Abstract
Synthesis of the ribosomal subunits from pre-rRNA requires a large number of trans-acting proteins and small nucleolar ribonucleoprotein particles to execute base modifications, RNA cleavages, and structural rearrangements. We have characterized a novel protein, RNA-binding domain-1 (RBD-1), that is involved in ribosome biogenesis. This protein contains six consensus RNA-binding domains and is conserved as to sequence, domain organization, and cellular location from yeast to human. RBD-1 is essential in Caenorhabditis elegans. In the dipteran Chironomus tentans, RBD-1 (Ct-RBD-1) binds pre-rRNA in vitro and anti-Ct-RBD-1 antibodies repress pre-rRNA processing in vivo. Ct-RBD-1 is mainly located in the nucleolus in an RNA polymerase I transcription-dependent manner, but it is also present in discrete foci in the interchromatin and in the cytoplasm. In cytoplasmic extracts, 20–30% of Ct-RBD-1 is associated with ribosomes and, preferentially, with the 40S ribosomal subunit. Our data suggest that RBD-1 plays a role in structurally coordinating pre-rRNA during ribosome biogenesis and that this function is conserved in all eukaryotes.
INTRODUCTION
In eukaryotic cells, a large number of different RNA-binding proteins bind to various RNA species to form functional ribonucleoprotein (RNP) complexes. A common RNA-binding motif in these proteins is the consensus RNA-binding domain (RBD), also called RNA recognition motif (reviewed by Varani and Nagai, 1998). The RBD is 80–90 amino acid residues in length, and within this loosely conserved region, two short sequences, RNP-1 and RNP-2, are more highly conserved. The structure of RBDs from different proteins has revealed a common βαββαβ fold, and the structure of several RBD–RNA complexes has demonstrated that the β-sheet is the main RNA interaction surface (Oubridge et al., 1994; Allain et al., 1996; Price et al., 1998; Deo et al., 1999; Ding et al., 1999; Handa et al., 1999; Allain et al., 2000; Inoue et al., 2000).
Several RBD-containing proteins are involved in ribosome biogenesis, and some of these proteins have multiple RBDs (Sun and Woolford, 1997; Ginisty et al., 1999). In the nucleolus, ribosome biogenesis starts with synthesis of the pre-rRNA transcript, which contains the 28S, 18S, and 5.8S rRNAs (Eichler and Craig, 1994). The maturation of this pre-rRNA follows essentially the same scheme in all eukaryotic cells (reviewed by Kressler et al., 1999; Venema and Tollervey, 1999). The pre-rRNA is assembled into an 80–90S nucleolar particle at the onset of maturation (Raué and Planta, 1991). Base modifications, cleavages, structural rearrangements, and stabilization followed by recruitment of specific ribosomal proteins will finally result in the 40S and 60S ribosomal subunits. At least in yeast, the final maturation of the pre-40S ribosomal subunit takes place in the cytoplasm (Udem and Warner, 1973; Valásek et al., 2001; Vanrobays et al., 2001). Intranuclear movement of the pre-60S ribosomal subunit requires specific proteins (Milkereit et al., 2001), and the final maturation of the pre-60S ribosomal subunit, possibly involving structural rearrangements, takes place in the cytoplasm (Venema and Tollervey, 1999; Senger et al., 2001). Nuclear export of the pre-60S ribosomal subunit involves the Ran-cycle (Hurt et al., 1999; Stage-Zimmermann et al., 2000) and the export factor Xpo1/Crm1, which binds to the ribosomal protein Rpl10p via Nmd3p (Ho et al., 2000; Gadal et al., 2001). Export of the pre-40S subunit also depends on the Ran-cycle (Moy and Silver, 1999), but no specific export factors have been identified.
A growing number of molecules are being identified that are involved in the biogenesis of the ribosomal subunits (reviewed by Kressler et al., 1999; Venema and Tollervey, 1999). Some molecules are necessary for the modifications of the rRNA nucleotides, whereas others are involved in endo- and exonucleolytic cleavages of the pre-rRNA. A large number of different snoRNPs serve as guides to target the modification and cleavage events (reviewed by Tollervey and Kiss, 1997; Weinstein and Steitz, 1999). In addition, many other trans-acting proteins are involved, for example, RNA helicases (de la Cruz et al., 1999) and several “assembly factors” of unknown function.
The maturation and assembly of the ribosomal subunits have to be strictly regulated processes. Comparatively little is known about how the coordinated action of all components involved in the biogenesis of the ribosome is brought about. Furthermore, little is known about the intranuclear transport of ribosomal subunits and about the molecular events that take place in the cytoplasm when ribosomal subunits are repeatedly recruited for translation. It is likely that additional proteins involved in the synthesis, assembly, export, and function of the ribosome remain to be identified (Lalev et al., 2000; Andersen et al., 2002).
In this study, we have characterized a novel protein, RBD-1, that has a unique domain organization and is conserved in eukaryotes. Our data suggest that RBD-1 is involved in ribosome biogenesis.
MATERIALS AND METHODS
Biological Material
Animals and Cells.
Chironomus tentans was cultured as described by Meyer et al. (1983). A C. tentans embryonic epithelial cell line was grown as described by Wyss (1982). Certain C. tentans cells were incubated with the transcriptional inhibitor actinomycin D (0.05 μg/ml) in standard medium.
C. elegans N2 was grown using standard conditions (Wood, 1988). Double-stranded RNA was injected into the gonad of young adult hermaphrodites (Fire et al., 1998), and offspring from the interval between 5 and 30 h after injection were analyzed.
Antibodies.
Anti-Ct-RBD-1 polyclonal rabbit antibodies were affinity purified by chromatography on cyanogen bromide-activated Sepharose 4B (Amersham Biosciences AB, Uppsala, Sweden). The affinity-purified antibodies detected a single, ∼95-kDa band in cytoplasmic and nuclear extracts in Western blots. This protein was partially purified by ion exchange chromatography and PAGE. The protein band was cut out from the gel and shown by mass spectrometry to be encoded by the cloned Ct-RBD-1 cDNA.
Anti-hrp45 (Kiseleva et al., 1994) and anti-hrp36 (Visa et al., 1996) antibodies were a gift from Prof. B. Daneholt. Anti-Sm (Y12) antibodies were a gift from Dr. I. Pettersson. Anti-FLAG-tag and anti-fibrillarin antibodies were purchased from Sigma-Aldrich (St. Louis, MO). The secondary antibodies were swine anti-rabbit Ig fluorescein isothiocyanate (FITC), rabbit anti-mouse Ig tetramethylrhodamine B isothiocyanate, swine anti-rabbit Ig horseradish peroxidase (HRP), and goat anti-mouse Ig HRP (DAKO, Glostrup, Denmark).
Cloning Procedures
RNA was extracted from C. tentans salivary gland cells as described previously (Edström et al., 1982). Poly(A+) RNA was purified by binding to oligo(dT) cellulose (Stratagene, La Jolla, CA). For cDNA synthesis, 5 μg of poly(A+) RNA was used. Degenerative deoxyoligonucleotides, representing conserved RNP-1 and RNP-2 motifs in the consensus RBD, were used for the reverse transcription-polymerase chain reaction (PCR) cloning as described by Kim and Baker (1993). Individual cDNA exhibiting sequence homology to RBDs was selected. The full-length Ct-RBD-1 cDNA sequence was obtained by screening a C. tentans tissue culture lambda Zap (Stratagene) cDNA library and by direct isolation of fragments extending in the 5′ or 3′ direction (CLONTECH, Palo Alto, CA).
Sequencing reactions were performed with the DYEnamic ET Terminator Cycling Sequencing Premix kit (Amersham Biosciences AB) and analyzed on a 373A automated DNA sequencer (Applied Biosystems, Union City, CA). DNA and protein sequences were analyzed by programs in the GCG package (Devereaux et al., 1984).
FLAG-tagged proteins were constructed by cloning PCR fragments encoding Ct-RBD-1 and the homologues from Saccharomyces cerevisiae and Homo sapiens into the vector pcDNA2 (Invitrogen), into which a sequence encoding the FLAG epitope had been inserted in frame 5′ of the cloning site. Green fluorescent protein (GFP)-fusion proteins were constructed using the pEGFP-C3 vector (CLONTECH).
Double-stranded RNA was synthesized from cDNA yk417f6, corresponding to the C. elegans open reading frame T23F6.4, by using T3 and T7 RNA polymerase (Ambion, Austin, TX).
Microdissection
For microdissection, C. tentans salivary glands from fourth instar larvae were fixed and mounted as described previously (Lambert and Daneholt, 1975). The cytoplasm was isolated by removing the nucleus from the cell by dissection well outside the nucleus to avoid nuclear contamination. Nuclei were carefully isolated to avoid cytoplasmic contamination. Intranuclear components were isolated from nuclei by further dissection to separate nucleoli from the chromosomes and interchromatin.
Protein Extraction and Western Blotting
Nuclear and cytoplasmic extracts of C. tentans tissue culture cells were prepared essentially as described by Wurtz et al. (1996). Nuclear extract of HeLa cells was prepared as described by Dignam et al. (1983). Cell extract from S. cerevisiae was prepared as described by Silve et al. (1991). Nuclear extract from Drosophila melanogaster was prepared as described by Petersen et al. (1995).
Proteins were separated on 12% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride filters. HRP-labeled secondary antibodies were detected by the enhanced chemiluminescence method (Amersham Biosciences AB).
Immunocytological Localization
Cells.
Cultured C. tentans diploid cells were prepared and stained with antibodies essentially as described previously (Baurén et al., 1996). For RNase treatment, cells were fixed in methanol for 2 min at −20°C, rinsed in phosphate-buffered saline (PBS), and incubated with RNase A (100 μg/ml) and RNase T1 (10 U/ml) for 2 h at room temperature. Cells were washed and processed for immunofluorescence as described above.
4,6-Diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) was used for chromatin staining in C. tentans tissue culture cells.
Isolated Polytene Chromosomes.
Chromosomes were isolated from C. tentans salivary glands and probed with antibodies essentially as described previously (Kiseleva et al., 1994). RNase treatment was performed by incubating with TKM (10 mM triethanolamine-HCl, pH 7, 100 mM KCl, 1 mM MgCl2) containing RNase A (200 μg/ml) and RNase T1 (10 U/ml) for 30 min at room temperature immediately after isolation. The chromosomes were washed in TKM and processed for immunofluorescence.
Preparations were mounted in antifade medium (VectaShield; Vector Laboratories, Burlingame, CA) and photographed in an Axioplan II microscope or in an LSM 510 confocal microscope (Carl Zeiss, Jena, Germany).
Expression of GFP- and FLAG-Fusion Proteins in HeLa cells
HeLa cells were transfected using LipofectAMINE (Invitrogen). Cells expressing FLAG-tagged proteins were fixed in 3.7% formaldehyde in PBS, washed, and treated with 0.2% Triton X-100 in PBS for 7 min. The cells were treated with blocking solution, probed with anti-FLAG antibodies, and incubated with secondary antibodies as described above for C. tentans cells. Cells expressing GFP-tagged proteins were fixed, mounted, and examined in the microscope.
RNA-Protein Binding
The coding part of the Ct-RBD-1 gene was cloned into the pET-15b expression vector (Novagen, Madison, WI) and expressed in Escherichia coli. His-tagged Ct-RBD-1 was purifed by Ni2+-NTA affinity chromatography (QIAGEN, Valencia, CA). To synthesize 32P-labeled RNA, PCR fragments or linearized plasmid DNA, pET-15b (Novagen), and pBluescript (Stratagene) were transcribed by T7 polymerase in vitro. The RNA was purified on polyacrylamide gels. Binding of Ct-RBD-1 to labeled RNA was investigated by a filter-binding assay, essentially as described by Ghisolfi-Neto et al. (1996). Then 2–3 fmol of RNA (in molecules) was heated at 60°C in 20 mM Tris-HCl, pH 7.5, 200 mM KCl, 5 mM MgCl2 for 15 min, cooled to 20°C, and incubated with different concentrations of purified protein in 60 μl of binding buffer (25 mM Tris-HCl, pH 7.5, 200 mM KCl, 5 mM MgCl2, 20% glycerol, 50 μg/ml tRNA, 10 μg/ml bovine serum albumin) for 30 min at 20°C. The reaction mixtures were filtered through wet nitrocellulose filters (0.45 μm HA; Millipore, Bedford, MA), followed by three washes with 300 μl of binding buffer. The RNA was essentially intact during the entire procedure as checked by electrophoresis in denaturing polyacrylamide gels. The percentage of bound RNA was determined by Cerenkov counting. The dissociation constant (Kd) was estimated as the protein concentration at which one-half of the RNA bound at saturation was retained on the filter (Carey et al., 1983).
Microinjection
Salivary glands were carefully dissected from C. tentans fourth instar larvae and placed in a drop of hemolymph surrounded by paraffin oil. Anti-Ct-RBD-1 antibodies (12.5 μg/μl) or a control antibody (12.5 μg/μl) in PBS was injected into individual nuclei (AIS Micro Systems; Carl Zeiss). Approximately 10 cells/gland were injected with ∼0.01 nl of antibody solution per nucleus. Each injected gland was incubated in hemolymph containing 3 μM α-[32P]ATP (400 Ci/mmol; Amersham Biosciences AB) for 60 min at 18°C. The gland was subsequently incubated in hemolymph containing 25 μM unlabeled ATP for 60 min. The glands were then fixed in 70% ethanol for 30 min on ice and prepared for microdissection. The nucleoli from ∼10 injected cells as well as the nucleoli from 10 uninjected control cells were isolated from each gland. RNA was extracted by incubation in 20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.5% SDS, 0.5 mg/ml proteinase K for 30 min at room temperature. After extraction with phenol:chloroform, the RNA was ethanol precipitated. The RNA was fractionated on 1% agarose gels, by using 20 mM Tris-HCl, pH 8, 20 mM NaCl, 2 mM EDTA, 0.2% SDS as running buffer. The gel was treated with cold 5% trichloroacetic acid, washed in water, dried, and exposed to x-ray film and to a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) screen for quantification analysis (Fujifilm FLA-3000, Image Gauge V3.45). In each experiment, RNA from injected cells was compared with RNA from noninjected cells from the same salivary gland. Injection of a control antibody did not affect the proportions of the pre-rRNA species.
Analysis of Polysomes, Ribosomes, and Ribosomal Subunits
C. tentans tissue culture cells were washed in PBS and UV irradiated (∼3 × 104 erg/mm2). The cells were resuspended in 10 mM Tris-HCl, pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 2 mM dithiothreitol, containing vanadyl ribonucleoside complex (50 μl/ml extract) and tRNA (0.5 μg/μl), and homogenized in a glass homogenizer. After centrifugation at 20,000 × g for 10 min, the supernatant was recovered. Deoxycholate and Triton X-100 were added to a final concentration of 1% each. The extract was layered onto a linear 15–50% sucrose gradient (wt/vol, in 20 mM Tris-HCl, pH 7.6, 30 mM KCl, 2 mM MgCl2, 6 mM β-mercaptoethanol) and centrifuged at 150,000 × g at 4°C for 1.5 h (AH650 rotor; Sorvall, Newton, CT). Fractions were collected and A260 nm was measured. The fractions were treated with RNase A (30 μg/ml), trichloroacetic acid precipitated, and analyzed by Western blotting.
For further analysis, the polysomes were concentrated by centrifugation at 85,000 × g at 4°C, overnight. The polysomes were dissolved in 20 mM Tris-HCl, pH 7.6, 30 mM KCl, 2 mM MgCl2, 6 mM β-mercaptoethanol. After addition of EDTA to 30 mM, the preparation was layered onto a linear 10–30% sucrose gradient (see above) and centrifuged for 2 h at 235,000 × g at 4°C. Fractions were RNase treated and analyzed as described above. The material in the initial 80S peak was also concentrated and analyzed in the same way.
To analyze the association of Ct-RBD-1 to ribosomes, extracts were prepared as described above and centrifuged through 1 M sucrose (in 20 mM Tris-HCl, pH 7.6, 30 mM KCl, 2 mM MgCl2, 6 mM β-mercaptoethanol). After centrifugation for 4 h at 235,000 × g at 4°C, the pellet and the supernatant were analyzed by Western blotting. In parallel, we analyzed extracts supplemented with 0.5 M KCl/5 mM MgCl. The extracts were layered onto 0.9 ml of 0.5 M sucrose [in 20 mM Tris-HCl, pH 7.6, 0.5 M NH4Cl, 5 mM Mg(OAc)2, 5 mM β-mercaptoethanol] with a 1.5-ml 1.5 M sucrose cushion [in 20 mM Tris-HCl, pH 7.6, 0.35 M KCl, 5 mM Mg(OAc)2, 5 mM β-mercaptoethanol] at the bottom.
RESULTS
Characterization of Ct-rbd-1 Gene
The coding region of the Ct-rbd-1 gene (the C. tentans rbd-1 gene) was isolated using a reverse transcription-PCR approach to search for genes encoding RBD-containing proteins. To investigate the exon-intron structure of the gene, genomic DNA and the cDNA were used as PCR templates, in combination with several different oligodeoxynucleotide primer pairs. We found a single 61-base pair-long intron that interrupts the RNP-1 sequence of the third RBD. The genomic organization of the gene was analyzed. A single band was seen in the Southern blots after cleavage of the genomic DNA with EcoRI, BamHI, or XbaI (our unpublished data). The gene was located to a single locus, 19 A, on chromosome II (our unpublished data). Combined, these results indicate that a single copy of the Ct-rbd-1 gene is present in the C. tentans genome.
Ct-RBD-1 Contains Six Conserved RNA Binding Domains
The cDNA contained a 2547-base pair open reading frame, encoding a protein of 849 amino acid residues. The most conspicuous property of the protein is that it contains six consensus RBDs spread out through the entire protein, together covering ∼60% of the protein (Figure 1A). Apart from several putative nuclear localization sequences, no other functional motifs could be identified in Ct-RBD-1.
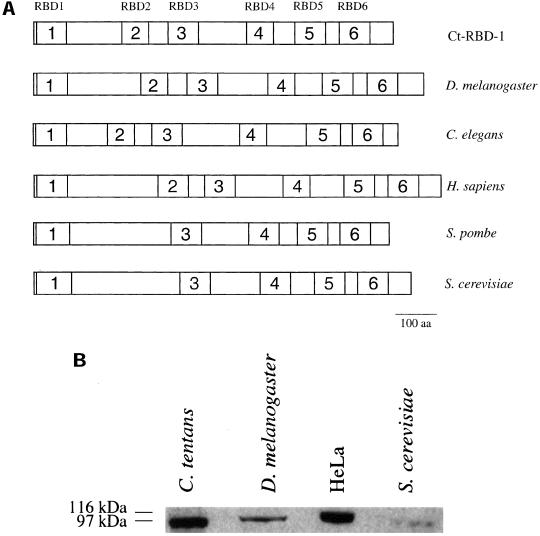
Figure 1.
Ct-RBD-1 is conserved in eukaryotes. (A) Comparison of the organization of the RBDs in Ct-RBD-1 (accession no. Q95ZH1) and in the sequence homologues in five different species. The proteins and the RBDs are drawn to scale. The six different RBDs in Ct-RBD-1 are marked by numbers. The proteins in D. melanogaster (accession no. Q9VT19), C. elegans (accession no. Q9XU67), H. sapiens (accession no. Q96E42), S. pombe (accession no. O13620), and S. cerevisiae (accession no. Q06106) have the same domain organization. Sequence comparisons between all the RBDs in the six proteins showed that there is a position-specific conservation of the six RBDs between species, shown by marking homologues RBDs with the same number. In S. pombe and S. cerevisiae, RBD2 is lacking according to the sequence comparisons. (B) Ct-RBD-1–specific antibodies detect similar sized proteins in C. tentans, D. melanogaster, HeLa cells, and S. cerevisiae. Extracts from each species were analyzed by Western blotting and probed with anti-Ct-RBD-1 antibodies.
Ct-RBD-1 is similar along its entire length to proteins encoded by single copy genes sequenced in D. melanogaster, C. elegans, H. sapiens, Schizosaccharomyces pombe, and S. cerevisiae. These proteins are 918, 872, 960, 833, and 887 amino acids in length, respectively. Nothing is so far known about the function of any of these proteins.
The organizations of the RBDs in the six proteins are compared in Figure 1A. The overall amino acid residue identity is 43–52% among the C. tentans, D. melanogaster, C. elegans, and H. sapiens proteins. These proteins all have six RBDs at similar positions. The sequence similarity is greatest within the RBDs, but it is also extensive outside the RBDs. We will therefore name the protein in D. melanogaster Dm-RBD-1, in C. elegans Ce-RBD-1, and in H. sapiens Hs-RBD-1. Ct-RBD-1 and the yeast proteins are 32–34% identical. The two yeast proteins have five RBDs.
Comparisons of all the individual RBDs in Ct-RBD-1, Dm-RBD-1, Ce-RBD-1, and Hs-RBD-1 revealed that each of the six RBDs in the proteins is more similar to the RBDs at the corresponding positions across species than to the other RBDs within the same protein (Figure 1A). This was also true for the five RBDs in the yeast proteins, where the second RBD seems to be missing compared with the other species.
In agreement with the sequence similarities between the proteins in the different species, Western blot analysis showed that the antibodies raised against Ct-RBD-1 recognize a protein of approximately the expected size in extracts from D. melanogaster, HeLa cells, and S. cerevisiae (Figure 1B).
In summary, the sequence comparisons show that Ct-RBD-1 is conserved in higher eukaryotes. The protein contains six RBDs spaced throughout the entire protein. The yeast proteins lack one RBD, but because of the overall sequence similarity and the localization studies (see below), we propose that Ct-RBD-1 is conserved from yeast to human.
Localization of Ct-RBD-1
Fractionation of C. tentans cells showed that Ct-RBD-1 is present mainly in the nucleus but also in the cytoplasm (Figure 2A). Anti-Ct-RBD-1 antibodies stained the nucleoli brightly in diploid C. tentans tissue culture cells (Figure 2B). They also stained the remaining nucleus in a punctated pattern against a more diffuse overall staining. The punctated pattern did not coincide with the pattern seen after staining with antibodies against fibrillarin, the SR protein hrp45, the Sm epitope in small nucleolar ribonucleoprotein particles, or the heterogeneous nuclear ribonucleoprotein protein hrp36 (our unpublished data). In addition, the anti-Ct-RBD-1 antibodies stained the cytoplasm, but weaker than the nucleus. Similar foci as in the nucleus were observed. The staining of the nucleolus, nucleus, and cytoplasm was drastically reduced after RNase treatment, indicating that Ct-RBD-1 is associated with RNA (our unpublished data).
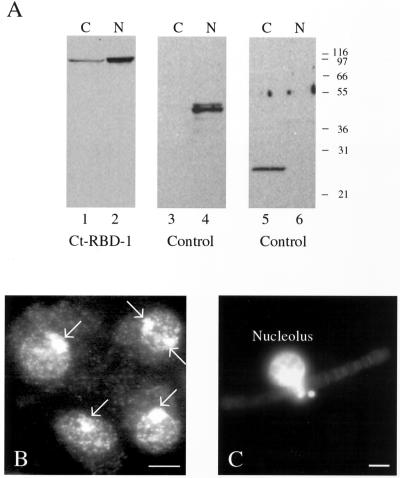
Figure 2.
Ct-RBD-1 is present in nucleoli, nucleoplasm, and cytoplasm. (A) C. tentans diploid tissue culture cells were homogenized and nuclei and cytoplasm were separated by centrifugation. Extracts of the two fractions were analyzed by Western blotting. Ct-RBD-1, migrating at ∼95 kDa, is present both in the cytoplasm and in the nucleus (lane 1 and 2, respectively). Cross-contamination of the fractions was checked using an antibody against the nuclear protein hrp45 (Kiseleva et al., 1994) (lanes 3 and 4) and an antibody recognizing a cytoplasmic protein of unknown function (lanes 5 and 6). (B) Immunocytology of C. tentans diploid tissue culture cells. The cells were centrifuged onto slides, fixed, permeabilized, and stained with the anti-Ct-RBD-1 antibodies and an FITC-conjugated secondary antibody. The nucleoli (marked by arrows) are most heavily stained. In the remaining nucleus and in the cytoplasm, discrete foci are detected against a diffuse overall staining. Bar, 5 μm. (C) Immunostaining of an isolated polytene chromosome III of a C. tentans salivary gland cell. The chromosome was isolated from a fixed salivary gland cell by pipetting and incubated with the anti-Ct-RBD-1 antibodies and a FITC-conjugated secondary antibody. The nucleolus is brightly stained. Bar, 10 μm.
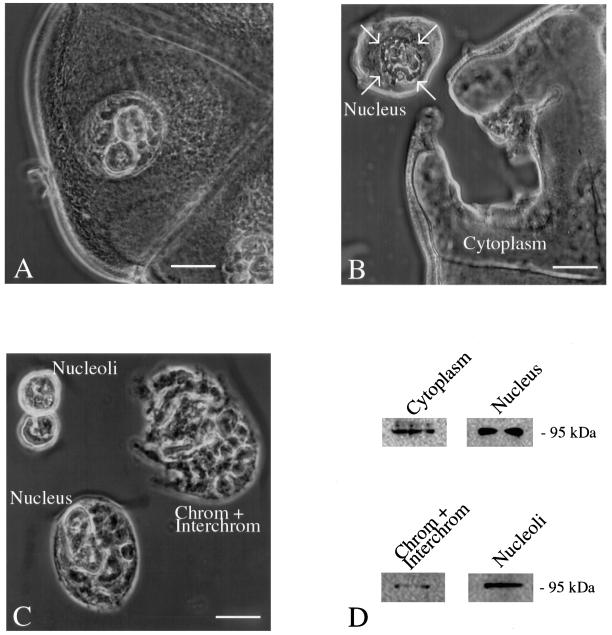
The location of Ct-RBD-1 was confirmed by analyzing salivary gland cells. First, polytene chromosomes were isolated, and in Figure 2C it is shown that the antibodies specifically stained the nucleolus. Little if any staining of active gene loci or chromatin could be detected. Second, we analyzed individual cellular compartments; cytoplasm, nuclei, nucleoli, and the combined chromosomes and interchromatin was isolated from fixed salivary gland cells by microdissection (Figure 3, A–D). Western blot analyses showed that Ct-RBD-1 is present in the analyzed cellular compartments.
Figure 3.
Microdissection analysis of C. tentans salivary gland cells. Salivary glands were isolated, fixed, and prepared for microdissection. The gland and the cellular components were viewed by phase contrast microscopy. (A) Salivary gland cell. The nucleus with the polytene chromosomes and the two nucleoli are seen. Bar, 40 μm. (B) Isolation of the cytoplasm. The nucleus was removed by dissecting well outside the nuclear membrane to avoid nuclear contamination of the cytoplasm. The arrows show the position of the nuclear membrane. Bar, 60 μm. (C) Salivary gland nuclei were carefully dissected to avoid cytoplasmic contamination. Subsequently the two nucleoli and the chromosomes plus interchromatin were isolated. Bar, 30 μm. (D) Western blot analysis of extracted proteins from the cytoplasm (isolated as shown in B), the nucleus, chromosomes plus interchromatin, and nucleoli (isolated as shown in C). Ct-RBD-1 is present in all the analyzed compartments.
On the basis of the combined results of our biochemical fractionation, immunocytology, and microdissection analyses, Ct-RBD-1 is located mainly in the nucleus, but also in the cytoplasm. In the nucleus, Ct-RBD-1 is concentrated in the nucleolus and is also present in the interchromatin.
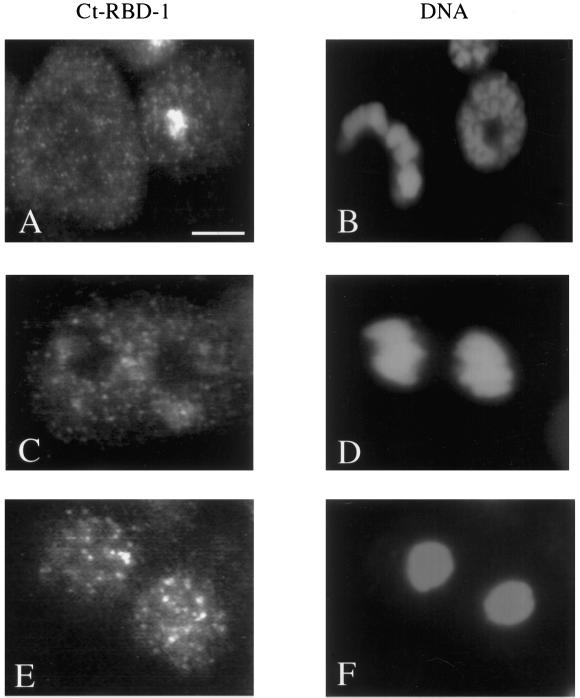
The location of Ct-RBD-1 during mitosis is shown in Figure 4. Ct-RBD-1 behaves differently compared with several proteins involved in rRNA transcription (reviewed by Scheer and Weisenberger, 1994) and compared with several rRNA-processing factors (Dundr et al., 1997, 2000; Dousset et al., 2000). In metaphase, Ct-RBD-1 is located in a large number of small granules in the entire cell and does not accumulate around the metaphase chromosomes (Figure 4A). These granules are similar to the granules seen in the nucleus and in the cytoplasm in interphase (compare the cells in Figure 2A). In late anaphase, Ct-RBD-1 is still found in numerous small granules throughout the entire cell, except at the condensed chromosomes (Figure 4C). In late telophase, larger granules were present in the nuclei, and in some cells, an initial nucleolar accumulation could be detected (Figure 4E), but we could not decide whether Ct-RBD-1 enters prenucleolar bodies in telophase nuclei. We observed that Ct-RBD-1 is recruited to the newly reformed nucleoli at the end of telophase.
Figure 4.
Localization of Ct-RBD-1 during mitosis. C. tentans tissue culture cells were fixed, permeabilized, and stained with the anti-Ct-RBD-1 antibodies. The cells were also stained for DNA with DAPI to allow identification of cells in different stages of mitosis. (A) Immunostaining of an interphase cell (right) and a metaphase cell (left). (B) DNA in the same cells as in A, stained with DAPI. (C) Immunostaining of a cell in late anaphase. (D) Same cell as in C, stained with DAPI. (E) Immunostaining of telophase cells. (F) Same cells as in E, stained with DAPI. Bar, 5 μm.
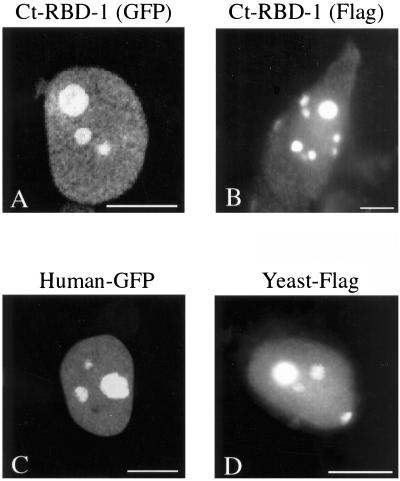
We further studied the cellular location of the D. melanogaster, H. sapiens, and S. cerevisiae sequence homologues to Ct-RBD-1. By using anti-Ct-RBD-1 antibodies, Dm-RBD-1 had exactly the same location in D. melanogaster cells (our unpublished data) as seen for Ct-RBD-1 in C. tentans cells (Figure 2B). The anti-Ct-RBD-1 antibodies also stained preferentially the nucleoli of HeLa cells, although the immunoreaction was weak (our unpublished data). We therefore transformed HeLa cells with GFP- and FLAG-tagged cDNA constructs. In Figure 5, A–C, it is shown that Ct-RBD-1 and Hs-RBD-1 are present in the nucleus and that they are highly concentrated in the nucleoli. Ct-RBD-1 could also be detected in the cytoplasm in a punctated pattern (Figure 5B). The signal in the cytoplasm was much lower than the signal in the nucleus. The same result was obtained for Hs-RBD-1 (our unpublished data). A control showed that GFP alone was found homogeneously throughout the entire cell (our unpublished data). The FLAG-tagged S. cerevisiae protein was also present in the entire nucleoplasm and concentrated in the nucleolus (Figure 5D). These results are in agreement with the location of Ct-RBD-1 in C. tentans (Figures 2 and 3) and show that Ct-RBD-1, Hs-RBD-1, and the S. cerevisiae protein have very similar nuclear locations.
Figure 5.
Localization of Ct-RBD-1 and the H. sapiens and S. cerevisiae homologues in HeLa cells. HeLa cells were transfected with cDNA constructs encoding RBD-1 proteins from the different species, tagged with GFP or the FLAG-epitope. Cells expressing GFP-fusion proteins were fixed and directly viewed in the fluorescence microscope, whereas cells expressing the FLAG-tagged proteins were first fixed, permeabilized, and stained with an anti-FLAG antibody. (A) Ct-RBD-1 (GFP) is located throughout the nucleus and highly concentrated in the nucleoli. (B) Ct-RBD-1 (FLAG) has the same location as Ct-RBD-1 (GFP). Many cells also exhibited a weak cytoplasmic staining, and some cells were more strongly stained as shown herein. (C) Hs-RBD-1 (GFP) was located throughout the nucleoplasm but showed the highest concentration in the nucleoli. (D) S. cerevisiae RBD-1 homologue (FLAG) is concentrated in the nucleolus. The nucleoplasm was also stained. Bars, 10 μm.
Nucleolar Location of Ct-RBD-1 Is Dependent on RNA Polymerase I Transcription
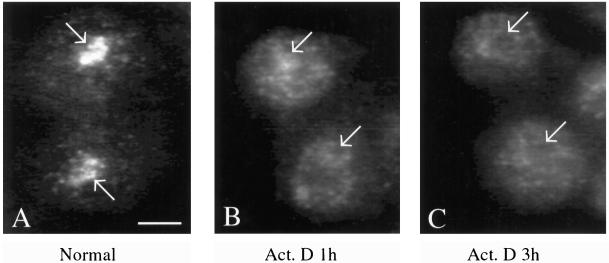
We investigated whether the presence of Ct-RBD-1 in the nucleolus is dependent on pre-rRNA synthesis. Tissue culture cells were grown in the presence of low doses of actinomycin D, known to preferentially inhibit the activity of RNA polymerase I. The amount of Ct-RBD-1 was very much reduced in the nucleolus, 1 h after the addition of actinomycin D (compare Figure 6, A and B). The staining of the nucleus outside the nucleolus, and of the cytoplasm was largely unaltered, but there was a tendency that the punctated pattern was reduced and the staining was more evenly distributed. After 3 h of actinomycin D treatment, the changes were more accentuated and the nucleolus was almost devoid of Ct-RBD-1 (Figure 6C). In phase contrast, the nucleolus was still detectable after both 1 and 3 h. Ct-RBD-1 is therefore only present in the nucleolus when pre-rRNA is synthesized. Together with the result that the location of Ct-RBD-1 in the nucleolus is sensitive to RNase treatment, this indicates that Ct-RBD-1 associates with newly transcribed pre-rRNA and that there is no extensive storage of Ct-RBD-1 in the nucleolus.
Figure 6.
Nucleolar location of Ct-RBD-1 is dependent on RNA polymerase I transcription. C. tentans tissue culture cells were grown in the presence of actinomycin D (0.05 μg/ml). Cells were fixed, permeabilized, and immunostained with the anti-Ct-RBD-1 antibodies at time 0 (A), after 1 h (B) and after 3 h (C) of actinomycin D treatment. Arrows mark the nucleoli. Bar, 5 μm.
Ct-RBD-1 Binds to pre-rRNA
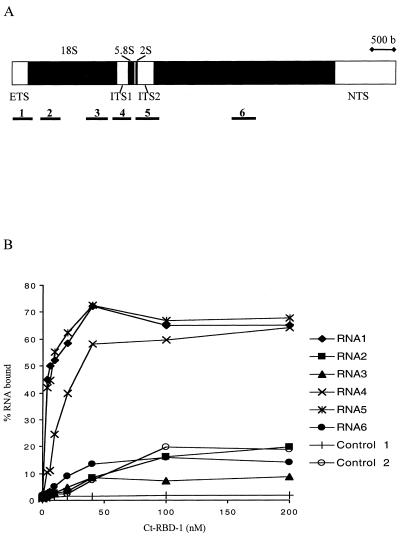
The transcription-dependent and RNase-sensitive nucleolar location suggested that Ct-RBD-1 binds to pre-rRNA. Data from sucrose gradient centrifugation of nuclear extracts also indicated that Ct-RBD-1 is present in large complexes compatible with ribosomal precursors (our unpublished data). We therefore investigated whether Ct-RBD-1 is able to bind to pre-rRNA. In vitro transcripts corresponding to defined regions of C. tentans pre-rRNA were analyzed (Figure 7A). In Figure 7B, it is shown that Ct-RBD-1 efficiently binds to pre-rRNA. We observed approximately the same efficient binding to regions of the pre-rRNA containing the transcribed spacers, the 5′ external transcribed spacer (5′ETS), the internal transcribed spacer (ITS) 1, and the ITS 2, all containing presumed processing cleavage sites. Binding to two regions within the 18S rRNA and to one region of the 28S rRNA was inefficient as was binding to non-rRNA control RNAs.
Figure 7.
Ct-RBD-1 interacts with pre-rRNA. (A) Schematic representation of the C. tentans rRNA gene. The regions of the pre-rRNA used for binding studies with Ct-RBD-1 are shown below the gene and numbered 1–6. 32P-Labeled RNAs were obtained by in vitro transcription of PCR fragments representing the different regions. The lengths of the RNAs were 438, 442, 477, 412, 519, and 536 nucleotides for RNA 1 to RNA 6, respectively. (B) Filter binding assay with Ct-RBD-1 and RNAs representing different pre-rRNA regions and control RNAs. Then 2–3 fmol of each RNA (in molecules) was incubated with increasing amounts of Ct-RBD-1 and filtered through nitrocellulose filters. The percentage retained RNA was plotted against the concentration of protein. Control RNAs were obtained by transcribing restriction enzyme cleaved pET-15b plasmid DNA (control 2, 430 nucleotides) or a PCR fragment representing a globin gene construct (control 1, 353 nucleotides). A third control RNA (transcribed from the plasmid pBluescript) behaved similarly to control RNA 2 (our unpublished data). The values shown are the averages of two separate measurements. Binding of RNA containing part of the 5′ ETS or ITS 2 (RNA 1 and RNA 5) bound Ct-RBD-1 with similar high affinities (Kd value of ∼5 nM). RNA representing the ITS 1 (RNA 4) bound with slightly less high affinity (Kd value of 10–15 nM). Binding of RNA containing 18S (RNA 2 and 3) or 28S (RNA 6) rRNA sequences was not different compared with the control RNAs.
Anti-Ct-RBD-1 Antibodies Repress pre-rRNA Processing In Vivo
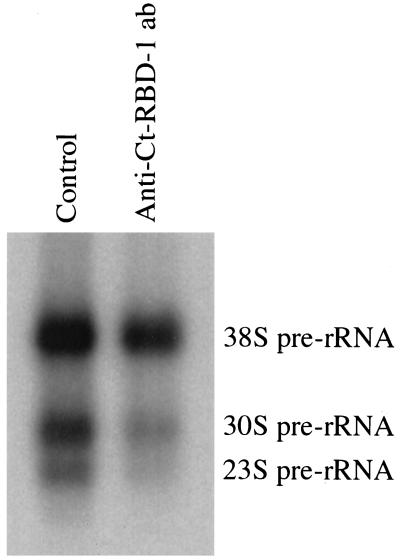
To establish that Ct-RBD-1 is involved in pre-rRNA synthesis and/or processing in vivo, we injected anti-Ct-RBD-1 antibodies into living C. tentans salivary gland cells and analyzed the effect on pre-rRNA. It has previously been shown that in the nucleolus in C. tentans, a 38S rRNA precursor is processed into 30S rRNA, a precursor to 28S rRNA, and into 23S rRNA, a precursor to 18S rRNA (Lambert and Daneholt, 1975). As shown in Figure 8, we observed that injection of anti-Ct-RBD-1 antibodies resulted in reduced relative levels of both 30S and 23S pre-rRNA intermediates. In six experiments, the average relative reduction in 30S pre-rRNA was 22% (range 0–29%) and for 23S pre-rRNA 56% (range 14–83%). An average relative increase in 38S pre-rRNA of 57% was also seen (range 33–165%). Injection of a control antibody did not affect the relative levels of 38S, 30S, and 23S pre-rRNA. The absolute amount of 38S, 30S plus 23S pre-rRNA was not significantly different in cells injected with antibodies compared with controls. These data suggest that when we interfere with Ct-RBD-1 function in vivo by injecting specific antibodies against Ct-RBD-1, there is a disturbance of pre-rRNA processing, with the greatest effect observed for 23S pre-rRNA.
Figure 8.
Anti-Ct-RBD-1 antibodies repress pre-rRNA processing in vivo. Anti-Ct-RBD-1 antibodies were injected into the nuclei of living C. tentans salivary gland cells. RNA was labeled by incubation of the glands in medium containing α-[32P]ATP. The gland cells were fixed and the nucleoli were isolated by microdissection from injected cells and from noninjected cells as controls. The RNA was extracted and separated in agarose gels. The gels were dried and the relative amounts of the 38S, 30S, and 23S pre-rRNA were determined using a PhosphorImager.
Ct-RBD-1 Is Associated with Ribosomes in Cytoplasm
We wished to further investigate the cytoplasmic distribution of Ct-RBD-1. First, we found that 20–30% of Ct-RBD-1 in cytoplasmic extracts sedimented together with ribosomes and polysomes through 1.5 M sucrose. By using the same conditions, but in the presence of 0.5 M KCl, little if any Ct-RBD-1 could be pelleted. The Ct-RBD-1–containing pellet could also be resuspended, washed in 0.5 M KCl, and resedimented through 1.5 M sucrose. Ct-RBD-1 was then released and was not present in the pelleted material (our unpublished data). Ct-RBD-1 is therefore associated with large molecular weight complexes in the cytoplasm in a salt-dependent manner.
In Figure 9, A and B, it is shown that Ct-RBD-1 sediments with polysomes and in fractions containing free ribosomes. Disruption of the polysomes with EDTA before centrifugation shifted the position of Ct-RBD-1 away from the polysome part of the gradient (our unpublished data).
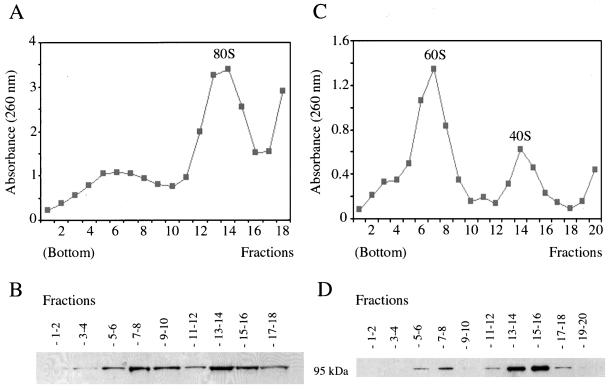
Figure 9.
Ct-RBD-1 is associated with ribosomes and preferentially with 40S ribosomal subunits. C. tentans tissue culture cells were homogenized and cytoplasmic extracts, treated with deoxycholate and Triton X-100, were centrifuged in sucrose gradients. After fractionation, the fractions were analyzed by measuring the absorbance at 260 nm and by Western blotting. (A) Polysome profile obtained after separation of cytoplasmic extract by using a 15–50% sucrose gradient. (B) Fractions from the gradient shown in A were pooled as indicated and analyzed by Western blotting, by using anti-Ct-RBD-1 antibodies. (C) Profile of ribosomal subunits obtained by EDTA treatment of isolated C. tentans polysomes and subsequent separation in a 10–30% sucrose gradient. (D) Fractions from the gradient shown in C were pooled as indicated and analyzed by Western blotting, with anti-Ct-RBD-1 antibodies.
We also recovered the polysomes from sucrose gradients (Figure 9A). After EDTA treatment of the isolated polysomes, we separated the ribosomal subunits by sucrose gradient centrifugation. In Figure 9, C and D, it is shown that Ct-RBD-1 mainly cosedimented with the 40S ribosomal subunit. We obtained the same result when we analyzed the 80S peak from the initial gradient (our unpublished data).
rbd-1 Is Essential in C. elegans
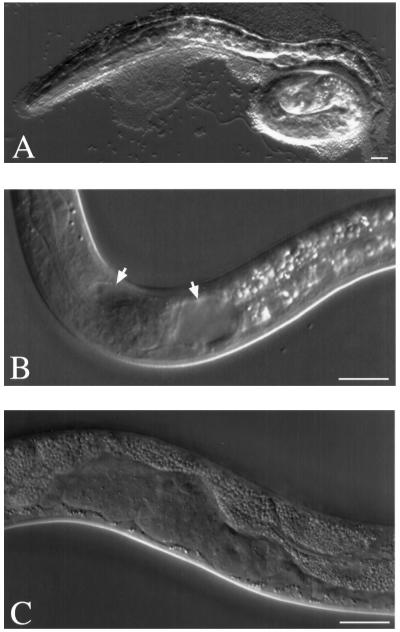
A cDNA for rbd-1 was sequenced to confirm the predicted open reading frame. Double-stranded RNA was used for RNA-mediated gene interference (RNAi) to knockdown rbd-1 gene function in C. elegans. Offspring of injected animals hatch but are arrested at the L1 larval stage. After 3–4 d, 50–70% of the arrested animals have died, showing necrotic features in many parts of their bodies (Figure 10A). Approximately 20% of the offspring can survive longer, sometimes reaching what seems to be an adult stage. Invariably, these animals display various abnormalities such as dumpy, incomplete molting, and incomplete and defective gonadal and vulval development (Figure 10, B and C). We conclude that rbd-1 is an essential gene; in the RNAi experiments the lethality becomes only apparent at the L1 stage, when feeding is necessary for further growth.
Figure 10.
RNAi phenotype of rbd-1. Double-stranded RNA corresponding to the Ce-RBD-1 mRNA was injected into the gonad of young adult hermaphrodites. The offspring were analyzed 5–30 h after the injection. (A) L1 larva, dying, with detached phenotype, i.e., pseudocoel filled with liquid. (B) L1 larva, vacuoles behind the terminal bulb of pharynx and anterior to it (arrows). (C) L2 larva, abnormal developing gonad with vacuoles. Bars, 10 μm.
DISCUSSION
RBD-1 Has a Unique Six × RBD Structure That Is Conserved in Eukaryotes
RBD-1 is a novel protein. The similarity in sequence, including linker regions, and domain organization as well as cellular location between proteins in yeast, Diptera, nematodes, and human, suggest that RBD-1 is conserved in all eukaryotes. The presence of as many as six consensus RBDs is exceptional. The RBD is common in other RNA-binding proteins, but it is usually present in one or two copies (Burd and Dreyfuss, 1994; Varani and Nagai, 1998), although some proteins have up to four RBDs (Burd et al., 1991; Gil et al., 1991; Patton et al., 1991; Ghetti et al., 1992; Sun and Woolford, 1997; Ginisty et al., 1999).
RBD-1 is further unusual because its RBDs are spread out from the N terminus to the C terminus of the protein. In other proteins, the RBDs are usually clustered in one region of the protein. This region is in most cases connected to one or more auxiliary domains that contribute specific properties, such as protein–protein interaction, RNA helicase activity, or additional RNA binding properties (reviewed by Biamonti and Riva, 1994; Varani and Nagai, 1998). No known sequence motifs, such as, for example, RGG repeats present in several characterized nucleolar proteins (Ginisty et al., 1999), are found in RBD-1.
The structure of two RBDs in tandem, with the connecting linker, have been determined in several different proteins (Shamoo et al., 1997; Xu et al., 1997; Crowder et al., 1999; Deo et al., 1999; Handa et al., 1999; Ito et al., 1999; Allain et al., 2000; Conte et al., 2000). In all cases, the individual RBDs have the βαββαβ fold, originally described for the RBD1 in U1A (Nagai et al., 1990). It is therefore most likely that each of the six RBDs in RBD-1 is folded into this common three-dimensional structure.
The fact that the six RBDs are spread out in RBD-1 results in four of the regions between consecutive RBDs being comparatively long. These linker regions are, for example, between 40 and 144 amino acid residues in Ct-RBD-1. In the complexes between two tandem RBDs and RNA for which the structures are known (Deo et al., 1999; Handa et al., 1999; Allain et al., 2000), the 10–25 amino acid-residue-long linker regions contribute to RNA binding. The much longer linker regions in RBD-1, in particular the first and third linker region, could, apart from contributing to RNA binding, potentially interact with other proteins.
Ct-RBD-1 binds with high affinity to pre-rRNA (Figure 7). From studies of other proteins, RNA recognition by RBDs is known to be influenced by several parameters, such as the specific amino acid sequence of the RBD itself, the immediate surrounding N- and C-terminal regions, RNA intramolecular interactions, and protein–protein interactions. A single RBD is sufficient for specific RNA binding, but it has been shown that two RBDs confer higher specificity and stability (reviewed by Varani and Nagai, 1998). RBDs in separate proteins can also interact with each other and potentially bring distant binding sites in a single RNA or binding sites in separate RNA molecules together (Ding et al., 1999). Two RBDs in tandem can bind to short RNA sequences in stem-loops and induce or stabilize the stem-loop (Allain et al., 2000) or bind to a longer nucleotide sequence and stretch out the RNA (Deo et al., 1999; Handa et al., 1999). All RBDs in a multi-RBD protein may be needed for binding as in the case of U2AF65 (Zamore et al., 1992). In contrast, only the two first of the four RBDs in poly(A)+ binding protein seem sufficient for binding to poly(A) (Nietfield et al., 1990; Burd et al., 1991; Kuhn and Pieler, 1996), and the second RBD in U1A seems not to bind RNA (Lu and Hall, 1995). Furthermore, different combinations of RBDs can be used to bind to different RNA sequences (Bouvet et al., 1997; Serin et al., 1997; Ginisty et al., 2001).
Because there is not a single, but a range of potential binding modes, we cannot predict how RBD-1 recognizes and binds pre-rRNA. The fact that the individual RBDs in RBD-1 have a conserved position-specific sequence (Figure 1A) suggests that the individual RBDs contribute specific properties. Because RBD-1 has six RBDs, it is conceivable that it can simultaneously bind to several different RNA sequences present in either the same RNA molecule and/or in separate RNA molecules.
Ct-RBD-1 Binds pre-rRNA and Is Involved in Ribosome Biogenesis
Ct-RBD-1 binds to pre-rRNA with high affinity in vitro (Figure 7), and we could only detect efficient binding to regions containing the 5′ ETS, the ITS 1, and the ITS 2. Anti-Ct-RBD-1 antibodies repress processing of pre-rRNA in vivo, preferentially the formation and/or stability of 23S rRNA (Figure 8), an intermediate in the 18S rRNA pathway. Furthermore, Ct-RBD-1 is concentrated in the nucleolus, where it is bound to RNA and the presence in the nucleolus is dependent on transcription by RNA polymerase I (Figure 6). Hs-RBD-1 was also recently found to be present in isolated nucleoli, designated NNP64 (Andersen et al., 2002). Collectively, our data argue that RBD-1 is involved in ribosome biogenesis. The conserved nature of RBD-1, including the same cellular location (Figure 5), suggests that the function of RBD-1 is of fundamental importance in all eukaryotes. This conclusion is in agreement with our result that RBD-1 is essential in C. elegans. This suggestion is also supported by studies of the S. cerevisiae sequence homologue. This previously unknown protein, named Mrd1p, is essential and required for the early cleavages of the pre-rRNA, which are necessary for formation of 40S ribosomal subunits (Jin et al., 2002). Mrd1p is also concentrated in the nucleolus and present in the remaining part of the nucleus.
Ct-RBD-1 is present in distinct nuclear foci outside the nucleolus, presumably in the interchromatin, because Ct-RBD-1 was not found in gene loci (Figure 2C). The pattern of small foci does not coincide with the speckled pattern seen for splicing factors. Therefore, Ct-RBD-1 is not likely to be involved in pre-mRNA processing. The interchromatin location for Ct-RBD-1 may reflect the intranuclear storage and turnover of the protein. On the basis of the likely role in ribosome biogenesis, the pattern could also reflect association of Ct-RBD-1 with mainly the 40S ribosomal subunits in transit through the interchromatin.
Ct-RBD-1 is present also in the cytoplasm in small foci. Our immunolocalization studies of the endogenous Ct-RBD-1 in C. tentans diploid cells (Figure 2B) and of tagged Ct-RBD-1 in transformed HeLa cells (Figure 5, A and B) and the combined microdissection and Western blot analysis of polytene C. tentans cells (Figure 3) confirm that Ct-RBD-1 is present in the cytoplasm. Our analysis of cytoplasmic extracts of C. tentans diploid cells showed that ∼20–30% of the Ct-RBD-1 present in the cytoplasmic extract was associated with ribosomes in a salt-dependent manner. We also showed that Ct-RBD-1 is preferentially associated with the 40S ribosomal subunit (Figure 9), a striking coincidence with the fact that the S. cerevisiae Mrd1p is required for synthesis of 40S ribosomal subunits (Jin et al., 2002).
In general, processing of RNA requires the RNA to adopt specific structures and such structures need to be stabilized or induced by binding to specific proteins (Herschlag, 1995). Structural analyses of ribosomes in particular, have highlighted the necessity of a high degree of coordination between formation of the compact ribosomal subunit structure and modification and cleavage reactions during ribosome biogenesis (reviewed in Venema and Tollervey, 1999; Lafontaine and Tollervey, 2001). The domain structure of RBD-1 indicates that RNA-binding is central for function. There are no domains suggesting that RBD-1 has other functions, such as nuclease or modification activity. It is therefore possible that RBD-1 is involved in structural coordination of the pre-rRNA and perhaps also in guiding other involved proteins and RNP complexes, during processing and ribosomal subunit assembly. An RNA chaperone activity has been proposed for the abundant nucleolar protein nucleolin (Allain et al., 2000). Our in vitro RNA-protein–binding data suggest that Ct-RBD-1 interacts with the pre-rRNA at sites present in the transcribed spacer regions. Several other proteins may bind specifically to the spacer regions (Lalev et al., 2000; Lalev and Nazar, 2001). Because preribosomal particles are converted to mature ribosomal subunits outside the nucleolus, RBD-1 could be important for 40S ribosomal subunit maturation throughout the transport to the cytoplasm and possibly also for some aspect of 40S ribosomal subunit function.
ACKNOWLEDGMENTS
We thank Kerstin Bernholm for excellent technical assistance. This work was supported by grants from Carl Tryggers Stiftelse and the Swedish Research Council (Natural and Engineering Sciences). We thank Prof. Odd Nygård for useful advice, Prof. Uno Lindberg for use of equipment, and Dr. Yui Kohara for cDNA clone yk417f6. T.R.B. was supported by a grant from the SSF.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0138. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0138.
REFERENCES
- Allain FH-T, Bouvet P, Diecmann T, Feigon J. Molecular basis of sequence specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH-T, Gubser CC, Howe PWA, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Baurén G, Jiang W-Q, Bernholm K, Gu F, Wieslander L. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J Cell Biol. 1996;133:929–941. doi: 10.1083/jcb.133.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Riva S. New insights into the auxiliary domains of eukaryotic RNA binding proteins. FEBS Lett. 1994;340:1–8. doi: 10.1016/0014-5793(94)80162-2. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Jain C, Belasco JG, Amalric F, Erard M. RNA recognition by the joint action of two nucleolin RNA-binding domains: genetic analysis and structural modeling. EMBO J. 1997;16:5235–5246. doi: 10.1093/emboj/16.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Burd CG, Matunis EL, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J, Cameron V, de Haseth PL, Ohlenbeck OC. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983;22:2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Conte MR, Grüne T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding proteins reveals novel features of the RRM fold. EMBO J. 2000;19:3132–3141. doi: 10.1093/emboj/19.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder SM, Kanaar R, Rio DC, Alber T. Absence of interdomain contacts in the crystal structure of the RNA recognition motifs of Sex-lethal. Proc Natl Acad Sci USA. 1999;96:4892–4897. doi: 10.1073/pnas.96.9.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A) binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu R-M. Crystal structure of the two RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset T, Wang C, Verheggen C, Chen D, Hernandez-Verdun D, Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Meier UT, Lewis N, Rekosh D, Hammarskjöld ML, Olson MOJ. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma. 1997;105:407–417. doi: 10.1007/BF02510477. [DOI] [PubMed] [Google Scholar]
- Dundr M, Mistelli T, Olson MOJ. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol. 2000;7:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edström J-E, Sierakowska H, Burvall K. Dependence of Balbiani ring induction in Chironomus tentans salivary gland cells on inorganic phosphate. Dev Biol. 1982;91:131–137. doi: 10.1016/0012-1606(82)90016-1. [DOI] [PubMed] [Google Scholar]
- Eichler DC, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acids Res Mol Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–810. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gadal O, Straub D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rp110p. Mol Cell Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNP-I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisolfi-Neto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- Gil A, Sharp PA, Jamison SF, Garcia Blanco MA. Characterization of cDNAs encoding the polypyrimidinetract binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Two different combinations of RNA-binding domains determine the RNA-binding specificity of nucleolin. J Biol Chem. 2001;276:14338–14343. doi: 10.1074/jbc.M011120200. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Benoit R, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in Ran-cycle and nucleoporin mutants. J Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Muto Y, Sakamoto H, Yokoyama S. NMR studies on functional structures of the AU-rich element-binding domains of HU antigen C. Nucleic Acids Res. 2000;28:1743–1750. doi: 10.1093/nar/28.8.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Muto Y, Green MR, Yokoyama S. Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF65) EMBO J. 1999;18:4523–4534. doi: 10.1093/emboj/18.16.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S-B, Zhao J, Björk P, Schmekel S, Ljungdahl PO, Wieslander L. Mrd1p is required for processing of pre-rRNA and for maintenance of steady-state levels of 40S ribosomal subunits in yeast. J Biol Chem. 2002;277:18431–18439. doi: 10.1074/jbc.M112395200. [DOI] [PubMed] [Google Scholar]
- Kim Y-J, Baker BS. Isolation of RRM-type RNA binding protein genes and the analysis of their relatedness by using a numerical approach. Mol Cell Biol. 1993;13:174–183. doi: 10.1128/mcb.13.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Wurtz T, Visa N, Daneholt B. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J. 1994;13:6052–6061. doi: 10.1002/j.1460-2075.1994.tb06952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Linder P, de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis. Mol Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interactions. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Tollervey D. The function and synthesis of ribosomes. Nature Mol. Cell Biol. 2001;2:514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- Lalev AI, Abeyrathne PD, Nazar RN. Ribosomal RNA maturation in Schizosaccharomyces pombe is dependent on a large ribonucleoprotein complex of the internal transcribed spacer I. J Mol Biol. 2000;302:65–77. doi: 10.1006/jmbi.2000.4015. [DOI] [PubMed] [Google Scholar]
- Lalev AI, Nazar RN. A chaperone for ribosome maturation. J Biol Chem. 2001;276:16655–16659. doi: 10.1074/jbc.M101157200. [DOI] [PubMed] [Google Scholar]
- Lambert B, Daneholt B. Microanalysis of RNA from defined cellular compartments. Methods Cell Biol. 1975;10:17–47. doi: 10.1016/s0091-679x(08)60728-1. [DOI] [PubMed] [Google Scholar]
- Lu J, Hall KB. An RBD that does not bind RNA: NMR secondary structure determination and biochemical properties of the C-terminal RNA binding domain from the human U1A protein. J Mol Biol. 1995;247:739–752. doi: 10.1006/jmbi.1995.0177. [DOI] [PubMed] [Google Scholar]
- Meyer B, Mähr R, Eppenberger H-M, Lezzi M. The activity of Balbiani rings 1 and 2 in salivary glands of Chironomus tentans larvae under different modes of development and after pilocarpine treatment. Dev Biol. 1983;98:265–277. doi: 10.1016/0012-1606(83)90357-3. [DOI] [PubMed] [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H. Maturation and intranuclear transport of pre-ribosomes require Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Nietfield W, Mentzel H, Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Patton JG, Mayer SA, Tempst P, Nadalginard B. Characterization and molecular cloning of polypyrimidine tract binding protein. A component of a complex necessary for pre-messenger-RNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Petersen U-M, Björklund G, Ip YT, Engström Y. The dorsal-related immunity factor, Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. EMBO J. 1995;13:3146–3158. doi: 10.1002/j.1460-2075.1995.tb07317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B"-U2A protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- Raué HA, Planta RJ. Ribosome biogenesis in yeast. Prog Nucleic Acid Res Mol Biol. 1991;41:91–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Senger B, Lafontaine DLJ, Graindorge J-S, Gadal O, Camasses A, Sanni A, Garnier J-M, Breitenbach M, Hurt E, Fasiolo F. The nucle(o)lar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- Serin G, Joseph G, Ghisolfi L, Bauzan M, Erard M, Amalric F, Bouvet P. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J Biol Chem. 1997;272:13109–13116. doi: 10.1074/jbc.272.20.13109. [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 Å resolution. Nat Struct Biol. 1997;4:215–222. doi: 10.1038/nsb0397-215. [DOI] [PubMed] [Google Scholar]
- Silve S, Volland C, Garnier C, Jund R, Chevallier MR, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol Biol Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Woolford JL., Jr The yeast nucleolar protein Nop4p contains four RNA recognition motifs necessary for ribosome biogenesis. J Biol Chem. 1997;272:25345–25352. doi: 10.1074/jbc.272.40.25345. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- Valásek L, Hasek J, Nielsen KH, Hinnebusch AG. Dual function of eIF3j/Hcr1p in processing 20S pre-rRNA and translation initiation. J Biol Chem. 2001;276:43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- Vanrobays E, Gleizes P-E, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M, Célugne J-P. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 2001;20:4204–4213. doi: 10.1093/emboj/20.15.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Biochem. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Visa N, Izzauralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds the Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- Wood WB. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Wurtz T, Kiseleva E, Nacheva G, Alzhanova-Ericsson A, Rosén A, Daneholt B. Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1996;16:1425–1435. doi: 10.1128/mcb.16.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp Cell Res. 1982;139:297–307. doi: 10.1016/0014-4827(82)90255-5. [DOI] [PubMed] [Google Scholar]
- Xu R-M, Jokhan L, Cheng X, Mayeda A, Krainer AR. Crystal structure of the human UP1, the domain of hnRNA A1 that contains two RNA-recognition motifs. Structure. 1997;5:559–570. doi: 10.1016/s0969-2126(97)00211-6. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]