Abstract
Background
Endoplasmic reticulum retention of misfolded cystic fibrosis transmembrane conductance regulator (CFTR) mutants and their rapid degradation is the major cause of cystic fibrosis (CF). An important goal is to understand the mechanism of how the misfolded proteins are recognized, retained, and targeted for degradation.
Results
Using a web-based algorithm, PESTFind, we found a PEST sequence in the regulatory (R) domain of CFTR. The PEST sequence is found in many short-lived eukaryotic proteins and plays a role in their degradation. To determine its role in the stability and degradation of misprocessed CFTR, we introduced a number of site-directed mutations into the PEST sequence in the cDNA of ΔF508 CFTR, the most prevalent misprocessed mutation found in CF patients. Analysis of these mutants showed that the disruption of the PEST sequence plays a minor role in the degradation of the CFTR mutants. Multiple mutations to the PEST sequence within the R domain of CFTR inhibit maturation of CFTR and prevent the formation of a 100 kDa degradation product. The mutations, however, do not improve the stability of the mutant ΔF508 CFTR.
Conclusion
These observations show that disruption of the structure of the R domain of CFTR can inhibit maturation of the protein and that the predicted PEST sequence plays no significant role in the degradation of CFTR.
Background
Mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene cause cystic fibrosis (CF), the most prevalent fatal recessive genetic disease in the Caucasian population [1]. CFTR is a polytopic integral membrane protein synthesized in the endoplasmic reticulum (ER) and normally expressed on the apical surface of epithelial cells where it functions as a phosphorylation-stimulated and ATP-dependent chloride channel. The majority of CF patients express processing defective CFTRs that fail to mature to the cell surface; instead, the processing defective CFTRs are retained in the ER and are targeted for rapid degradation [2,3].
The retention of processing defective CFTR is a response of the ER quality control system to misfolded proteins, which prevents the progression of misfolded or misassembled membrane and secretory proteins into later compartments of the secretory pathway [3]. During synthesis, nascent CFTR polypeptide chains are translated from ER membrane-bound ribosomes and are inserted into the ER membrane [3]. Various classes of chaperones associate with the nascent polypeptide both in the lumen of the ER and in the cytosol to aid in folding [4-7]. Upon proper folding, the properly folded CFTR dissociate from the chaperones and are packaged into transport vesicles for export to a post-ER compartment in the secretory pathway, the Golgi. Many of the missense mutations in CFTR retard the folding process. This leads to prolonged association of the nascent chains with the molecular chaperones and prevents the nascent chains from exiting the ER through the default secretory pathway. Instead, the misfolded polypeptides are retrotranslocated across the ER membrane, into the cytosol, and targeted for degradation by the ubiquitin-proteasome pathway [8].
Although much of the molecular mechanism of the ubiquitin-proteasome system has now been elucidated (reviewed in [9]), the precise mechanism and determinants of recognition of the misfolded polypeptides remain unclear [10]. As proposed by Chang et al. [11], the retention of misfolded CFTR is most likely due to the exposure of short sequence motifs specifically recognized by components of the ER quality control system or vesicular transport system; the mutations may cause localized misfolding leading to global misfolding to expose or bury motifs that signal for degradation, retention or exportation from the ER. Indeed, it has been shown that the removal of multiple arginine-framed ER retention/retrieval trafficking signals overcomes misprocessing of ΔF508 CFTR, the most prevalent processing defective CF mutation [11].
Furthermore, attempts to promote maturation of the processing defective mutants by shutting down the cytosolic proteasomes via proteasome inhibitors have led to the speculation of the existence of other systems responsible for the retention and degradation of these processing defective CFTR [12]. Treatment of cells expressing wild-type (WT) CFTR with MG-132, an inhibitor of the 26S proteasome in the ubiquitin-proteasome pathway, leads to inhibition of maturation of the CFTR polypeptide [12,13]. The resulting maturation-hindered WT CFTR polypeptide exhibit similar stability, structural, and functional properties to misprocessed CFTR mutants such as the prevalent ΔF508 CFTR [12,14].
PEST sequences are found in many rapidly degraded proteins. These sequences have been suggested to serve as signals for proteolytic degradation. From a survey of the amino acid sequences of 10 short-lived eukaryotic proteins, Rogers et al. [15] found the proteins to contain one or more regions rich in proline (P), glutamic acid (E), serine (S), and threonine (T). These regions are often flanked by positively charged amino acids. They named these regions PEST regions. Based on their observation, they developed an algorithm that would search for such regions in a given protein sequence.
In the present study, we examine the role of a PEST sequence, found in CFTR, in the stability and degradation of CFTR.
Results
PEST sequence of CFTR
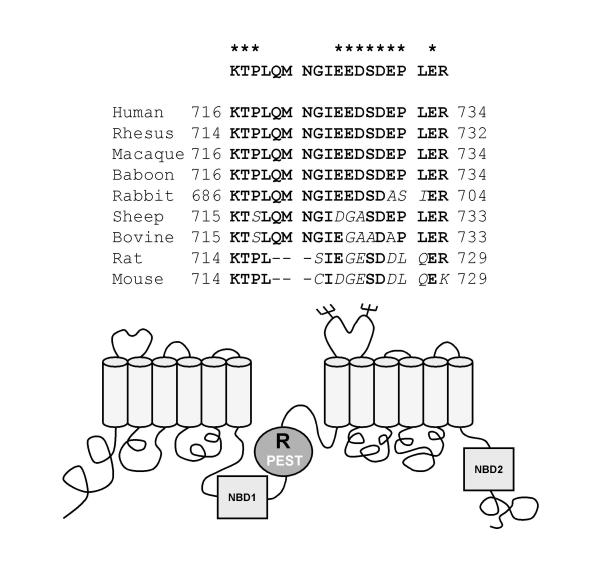
The algorithm, PESTFind, searches for hydrophilic regions of 12 or greater amino acids that contain at least one P (proline), one E (glutamic acid) or D (aspartic acid), and one S (serine) or T (threonine), flanked by K (lysine), R (arginine), or H (histidine) residues. The algorithm assigns a score to each possible PEST sequence found. The score ranges from -50 to +50, with a score above zero denoting a possible PEST region while a value greater than + 5 being of particular interest. Using the algorithm, PESTFind http://www.at.embnet.org/embnet/tools/bio/PESTfind/, we found a highly conserved PEST region in the regulatory domain of the CFTR protein (figure 1). The CFTR PEST region (residues 716 – 734) scored + 6.91 and had a hydrophobicity index of 32.63. Within the 19 residues PEST region, 8 are charged (i.e. 1 lysine, 1 arginine, 4 glutamic acid, and 2 aspartic acid), 4 polar (i.e. 1 threonine, 1 serine, 1 glutamine, and 1 asparagine), and 7 non-polar (i.e. 2 proline, 2 leucine, 1 methionine, 1 glycine, and 1 isoleucine). The low hydrophobicity suggests the region may be surface accessible to proteases or for protein-protein interaction with other proteins such as molecular chaperones, trafficking proteins, or components of proteolytic systems.
Figure 1.
Alignment of the PEST region of CFTR. The top line is the human PEST sequence used in the study. A cross-species alignment of the CFTR amino acid sequences from residues 716 to 734 of the human CFTR shows great conservation in the region. Discrepancies with the human CFTR are noted in italics. Significant PEST residues are noted with asteriks. The PEST region is in the Regulatory (R) domain of CFTR. Partial sequences were found using the PubMed http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed. Accession number and references for each species are as follows: Human (P13569; [1]), Rhesus (AAC14012; [56]), Macaque (AAF80467; unpublished; direct submission), Baboon (AAD46907; unpublished; direct submission), Rabbit (Q00554; [57]), Sheep (Q00555; [58]), Bovine (P35071; [58]), Rat (1901178A; unpublished; direct submission), Mouse (P26361; [59]). The lower panel shows a model of CFTR and location of the predicted PEST region in the R-domain (R). The cylinders represent predicted transmembrane domains and the squares represent the nucleotide-binding domains (NBD1 and NBD2).
Construction of PEST mutants
To examine whether the PEST sequence directly affects the processing or proteolytic degradation of CFTR, various CFTR PEST disrupting mutants were constructed into the ΔF508 CFTR cDNA using site-directed mutagenesis (table 1). An A52 epitope was also appended at the COOH-terminus for detecting transfected CFTR as opposed to endogenous CFTR. Table 1 lists the mutants with their sequences and PEST scores as determined by the algorithm, PESTFind. The mutants' PEST scores range from being poor PEST candidates (+4.07 for S728A and -26.19 for poly-valine) to being invalid PEST candidates (i.e. no score available). We were particularly interested in the stretch of polar and PEST-significant region, residues 725 to 731. For that region, we made a poly-valine mutant where we mutated the 6 consecutive polar/charged residues (725 – 730) to valine, a relatively small, neutral residue. This minimized the hydrophobicity of the PEST region and removed the majority of the significant PEST residues; the proline residue 731 was not altered because it may be important structurally (i.e. for formation of a β-turn). The E725K/E726K mutant was included to test if alteration of charges would affect the function of the PEST sequence. Since serine and threonine are often implicated in functional group modification such as phosphorylation, their role in the PEST sequence was examined. S728 and T717 were both altered to alanine, a small, neutral residue; S728A has PEST score of +4.07. However, since both S728A and T717A mutant have a high PEST score (T717A mutant alone has PEST score of +3.87), we added an additional E725K mutation to the T717A mutant just to further disrupt the PEST sequence. The mutants were also made in WT background. The WT-background constructs were included to ensure that the mutations themselves did not contribute to additional deleterious effects on the protein.
Table 1.
Mutations introduced into the predicted PEST sequence of CFTR
| Mutant | Sequence | PEST Score |
| Wild-type (WT) | 716 – KTPLQMNGIEEDSDEPLER – 734 | +6.91 |
| Poly-Valine | 716 – KTPLQMNGIVVVVVVPLER – 734 | -26.19 |
| E725K/E726K | 716 – KTPLQMNGIKKDSDEPLER – 734 | N/A |
| S728A | 716 – KTPLQMNGIEEDADEPLER – 734 | +4.07 |
| T717A/E725K | 716 – KAPLQMNGIKEDSDEPLER – 734 | N/A |
* Residues in bold are the mutations introduced.
Expression of PEST mutants
The cDNAs encoding the PEST mutants were transiently transfected into COS-1 cells. The transfected cells were harvested and lysed 3 days post-transfection, separated by electrophoresis and subjected to Western blotting using a mouse monoclonal antibody against the A52 tag [16,17] attached at the COOH-terminus of the protein. The two N-glycosylation sites on the putative extracellular loop 4 of CFTR (figure 1) enable us to determine the maturation state of CFTR. During its synthesis in the ER, two carbohydrate moieties are covalently attached onto the two N-glycosylation sites; this is the immature, core-glycosylated CFTR. The carbohydrate moieties are modified only upon the nascent polypeptide's maturation and progression through the Golgi apparatus onto the cell surface; this is the mature, fully-glycosylated CFTR. The modification of the carbohydrate moieties lead to a shift in the protein's apparent molecular mass: the core-glycosylated ER-resident CFTR has an apparent molecular mass of ~140 kDa while the fully-glycosylated CFTR expressed on the cell surface has an apparent molecular weight of ~170 kDa [18]. While glycosylation of CFTR is not crucial in the maturation CFTR, it serves as an indicator of CFTR's maturation state [3].
As shown in figure 2A, the mutants had no apparent effect on the maturation of CFTR with the exception of the poly-valine mutant. The WT background constructs, except the poly-valine mutant, all have similar levels of expression and maturation as the WT CFTR. The poly-valine construct in WT background is processing defective, similar to the ΔF508 mutant. All of the ΔF508 background mutants had no apparent effect on the maturation state of ΔF508 CFTR and are all immature. An over-exposure of the immunoblot (figure 2B), however, suggests removal of a protease site by the poly-valine mutations. Since we do not see any larger fragments (>100 kDa) in the poly-valine mutants, it is also possible that the poly-valine mutations introduced additional cleavage sites in the R domain. As shown in figure 2B, the 100 kDa degradation product that is present in ΔF508 is absent in the poly-valine constructs, both of the WT and ΔF508 background. There is, however, no improvement in maturation or in the level of expression.
Figure 2.
Expression of PEST Mutants in COS-1 Cells. A) COS-1 cells were transiently transfected with cDNA encoding the various PEST mutants in both WT and ΔF508 background. Whole cell extracts were subjected to SDS-PAGE and immunoblot analysis (monoclonal antibody against the A52 tag) as described in 'Materials and Methods'. B) An over-exposure of the immunoblot showed the disappearance of the CFTR 100 kDa degradation product with the introduction of the poly-valine constructs.
Biogenesis of PEST mutants
It is possible that the effect of disruption of the PEST region may be too subtle to be detected from a simple expression blot. Effects of the disruption of the PEST region may be only apparent by examining its stability in a pulse-chase experiment.
To examine whether the disruption of the PEST sequence affected the stability of the various mutants, the mutants' biosynthetic maturation were examined kinetically in pulse-chase experiments (figure 3). COS-1 cells were transiently transfected with the various constructs for 3 days prior to the pulse-chase experiments. For the pulse-chase experiment, the cells were starved for 30 minutes in sulfur-minus (S-) media depleted of methionine and cysteine. The cells were then pulsed in S- media supplemented with radiolabeled methionine for 30 minutes. The cells were then chased for 0, 2, 4, 6, 8, 12, and 24 hours in plain media. At each chase point, the cells were harvested and frozen in plain media containing 10% DMSO and stored in a -70°C freezer. After harvesting all the time points, the cells were lysed, immunoprecipitated with mouse anti-A52 antibody, separated by electrophoresis and detected by autoradiograph. As shown in figure 3A, all of the WT constructs, with the exception of the poly-valine constructs, were still stable after 12 hours of chase. All of the misprocessed constructs (ΔF508 background and the poly-valine in WT background), however, were completely degraded by the end of the 6 hours chase. There was no apparent difference in the stability of the constructs.
Figure 3.
Biosynthetic Maturation of CFTR PEST Mutants. A) Pulse-chase radiograph for non-processing defective constructs: WT, WT/E725K/E726K, and WT/T717A/E725K; results for WT/S728A not shown, but is similar to that of WT. B) Pulse-chase radiograph for processing defective constructs: ΔF508, ΔF508/poly-valine, ΔF508/E725K/E726K, and ΔF508/T717/E725K; results for WT/Poly-valine and ΔF508/S728A not shown but they show similar results as ΔF508. Pulse-chase experiments and immunoprecipitation were performed as described in the "Materials and Methods" section.
Global structural comparison of PEST mutants
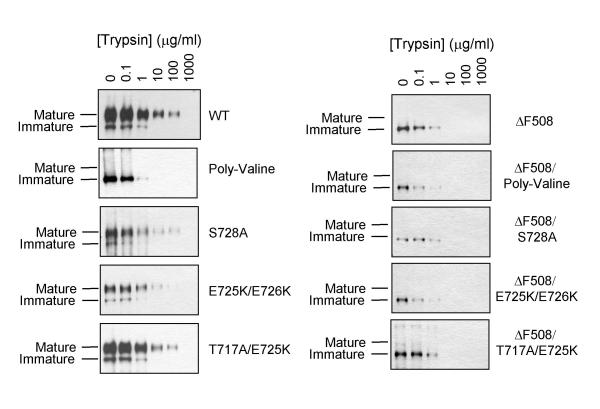
To ensure that the disruption of the PEST region did not cause significant structural differences in the protein, we compared the global structures by examining their sensitivity to trypsin digestion (figure 4). The trypsin-sensitivity assay is based on the rationale that trypsin-sensitivity of the various constructs depend on the exposure of trypsin-sensitive sites. It has been previously established that neither the subcellular location nor the glycosylation state of CFTR has a significant effect on its trypsin sensitivity [14]. Crude membranes were made from transiently transfected cells. The membranes were treated with various concentrations of trypsin at room temperature for 5 minutes. The reactions were stopped by the addition of lima bean trypsin inhibitor. The samples were separated by SDS-PAGE and subjected to Western blot with anti-A52 antibody. As shown in figure 4, mature CFTR were about 100–1000 fold more resistant to trypsin than the immature ER-resident CFTR. Thus, with the exception of the poly-valine mutant with WT background, the PEST sequence-disrupting mutants did not have any apparent effect on the folding pattern of the CFTR protein.
Figure 4.
Trypsin Sensitivity of CFTR PEST Mutants. Membranes were prepared from COS-1 cells transiently transfected with the cDNA of the various constructs. The membranes were treated with various concentrations (0–1000 μg/ml) of TPCK-treated trypsin. The reactions were stopped by the addition of lima bean trypsin inhibitor. Equivalent amounts of protein were subjected to immunoblot analysis (monoclonal antibody against the A52 tag).
Effect of MG-132 on PEST mutants
We then examined the effects of proteasome inhibitors on the expression of these mutants to test our hypothesis that the PEST region may act as a backup proteolytic system to the ubiquitin-proteasome system; as the ubiquitin-proteasome system has been shown to be the prevalent proteolytic system of misprocessed CFTR, it is possible for it to overshadow whatever proteolytic effects of the PEST region may possess. COS-1 cells transiently transfected with various cDNA of the PEST constructs were treated with 2 μM of MG-132. MG-132 is a potent peptide aldehyde inhibitor designed to enter mammalian cells to inhibit the ubiquitin-proteasome pathway [19,20]. As shown in figure 5, MG-132 blocks the maturation of WT CFTR and WT-background mutants. Inhibition of maturation by MG-132 has also been observed with P-glycoprotein [21]. The level of expression, however, remains similar to that of the immature CFTR in untreated cells. With the ΔF508 CFTR, poly-valine mutant and the other ΔF508-backgound mutants, however, there was lower expression in the presence of MG-132.
Figure 5.
Treatment with Proteasome Inhibitors. Transiently transfected COS-1 cells were treated with (+) or without (-) 2 μM MG-132. The cells were subsequently lysed and subjected to immunoblot analysis (monoclonal antibody against the A52 tag).
Discussion
Increasing the stability and promoting the maturation of misprocessed CFTR mutants has been a key interest in CF research. So far, not much is known about the actual mechanism of how these misprocessed CFTR are retained in the ER and targeted for degradation. Many lines of evidence have suggested the ER-associated degradation (ERAD) pathway, which includes the ubiquitin-protease pathway, as the dominant pathway for the disposal of misfolded CFTR in mammalian cells [4,22,23]. Misfolded or misassembled proteins, such as processing defective ΔF508 mutant CFTR, are recognized and retained in the ER by the quality control system in the ER; although the exact mechanism for recognition is yet to be elucidated, evidence indicate that the prolonged association with molecular chaperones may play a role in the retention of CFTR [5,24]. Ubiquitination, the covalent attachment of ubiquitin to a lysyl ε-amino group of the polypeptide, is necessary for degradation by the 26S proteasome [25]. Ubiquitin is a highly conserved polypeptide of 76 amino acids. CFTR may be both co- and post-translationally ubiquitinated in cell-free systems [26,27]. Although the precise mechanism is still unclear, ubiquitination and ER retention eventually lead to the retrotranslocation of the misfolded protein back into the cytosol via the sec61p translocon [27,28]. The misfolded protein is then targeted for degradation by the 26S proteasome [25].
There are still many questions about the ERAD pathway and the maturation process of WT CFTR: 1) Although the mechanism for the ubiquitination reaction is well understood [10], the recognition determinants for ubiquitination are still unknown. 2) How does the ERAD pathway recognize the misfolded protein? 3) Although both WT and ΔF508 CFTR are ubiquitinated, how is some WT CFTR still able to progress onto the post-ER compartments of the secretory pathway? In a recent report, CHIP, an Hsc70 co-chaperone was actually found to target immature CFTR for proteasomal degradation [5]. It is likely that there are other chaperones that function in a similar manner. So, what are the selective molecular determinants for chaperones such as the CHIP to target only immature CFTR but not the mature form? Furthermore, it is the general consensus that there must be a number of criteria for exiting the ER and progressing through the secretory pathway for expression on the cell surface. Peptide motifs and signals may be exposed or buried depending on the folding state of the protein, thus providing the cell with a means of quality control. The perfect example is the four arginine-framed trafficking signals in CFTR discovered by Chang et al. [11]. The arginine-framed trafficking signal, however, may just be one of the many types of signals.
There are a number of peptide motifs identified in rapidly degraded proteins that target proteins for rapid degradation: PEST regions [29], lysosome-targeting KFERQ motifs [30], and the cyclin destruction box responsible for eukaryotic cell cycle [31]. The PEST region is of particular interest to us because it has been found in many rapidly-degraded proteins and shown to contribute to their degradation [15,29,32]. Although the presence of PEST motifs does not necessarily lead to constitutive degradation of the protein, there were a number of reasons why we thought it was worth investigating. Aside from its proteolytic roles, PEST motifs have also been found to be involved in protein-protein interactions: direct interaction with ubc9 [33] and ligand recognition [34,35]. The PEST region of CFTR lies within its R domain, which is not present in its processing efficient sister protein, P-glycoprotein [36]. Thus, apart from a potential proteolytic site, the PEST region in CFTR also has the potential to interact with ubiquitination proteins, other proteases, or molecular chaperones. We were interested in seeing if it affects the maturation, folding, and stability of the CFTR protein. To this end, a number of constructs were made via site-directed mutagenesis aiming to disrupt the PEST region and examine the stability and folding pattern of the constructs.
To narrow the scope of our study in this paper, we tested the PEST sequence's potential role as a proteolytic site in misprocessed CFTR. A number of mutants were made to disrupt the PEST sequence in the processing defective ΔF508 CFTR mutant in an attempt to rescue it. The mutants' stability and maturation was examined via pulse-chase experiments and their global structure compared by testing their trypsin sensitivity. Overall, it was found that perturbation to the PEST sequence (residues 716 – 734) of CFTR made no apparent improvement on the stability of ΔF508 CFTR.
In our survey of PEST-disrupting mutants, we included the WT-background mutants as well as the processing defective ΔF508-background mutants as a control to ensure the missense mutations themselves did not cause other deleterious effect on the protein. The misprocessed poly-valine construct proves this precaution to be necessary.
The observation that poly-valine caused defective processing came as a surprise since the R domain has been previously shown to be predominantly random coil [37]. Although the boundaries of the R domain are still not precisely defined, the R domain extends approximately from residues 634–708 at the NH2-terminus to approximately 835 at the COOH-terminus [37-39]. Various studies have shown that the deletion of the R-domain yields cell surface expressing, functional CFTR [40-45]. Furthermore, of the 19 disease-associated mutations in the R-domain examined by Vankeerberghen et al. [46], none of the clinical missense mutations studied within the boundaries of the R domain (residues 634 – 835) was processing defective. Thus, it appeared that the R-domain did not contain significant structural information.
However, the fact that the poly valine mutant used in our study leads to such dramatic disturbance as to cause the mutant protein to not mature suggests otherwise. Using a web-based secondary structure prediction program, NNPREDICT http://www.cmpharm.ucsf.edu/~nomi/nnpredict, the PEST region and surrounding regions (residues 712–737) contain turning elements, with 3 helical elements. With the PEST region's low hydrophobicity, the region is likely to be surface-accessible. The poly-valine mutant, however, introduces beta strand elements in the middle of the turning elements. This could have led to disruption of a turn, and thus disrupting global structure. If the primary sequence around the PEST region does indeed carry structural information, perturbation of the PEST region by the other mutations might have also led to structural change and the trypsin assay might not be sensitive enough to detect them. These structural changes might have led to exposure of other proteolytic sites to counter the loss of proteolytic effect caused by alternation of the PEST sequence, hence, no apparent improvement in stability. An alternative test of structural change would be to find cysteine cross-linking pairs in CFTR and test if they still exist in the mutants [47]. This would act as an additional structural check of the mutants.
The PEST region may still interact with other proteins. The c-Myb transcription factor is a highly regulated nuclear phosphoprotein involved in the regulation of proliferation, differentiation, and apoptosis of immature hematopoietic cells. Its activity and proteolytic stability are, in part, regulated by the ubiquitin-proteasome system. c-Myb contains a PEST motif in its regulatory domain at its COOH-terminus. The PEST motif was found to directly interact with Ubc9. Ubc9 covalently attaches SUMO-1 protein, an ubiquitin-like protein, onto the lysyl residue near the PEST motif [33]. Similarly, Ubc9 interacts with the PEST region of another nuclear SUMO-1 target protein, HIPK2 [48]. Deletion of the PEST motif, however, does not increase the stability of the protein, suggesting other additional factors may contribute to the proteolytic stability of the proteins.
There are a number of lysyl residues to the NH2-terminus of the PEST region and may act as potential candidates for ubiquitination. If CFTR's PEST motif also acts as a binding site for ubiquitin-conjugating enzymes like Ubc9 was for SUMO-1, that would explain why no difference was observed in the treatment of proteasome inhibitors between the WT and PEST mutants; the PEST motif would be part of the ubiquitin-proteasome pathway and not a separate proteolytic pathway. A potential candidate may be the Ubc6, an ER-resident ubiquitin-conjugating enzyme. In a recent report, mutation in Ubc6 was able increase the stability of ΔF508 CFTR by 2 fold [22].
Conclusions
Our results suggest the PEST region in the R domain of CFTR plays no discernible role in degradation of CFTR and that there may be structural information encoded in the R domain as mutations in the region may lead to misprocessing of the protein. The issue of whether CFTR's PEST motif can act as a recognition site, for other proteins, however, has not yet been addressed. As retention and degradation of misfolded CFTR is likely a collective work by many protein complexes, a survey of components of the proteolytic systems, molecular chaperones may yield more insight into the whether PEST motif plays a role in the degradation of CFTR.
Methods
Construction of mutants
The CFTR point mutations were constructed and inserted into the mammalian expression vectors pMT21 as previously described [18,49]. The full-length cDNA of all constructs were modified to encode the epitope for monoclonal antibody A52 [50] and 6 histidine residues at the C-terminus [51]. The sequence at the COOH terminus that would normally end DTRL then became DTRRAISLISNSCSPEFDDLPLAEQREACRRGD (His)6PRQ. Briefly, a BssHII restriction site was constructed at amino acid residue 1480 via site-directed mutagenesis using the DNA oligomer AAGATACAAGGCGCGCGAGAGCAGCAT to alter the Leu and stop codon to Arg and Ala. The DNA fragment encoding the A52 tag with a 5' BssHII site was then ligated to the 3' of the cftr gene.
Expression of wild-type CFTR and mutants
Subconfluent COS-1 cells were transfected with the cDNA constructs with 1 μg/ml of the various vector constructs using a calcium phosphate precipitation method adapted from Chen and Okayama [52]. The media was replaced with plain media at 20 hours post-transfection. The cells were harvested after another 48 hours.
Isolation of microsomal membrane vesicles for trypsin digestion
The crude membranes were prepared in the same manner as optimized by Loo and Clarke [53] for trypsin digestion assays. Briefly, for each CFTR construct, 20 (100 mm) tissue culture plates of transfected COS-1 cells were harvested and washed with PBS (phosphate-buffered saline). The cell pellets were subsequently resuspended in 3 ml of low ionic strength buffer (10 mM Tris-HCl, pH 7.5, 0.5 mM MgCl2). After incubating on ice for 20 min., the samples were homogenized using 40 strokes in a loose fitting Dounce homogenizer, followed by 20 strokes after addition of 3 ml of sucrose buffer (10 mM Tris-HCl, pH 7.5, 500 mM sucrose, 0.3 M KCl). The cell debris was removed by centrifugation at 3000 × g for 7 min. at 4°C. The microsomes were collected from the supernatant by centrifugation at 44,000 × g for 45 min. at 4°C and resuspended with 300 μl of TBS (Tris-buffered saline; 10 mM Tris-HCl, pH 7.5, 150 mM NaCl). The membrane vesicles were immediately used for the trypsin digestion.
Trypsin digestion
As described previously [54], membrane vesicles from various constructs were treated with different concentrations of TPCK-trypsin (L-1-tosyl-amido-2-phenylethylchloromethyl ketone-treated trypsin; Sigma, 12,000 BAEE units per mg) at room temperature for 5 min.; the final concentrations of TPCK-trypsin were: 0, 0.1, 1, 10, 100, and 1000 μg/ml. The reactions were stopped by the addition of 2 mg/ml lima bean trypsin inhibitor (Worthington). The vesicles were solubilized with 2X sample buffer (3% SDS, 5% β-mercaptoethanol, 10% glycerol, 62.5 mM Tris-HCl, pH 6.8). The samples were then subjected to SDS-PAGE and immunoblot analysis using mouse A52 monoclonal antibody as the primary antibody and a horseradish peroxidase labeled goat anti-mouse antibody as the secondary antibody. The signals were detected by chemiluminescence (enhanced chemiluminescence, Pierce).
Pulse chase
Using methods adapted from Loo and Clarke [55], COS-1 cells expressing various CFTR constructs were radiolabeled and biogenesis of CFTR was followed. For each CFTR construct, 8 (100 mm) tissue culture plates of subconfluent COS-1 cells were transfected. At 24 hours post-transfection, the transfection media were replaced with plain media. At 72 hours post-transfection, the cells were starved for 40 minutes in Dulbecco's Modified Eagle Medium (DMEM) without L-methionine or L-cystine (Invitrogene Life Technologies). After aspirating the sulfur-deficient medium, the cells were labeled for 30 minutes in sulfur-deficient DMEM supplemented with 40 μCi/ml [35S] L-methionine and [35S] L-cystine (ICN Radiochemicals). The samples were then chased with plain media. One (100 mm) tissue culture plate from each construct was harvested at various chase time point, resuspended in 10% dimethylsulfoxide-containing media, and frozen in -70°C freezer.
Immunoprecipitation of CFTR
After thawing, each harvested pulse-chase sample was washed in PBS and subsequently resuspended in 100 μl PBS. The samples were then lysed in 1 mL buffer I (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 % (v/v) Triton X-100, 0.5 % (w/v) deoxycholic acid, 1 mM EDTA, pH 8.0) with protease inhibitors (50 μg/ml AEBSF, 10 μg/ml aprotinin, 25 μg/ml benzamidine, 1 μg/ml E64, and 0.5 μg/ml leupeptin). After removing the cell debris via centrifugation at 16,000 × g for 5 minutes, the supernatants were incubated with 11 μg of monoclonal A52 antibody at 4°C for 1.5 hour. 50 μl 50 % (v/v) slurry of Protein A beads in TBS (Protein A sephrose 4 Fast Flow; Amersham Pharmacia) was added to each sample and incubated at 4°C for 2 hours. The Protein A beads were then washed 3 times with buffer I. The CFTR proteins were then eluted with 50 μl 2X Sample Buffer with 2% β-mercaptoethanol. The samples were then subjected to separation on 6% SDS-acrylamide gel via SDS-PAGE. The gels were fixed in 10% acetic acid for 30 minutes and the 35S signal amplified in Amplify solution (Amersham Pharmacia) for 30 minutes. The gels were then dried and exposed to X-OMAT AR film (Kodak) for 12 hours – 1 week @ -70°C.
Author's Contributions
E.Y.C. carried out the experimental aspects of the studies and the preparation of the manuscript. D.M.C. conceived of the study and participated in its design and coordination.
Acknowledgments
Acknowledgment
We thank Dr. Tip W. Loo for technical support and comments on the manuscript. The A52 epitope and antibody was a gift from Dr. David H. MacLennan. The pMT21 expression vector was a gift from Dr. Randal Kaufman (Boston, MA). This work was supported by grants from the Canadian Cystic Fibrosis Foundation, the Canadian Institutes for Health Research, and the National Institutes of Health (R01-CA80900). D.M.C. is a Scientist of the Canadian Institutes for Health Research and the Canadian Cystic Fibrosis Foundation Zellers Senior Scientist. E.Y.C. is a recipient of the Canadian Institutes for Health Research Doctoral Research Awards and the Canadian Cystic Fibrosis Foundation Doctoral Studentship.
Contributor Information
Eva Y Chen, Email: evayj.chen@utoronto.ca.
David M Clarke, Email: david.clarke@utoronto.ca.
References
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui L-C. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Ko YH, Pedersen PL. Cystic fibrosis: a brief look at some highlights of a decade of research focused on elucidating and correcting the molecular basis of the disease. J Bioenerg Biomembr. 2001;33:513–521. doi: 10.1023/A:1012831322753. [DOI] [PubMed] [Google Scholar]
- Riordan JR. Cystic fibrosis as a disease of misprocessing of the cystic fibrosis transmembrane conductance regulator glycoprotein. Am J Hum Genet. 1999;64:1499–1504. doi: 10.1086/302429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Loo MA, Jensen TJ, Cui L, Hou Y, Chang XB, Riordan JR. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pind S, Riordan JR, Williams DB. Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1994;269:12784–12788. [PubMed] [Google Scholar]
- Kopito RR. Biosynthesis and degradation of CFTR. Physiol Rev. 1999;79:S167–173. doi: 10.1152/physrev.1999.79.1.S167. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Wade RC. Elusive recognition determinants for ubiquitination. J Mol Recognit. 2002;15:3–5. doi: 10.1002/jmr.561. [DOI] [PubMed] [Google Scholar]
- Chang XB, Cui L, Hou YX, Jensen TJ, Aleksandrov AA, Mengos A, Riordan JR. Removal of multiple arginine-framed trafficking signals overcomes misprocessing of delta F508 CFTR present in most patients with cystic fibrosis. Mol Cell. 1999;4:137–142. doi: 10.1016/S1097-2765(00)80196-3. [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Chen EY, Bartlett MC, Clarke DM. Cystic fibrosis transmembrane conductance regulator has an altered structure when its maturation is inhibited. Biochemistry. 2000;39:3797–3803. doi: 10.1021/bi992620m. [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Functional consequences of phenylalanine mutations in the predicted transmembrane domain of P-glycoprotein. J Biol Chem. 1993;268:19965–19972. [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Functional consequences of proline mutations in the predicted transmembrane domain of P-glycoprotein. J Biol Chem. 1993;268:3143–3149. [PubMed] [Google Scholar]
- Seibert FS, Jia Y, Mathews CJ, Hanrahan JW, Riordan JR, Loo TW, Clarke DM. Disease-associated mutations in cytoplasmic loops 1 and 2 of cystic fibrosis transmembrane conductance regulator impede processing or opening of the channel. Biochemistry. 1997;36:11966–11974. doi: 10.1021/bi9712652. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–71. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/S0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. The human multidrug resistance P-glycoprotein is inactive when its maturation is inhibited: potential for a role in cancer chemotherapy. FASEB J. 1999;13:1724–1732. doi: 10.1096/fasebj.13.13.1724. [DOI] [PubMed] [Google Scholar]
- Lenk U, Yu H, Walter J, Gelman MS, Hartmann E, Kopito RR, Sommer T. A role for mammalian Ubc6 homologues in ER-associated protein degradation. J Cell Sci. 2002;115:3007–3014. doi: 10.1242/jcs.115.14.3007. [DOI] [PubMed] [Google Scholar]
- Gelman MS, Kannegaard ES, Kopito RR. A principal role for the proteasome in endoplasmic reticulum-associated degradation of misfolded intracellular cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2002;277:11709–11714. doi: 10.1074/jbc.M111958200. [DOI] [PubMed] [Google Scholar]
- Brodsky JL. Chaperoning the maturation of the cystic fibrosis transmembrane conductance regulator. Am J Physiol Lung Cell Mol Physiol. 2001;281:L39–L42. doi: 10.1152/ajplung.2001.281.1.L39. [DOI] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/S0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- Xiong X, Chong E, Skach WR. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J Biol Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- Bebok Z, Mazzochi C, King SA, Hong JS, Sorscher EJ. The mechanism underlying cystic fibrosis transmembrane conductance regulator transport from the endoplasmic reticulum to the proteasome includes Sec61beta and a cytosolic, deglycosylated intermediary. J Biol Chem. 1998;273:29873–29878. doi: 10.1074/jbc.273.45.29873. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. doi: 10.1016/0968-0004(96)10031-1. [DOI] [PubMed] [Google Scholar]
- Dice JF. Microinjected ribonuclease A as a probe for lysosomal pathways of intracellular protein degradation. J Protein Chem. 1988;7:115–127. doi: 10.1007/BF01025241. [DOI] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/S0959-437X(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Olmo MT, Urdiales JL, Pegg AE, Medina MA, Sanchez-Jimenez F. In vitro study of proteolytic degradation of rat histidine decarboxylase. Eur J Biochem. 2000;267:1527–1531. doi: 10.1046/j.1432-1327.2000.01153.x. [DOI] [PubMed] [Google Scholar]
- Bies J, Markus J, Wolff L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J Biol Chem. 2002;277:8999–9009. doi: 10.1074/jbc.M110453200. [DOI] [PubMed] [Google Scholar]
- Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat Struct Biol. 2001;8:998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloviev MM. Detection of a surface-exposed PEST like sequence in the metabotropic glutamate receptor mGluR1 alpha. Bioinformatics. 2000;16:837–838. doi: 10.1093/bioinformatics/16.9.837. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Correction of defective protein kinesis of human P-glycoprotein mutants by substrates and modulators. J Biol Chem. 1997;272:709–712. doi: 10.1074/jbc.272.2.709. [DOI] [PubMed] [Google Scholar]
- Ostedgaard LS, Baldursson O, Vermeer DW, Welsh MJ, Robertson AD. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc Natl Acad Sci USA. 2000;97:5657–5662. doi: 10.1073/pnas.100588797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard LS, Baldursson O, Welsh MJ. Regulation of the cystic fibrosis transmembrane conductance regulator Cl-channel by its R domain. J Biol Chem. 2001;276:7689–7692. doi: 10.1074/jbc.R100001200. [DOI] [PubMed] [Google Scholar]
- Csanady L, Chan KW, Seto-Young D, Kopsco DC, Nairn AC, Gadsby DC. Severed channels probe regulation of gating of cystic fibrosis transmembrane conductance regulator by its cytoplasmic domains. J Gen Physiol. 2000;116:477–500. doi: 10.1085/jgp.116.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Zhao J, Davis PB, Ma J. Conformation, independent of charge, in the R domain affects cystic fibrosis transmembrane conductance regulator channel openings. Biophys J. 2000;78:1293–1305. doi: 10.1016/S0006-3495(00)76685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankeerberghen A, Lin W, Jaspers M, Cuppens H, Nilius B, Cassiman JJ. Functional characterization of the CFTR R domain using CFTR/MDR1 hybrid and deletion constructs. Biochemistry. 1999;38:14988–14998. doi: 10.1021/bi991520d. [DOI] [PubMed] [Google Scholar]
- Ma J, Zhao J, Drumm ML, Xie J, Davis PB. Function of the R domain in the cystic fibrosis transmembrane conductance regulator chloride channel. J Biol Chem. 1997;272:28133–28141. doi: 10.1074/jbc.272.44.28133. [DOI] [PubMed] [Google Scholar]
- Rich DP, Gregory RJ, Anderson MP, Manavalan P, Smith AE, Welsh MJ. Effect of deleting the R domain on CFTR-generated chloride channels. Science. 1991;253:205–207. doi: 10.1126/science.1712985. [DOI] [PubMed] [Google Scholar]
- Rich DP, Gregory RJ, Cheng SH, Smith AE, Welsh MJ. Effect of deletion mutations on the function of CFTR chloride channels. Receptors Channels. 1993;1:221–232. [PubMed] [Google Scholar]
- Zhang L, Wang D, Fischer H, Fan PD, Widdicombe JH, Kan YW, Dong JY. Efficient expression of CFTR function with adeno-associated virus vectors that carry shortened CFTR genes. Proc Natl Acad Sci USA. 1998;95:10158–10163. doi: 10.1073/pnas.95.17.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankeerberghen A, Wei L, Jaspers M, Cassiman JJ, Nilius B, Cuppens H. Characterization of 19 disease-associated missense mutations in the regulatory domain of the cystic fibrosis transmembrane conductance regulator. Hum Mol Genet. 1998;7:1761–1769. doi: 10.1093/hmg/7.11.1761. [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. Introduction of the Most Common Cystic Fibrosis Mutation (Delta F508) into Human P-glycoprotein Disrupts Packing of the Transmembrane Segments. J Biol Chem. 2002;277:27585–27588. doi: 10.1074/jbc.C200330200. [DOI] [PubMed] [Google Scholar]
- Kim YH, Choi CY, Kim Y. Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc Natl Acad Sci USA. 1999;96:12350–12355. doi: 10.1073/pnas.96.22.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibert FS, Linsdell P, Loo TW, Hanrahan JW, Riordan JR, Clarke DM. Cytoplasmic loop three of cystic fibrosis transmembrane conductance regulator contributes to regulation of chloride channel activity. J Biol Chem. 1996;271:27493–27499. doi: 10.1074/jbc.271.44.27493. [DOI] [PubMed] [Google Scholar]
- Zubrzycka-Gaarn E, MacDonald G, Phillips L, Jorgensen AO, MacLennan DH. Monoclonal antibodies to the Ca2+ + Mg2+-dependent ATPase of sarcoplasmic reticulum identify polymorphic forms of the enzyme and indicate the presence in the enzyme of a classical high-affinity Ca2+ binding site. J Bioenerg Biomembr. 1984;16:441–464. doi: 10.1007/BF00743238. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. P-glycoprotein. Associations between domains and between domains and molecular chaperones. J Biol Chem. 1995;270:21839–21844. doi: 10.1074/jbc.270.45.27099. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Rapid purification of human P-glycoprotein mutants expressed transiently in HEK 293 cells by nickel-chelate chromatography and characterization of their drug-stimulated ATPase activities. J Biol Chem. 1995;270:21449–21452. doi: 10.1074/jbc.270.45.27099. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Superfolding of the partially unfolded core-glycosylated intermediate of human P-glycoprotein into the mature enzyme is promoted by substrate-induced transmembrane domain interactions. J Biol Chem. 1998;273:14671–14674. doi: 10.1074/jbc.273.24.14671. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Quality control by proteases in the endoplasmic reticulum. Removal of a protease-sensitive site enhances expression of human P-glycoprotein. J Biol Chem. 1998;273:32373–32376. doi: 10.1074/jbc.273.49.32373. [DOI] [PubMed] [Google Scholar]
- Wine JJ, Glavac D, Hurlock G, Robinson C, Lee M, Potocnik U, Ravnik-Glavac M, Dean M. Genomic DNA sequence of Rhesus (M. mulatta) cystic fibrosis (CFTR) gene. Mamm Genome. 1998;9:301–305. doi: 10.1007/s003359900753. [DOI] [PubMed] [Google Scholar]
- Hart P, Warth JD, Levesque PC, Collier ML, Geary Y, Horowitz B, Hume JR. Cystic fibrosis gene encodes a cAMP-dependent chloride channel in heart. Proc Natl Acad Sci USA. 1996;93:6343–6348. doi: 10.1073/pnas.93.13.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond G, Scanlin TF, Zasloff MA, Bevins CL. A cross-species analysis of the cystic fibrosis transmembrane conductance regulator. Potential functional domains and regulatory sites. J Biol Chem. 1991;266:22761–22769. [PubMed] [Google Scholar]
- Tata F, Stanier P, Wicking C, Halford S, Kruyer H, Lench NJ, Scambler PJ, Hansen C, Braman JC, Williamson R, Wainwright BJ. Cloning the mouse homolog of the human cystic fibrosis transmembrane conductance regulator gene. Genomics. 1991;10:301–307. doi: 10.1016/0888-7543(91)90312-3. [DOI] [PubMed] [Google Scholar]