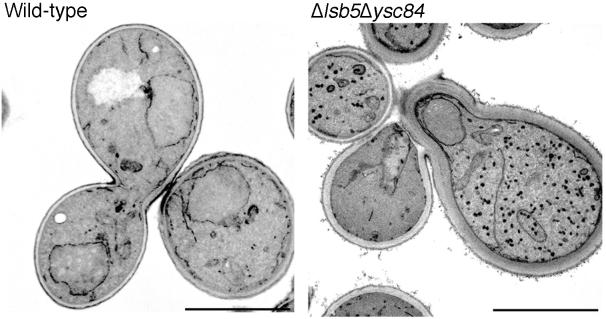
Finally, wild-type and Δysc84 Δlsb5 strains were fixed and processed for electron microscopy analysis. As shown in Figure 9 cells carrying the double deletion have a dramatic accumulation of vesicles or small organelles. All Δysc84 Δlsb5 cells observed contained these structures with a mean of 43.6 ± 5.8 per cell. This compares with 4.2 ± 0.7 structures per cell for the wild-type strain. Additionally, the cell walls of the mutant cells were significantly thicker than those of wild-type cells, which may also be an indication of inappropriate trafficking in these cells. These data provide further evidence that deletion of LSB5 and YSC84 in combination leads to a block in the normal membrane trafficking pathways.
Figure 9.
Δysc84 Δlsb5 cells accumulate vesicles and have thick cell walls. Wild-type (KA302) and Δysc84 Δlsb5 (KAY516) cells were grown to log phase and fixed for electron microscopy as described in MATERIALS AND METHODS. The mutant cells can be seen to have accumulated a large number of vesicle like structures. Quantitation revealed an average of 43 vesicles per cell in Δysc84 Δlsb5 cells compared with 4.2 similar vesicular structures in wild-type cells. Bar, 5 μm.
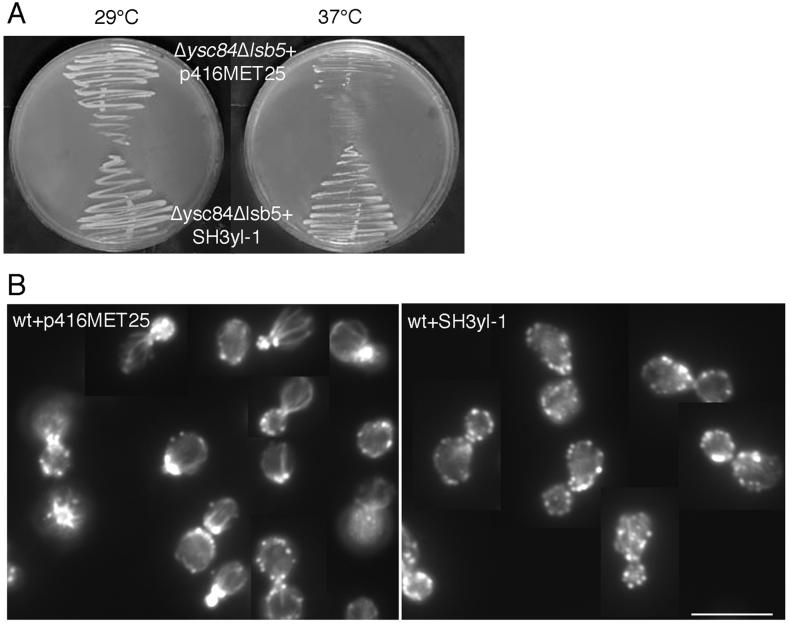
Expression of Mammalian Ysc84 (SH3yl-1) in Yeast Has Effects on Growth and the Actin Cytoskeleton
The human Ysc84 homologue, known as SH3yl-1, was obtained as a cDNA (generous gift from Dr. Hata, University of Tokyo). The cDNA was cloned into a yeast expression vector in which it can be expressed from a heterologous methionine promoter (pMET-SH3yl-1). This construct was then transformed into the Δysc84 Δlsb5 mutant, where expression of the mammalian gene was able to rescue the temperature sensitivity associated with the mutations (Figure 10A). Therefore, the mammalian Ysc84p homologue is at least partially functional in yeast. The growth rate for the transformed cells was, however, still less than observed for wild-type cells or the single Δlsb5 mutant, indicating that rescue is not complete. In addition, the actin phenotype appears to be similar to that of the untransformed double mutant, Δysc84 Δlsb5. In our hands the methionine promoter is leaky, and this rescue of temperature sensitivity was best observed when the strains were grown on media selecting for plasmid alone without further induction using media also lacking methionine.
Figure 10.
SH3yl-1, the human homologue of Ysc84p, can rescue the temperature-sensitive growth defect of Δysc84 Δlsb5 cells but disrupts the actin cytoskeleton of wild-type cells. (A) KAY516 cells lacking Δysc84 and Δlsb5 are temperature sensitive. Transformation of a plasmid carrying SH3yl-1 into these cells now permits their growth at 37°C. However, the disrupted actin phenotype of the cells is not rescued. (B) If the SH3yl-1 protein is able to interact with some but not all of its normal complement of binding proteins, it might have a disrupting effect on the actin cytoskeleton. This was observed when SH3yl-1 was expressed in wild-type (KAY447) yeast cells after a shift for 2 h into media lacking methionine, suggesting that some of the interactions of SH3yl-1 have been conserved across evolution. Bar, 10 μm.
To determine whether SH3yl-1 is able to interact with the actin cytoskeleton when expressed at increased levels, wild-type cells were transformed with either an empty or the pMET-SH3yl-1 plasmid. A dramatic effect on the actin cytoskeleton was observed in cells expressing SH3yl-1 when grown for 2 h in media lacking methionine (compared with our normal synthetic media, which contains 20 mg/l methionine). In these cells the cortical actin patches become almost completely depolarized (Figure 10B), demonstrating that SH3yl-1 is able to interact with the yeast actin cytoskeleton. Again, the association appears to be only partly functional giving rise to a dominant negative phenotype in which the actin cytoskeleton is disrupted.
DISCUSSION
In this study we have demonstrated the importance of two proteins that interact with Sla1p, for linking cortical actin to the process of endocytosis. One of the proteins, Ysc84p associates with actin patches, whereas Lsb5p is likely to be part of the membrane trafficking machinery. Loss of either single component does not have a marked phenotype, indicating that other proteins may compensate in their absence. However, the double deletion Δlsb5 Δysc84 has very severe growth defects, suggesting that the proteins function in distinct pathways, one of which must operate for endocytosis to occur. The Δlsb5 Δysc84 endocytosis defect is considerably more marked than that of a Δsla1 mutant strain, indicating that although both proteins might function in conjunction with Sla1p, they are likely to additionally associate with other components in this complex network.
Ysc84p, a Cortical Actin Patch Protein
The data presented here demonstrate that Ysc84p localizes to the actin cytoskeleton at the cell cortex and is dependent on F-actin for its localization. Its localization is distinct from that of Sla1p, which shows only partially overlapping localization with actin. We have previously suggested that the sites of Sla1p-actin colocalization mark sites of endocytosis, where components associated with the endocytic machinery are actively promoting actin dynamics in order to facilitate the endocytic process (Warren et al., 2002). The lack of Ysc84-Sla1p colocalization may indicate that the interaction between these proteins is transient and involved in the association of the larger complexes during endocytosis.
Two proteins were shown to be important in localizing Ysc84p. These are Abp1p and Las17p/Bee1p. Intriguingly, in the Δabp1 strain, the actin cytoskeleton is wild type in appearance but Ysc84p is unable to localize to this actin cytoskeleton. This indicates that Abp1p is a major factor localizing Ysc84p in cells. In the absence of las17 the actin cytoskeleton is extremely aberrant, so it is possible that the lack of localization of Ysc84p is due to an indirect consequence of a larger scale actin disruption. Both Abp1p and Las17p are important activators of Arp2/3 (and thereby actin polymerization) in yeast and so present important targets of regulation. Our data would indicate that Ysc84p is in a position to be regulating these proteins or recruiting other factors that themselves can lead to changes in actin polymerization.
Interestingly, the Ysc84 protein has been conserved across evolution, suggesting that its function is indeed important to cell function. To date the only studies reported in mammalian cells have suggested a possible role for SH3yl-1 in hair follicle development (Aoki et al., 2000). A wider role is suggested by the evolutionary conservation of this protein, and this awaits further investigation. Importantly, our data demonstrate a partial rescue of cell phenotypes when SH3yl-1 is expressed in Δlsb5 Δysc84 cells, indicating that some of the interactions this family of proteins have been maintained.
Lsbp5: A New Membrane-trafficking Adaptor Protein?
Our data show that Lsb5p is primarily localized to the plasma membrane of cells, but it is also found in compartments inside the cell. These compartments appear similar to previously described endosomes (Lewis et al., 2000). The localization of Lsb5p is not polarized and is similar to that observed for End3-HA (our unpublished observations), nor is it dependent on actin for its localization. These data, coupled with its domain structure, suggest a role for Lsb5p in membrane trafficking.
VHS domains are ∼150 amino acids long and are found most frequently at the N-termini of proteins associated with endocytosis and/or vesicular trafficking (Lohi et al., 2002). It is currently thought that VHS act as protein-binding domains, although the binding partners in most cases are unknown (Misra et al., 2000; Seykora et al., 2002). The most definitive results concerning the function of the domain has come from studies of the GGA proteins, in which the VHS domain has been shown to interact with sorting receptors such as sortilin that traffic between the TGN and endosomal compartments (Nielsen et al., 2001; reviewed in Lohi et al., 2002). The level of sequence identity between Lsb5p and other VHS domain-containing proteins is relatively low, although several factors support its identification as a bona fide domain of this type. First, the domain contains the majority of residues that are found in the consensus sequence for VHS domains. Second, like the majority of VHS domains, it is found at the extreme N-terminus of the protein. Third, the structure of two VHS domains have been solved (Mao et al., 2000; Misra et al., 2000) and have been shown to be largely comprised of a series of α-helices. Secondary structure prediction of the putative VHS domain of Lsb5p indicates that it is almost entirely α-helical in nature and that these helices are in approximately the same place within the domain (our unpublished observations). The GAT domain is also only weakly recognized as such by motif detection program, although currently the domain is less well defined than the VHS domain. However, a preliminary investigation of interactions between active and inactive mammalian Arfs and Lsb5 has allowed us to detect an interaction between Lsb5 and one of the activated Arf proteins (D.W., K.A., unpublished observations). Future studies will aim to investigate whether the putative GAT domain of Lsb5p is able to interact with specific Arf proteins in yeast. Finally, at the C terminus Lsb5p has an NPFXD motif. These have been demonstrated to bind to EH domains (de Beer et al., 2000; Kim et al., 2001), and as such there are several candidate proteins such as Pan1p and End3p, which associate with the endocytic machinery that could also be involved in localizing this protein (Tang et al., 1997). More recently however, Howard and colleagues (2002) have reported that Sla1p is able to bind directly to NPFXD motifs itself and that this interaction maybe important in uptake of specific proteins including Ste2p, However, in the two-hybrid interactions between Sla1p and Lsb5p reported in this study, the region of interaction does not encompass the NPFXD, suggesting that Sla1p binds to Lsb5p through sites other than at its NPFXD motif.
Complexes Linking Actin and Endocytosis
The body of evidence linking F-actin to endocytosis is growing, but despite a great number of reports, the mechanistic requirement for actin to facilitate endocytosis is still very unclear. In addition, there is little insight into the reason for the level of complexity. Why does the cell need multiple proteins to link the processes of actin dynamics and endocytosis? One possible model is that several pathways may have evolved to facilitate internalization of specific cargoes or to operate under certain conditions. This model would suggest that deletion of some genes may have no apparent phenotype, only because we are not investigating the right cargo; others may have mild phenotypes because other proteins are sufficiently similar to take over a secondary role, albeit less optimally; and relatively few proteins are absolutely required, perhaps those that are involved in all internalization events, or maybe these essential proteins have other critical roles within the cell.
Support for the idea of preferential cargo uptake has been demonstrated recently in studies investigating the role of Sla1p in endocytosis. These studies report that deletion of sla1 has only a mild effect on fluid phase endocytosis, whereas the NPFXD-mediated uptake of Ste2p in response to α-factor is dramatically reduced (Howard et al., 2002; Warren et al., 2002). Thus Sla1p may have a more specific function in receptor-mediated uptake of pheromone receptor and possibly a subsidiary role in general endocytic events. Further evidence for distinct pathways comes from the genetic interaction data, in which some combinations of mutations cause lethality, whereas other combinations have no additive effects. In some cases, synthetic lethality has been suggested to occur because of functional redundancy between the two proteins encoded by the genes. In the case of Δysc84 and Δlsb5, this seems less likely because the proteins do not share the same localization in the cell, Ysc84p appears to be localized to the actin patches, whereas Lsb5p seems to be more closely associated with the membrane trafficking machinery. In this case, lethality may occur because the two proteins operate in distinct actin–endocytosis coupling pathways. They are each required for the functioning of one pathway, but at different levels. However, one of the pathways must operate in order for endocytosis to proceed effectively.
Although there is evidence for distinct pathways of endocytosis, there are also much data demonstrating overlapping functions among some of the proteins involved in coupling actin to the endocytic machinery. Proteins such as Sla1p, Las17p, and Rvs167p have a similar set of interacting proteins identified by a number of genetic and biochemical approaches. For example, Sla1p and Rvs167p interact with Abp1 (Lila and Drubin, 1997; Warren et al., 2002); Sla1p and Las17p interact with Ysc84p, Lsb3p, and Lsb5p (Madania et al., 1999; this study); furthermore, all three components have been reported to interact with each other, even though they do not show complete colocalization (Li, 1997; Drees et al., 2001; Warren et al., 2002).
One model might suggest that Sla1p links a specific cargo to the endocytic machinery and then through an interaction with Ysc84p is able to couple the endocytic machinery to proteins involved in actin polymerization. Similarly, other proteins might link their specific cargoes to both the endocytic and actin machineries. Proteins such as Lsb5p might be recruited after the interaction to play a further role in cargo recognition or in vesicle formation. The studies in yeast, however, highlight the levels of complexity even in a relatively simple unicellular eukaryote. It is maybe not surprising that elucidating the roles of individual proteins in mammalian cells has been contentious.
To summarize, we have characterized two proteins, Ysc84p and Lsb5p, involved in coupling the processes of actin dynamics and endocytosis in yeast. The two proteins may now be useful markers to permit a study of the real time association of distinct cortical complexes involved in linking actin to the process of endocytosis. In addition, they are likely to be a major focus for other future studies, Ysc84p as a member of an evolutionarily conserved family of actin-associated proteins and Lsb5p as a putative new adaptor protein involved in membrane trafficking.
ACKNOWLEDGMENTS
We thank Steve Winder and Gwyn Gould for critical reading of the manuscript; Margaret Mullin for technical assistance with the electron microscopy; Phil Crews (UC Santa Cruz) for latrunculin-A; Dr. Hata (U. Tokyo) for the SH3yl-1 cDNA containing plasmid; David Drubin (UC Berkeley) for yeast strains; and Francis Barr (MPI, Munich) for the two-hybrid library used in this study. This work was supported by an MRC Senior Research Fellowship to K.R.A. (G117/394 Wellcome Trust award (050934/Z/97), BBSRC grant (17/C12769), a BBSRC studentship to F.G., and a Wellcome Trust Prize studentship to M.R.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0262. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0262.
REFERENCES
- Aoki N, Ito K, Ito M. A novel mouse gene, Sh3yl1, is expressed in the anagen hair follicle. J Invest Dermatol. 2000;114:1050–1056. doi: 10.1046/j.1523-1747.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DC. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Drubin DG. Immunofluorescence microscopy of yeast cells. In: Celis J, editor. Cell Biology: A Laboratory Handbook. Vol. 2. New York: Academic Press; 1998. pp. 477–485. [Google Scholar]
- Ayscough KR, Eby JJ, Lila T, Dewar H, Kozminski KG, Drubin DG. Sla1p is a functionally modular component of the yeast cortical actin cytoskeleton required for correct localization of both Rho1p-GTPase and Sla2p, a protein with talin homology. Mol Biol Cell. 1999;10:1061–1075. doi: 10.1091/mbc.10.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Zhang CJ, Zhu XJ, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M. Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol. 2000;7:1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees BL, et al. A protein interaction map for cell polarity development. J Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic V, Egerton M, Elgundi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AEY, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol. 2001;154:1209–1223. doi: 10.1083/jcb.200106089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- Goode BL, Rodal AA. Modular complexes that regulate actin assembly in budding yeast. Curr Opin Microbiol. 2001;4:703–712. doi: 10.1016/s1369-5274(01)00272-7. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Ayscough KR. Fluorescence microscopy in yeast. In: Allan VJ, editor. Protein Localization by Fluorescence Microscopy: A Practical Approach. New York: Oxford University Press; 2000. pp. 179–205. [Google Scholar]
- Hirst J, Lindsay MR, Robinson MS. GGAs: roles of the different domains and comparison with AP-1 and clathrin. Mol Biol Cell. 2001;12:3573–3588. doi: 10.1091/mbc.12.11.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JP, Hutton JL, Olson JM, Payne GS. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kessels MM, Engqvist-Goldstein AEY, Drubin DG, Qualmann B. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J Cell Biol. 2001;153:351–366. doi: 10.1083/jcb.153.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Cullis DN, Feig LA, Baleja JD. Solution structure of the Reps1 EH domain and characterization of its binding to NPF target sequences. Biochemistry. 2001;40:6776–6785. doi: 10.1021/bi002700m. [DOI] [PubMed] [Google Scholar]
- Lee E, De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci USA. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HRB. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila T, Drubin DG. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol Biol Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi O, Poussu A, Mao YX, Quiocho F, Lehto VP. VHS domain—a longshoreman of vesicle lines. FEBS Lett. 2002;513:19–23. doi: 10.1016/s0014-5793(01)03287-2. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Madania A, Dumoulin P, Grava S, Kitamoto H, Scharer-Brodbeck C, Soulard A, Moreau V, Winsor B. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol Biol Cell. 1999;10:3521–3538. doi: 10.1091/mbc.10.10.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YX, Nickitenko A, Duan XQ, Lloyd TE, Wu MN, Bellen H, Quiocho FA. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell. 2000;100:447–456. doi: 10.1016/s0092-8674(00)80680-7. [DOI] [PubMed] [Google Scholar]
- May RC, Caron E, Hall A, Machesky LM. Involvement of the Arp2/3 complex in phagocytosis mediated by FcγR or CR3. Nat Cell Biol. 2000;2:246–248. doi: 10.1038/35008673. [DOI] [PubMed] [Google Scholar]
- Misra S, Beach BM, Hurley JH. Structure of the VHS domain of human Tom1 (target of myb 1): insights into interactions with proteins and membranes. Biochemistry. 2000;39:11282–11290. doi: 10.1021/bi0013546. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Madsen P, Christensen EI, Nykjaer A, Gliemann J, Kasper D, Pohlmann R, Petersen CM. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001;20:2180–2190. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM, Kelly RB. Molecular links between endocytosis and the actin cytoskeleton. J Cell Biol. 2000;150:F111–F116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA. Coupling actin dynamics and membrane dynamics during endocytosis. Curr Opin Cell Biol. 2002;14:76–81. doi: 10.1016/s0955-0674(01)00297-6. [DOI] [PubMed] [Google Scholar]
- Seykora JT, Mei L, Dotto GP, Stein PL. Srcasm: a novel Src activating and signaling molecule. J Biol Chem. 2002;277:2812–2822. doi: 10.1074/jbc.M106813200. [DOI] [PubMed] [Google Scholar]
- Shurety W, Bright NA, Luzio JP. The effects of cytochalasin D and phorbol myristate acetate on the apical endocytosis of ricin in polarised Caco-2 cells. J Cell Sci. 1996;109:2927–2935. doi: 10.1242/jcs.109.12.2927. [DOI] [PubMed] [Google Scholar]
- Tang HY, Munn A, Cai MJ. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HY, Xu J, Cai MJ. Pan1p, End3p, and Sla1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebar F, Sorkina T, Sorkin A, Ericsson M, Kirchausen T. Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J Biol Chem. 1996;271:28727–28730. doi: 10.1074/jbc.271.46.28727. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vida T, Emr S. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;118:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle J, Karpova T, Waterson R, Cooper J. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DT, Andrews PD, Gourlay CG, Ayscough KR. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J Cell Sci. 2002;115:1703–1715. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn AL, Riezman H. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdankina O, Strand NL, Redmond JM, Boman AL. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast. 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]