Abstract
The LOV1 domain of the blue light Phot1-receptor (phototropin homolog) from Chlamydomonas reinhardtii has been studied by vibrational spectroscopy. The FMN modes of the dark state of LOV1 were identified by preresonance Raman spectroscopy and assigned to molecular vibrations. By comparing the blue-light-induced FTIR difference spectrum with the preresonance Raman spectrum, most of the differences are due to FMN modes. Thus, we exclude large backbone changes of the protein that might occur during the phototransformation of the dark state LOV1-447 into the putative signaling state LOV1-390. Still, the presence of smaller amide difference bands cannot be excluded but may be masked by overlapping FMN modes. The band at 2567 cm−1 is assigned to the S-H stretching vibration of C57, the residue that forms the transient thio-adduct with the chromophore FMN. The occurrence of this band is evidence that C57 is protonated in the dark state of LOV1. This result challenges conclusions from the homologous LOV2 domain from oat that the thiolate of the corresponding cysteine is the reactive species.
INTRODUCTION
Blue-light reception in plants leads to phototropic plant movement, chloroplast relocation, and opening of stomatal guard cells (see Briggs and Christie, 2002, for a recent review). The light receptor for these tasks has recently been identified and later named phototropin (Phot) (Christie et al., 1999). All Phot proteins comprise a so-called LOV-sensitive domain and a downstream kinase. Most of the phototropins have two closely related LOV domains (LOV1 and LOV2) and a serine/threonine kinase. However, a phototropin related pigment (Ytva) from Bacillus subtilis has been characterized that contains only one LOV domain (Losi et al., 2002). Recently, the first Phot gene has been discovered in the green alga Chlamydomonas reinhardtii. It was found that under nitrogen starvation, the formation of sexually competent gametes, which is a blue-light-dependent process in wild-type cells, is inhibited in Phot1-deprived antisense transformants. Thus, at least one function of Phot1 is the regulation of gametogenesis under low nutrient conditions (Huang et al., 2002).
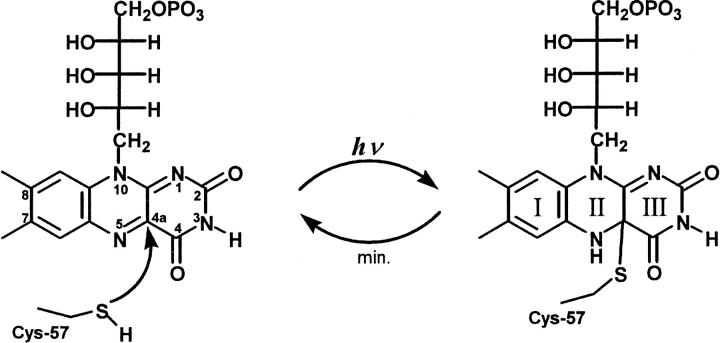
The chromophore of the LOV domains is FMN (Fig. 1), noncovalently bound to the apoprotein (Christie et al., 1999). The three-dimensional structure of the LOV2 domain from Adiantum capillus-veneris (Crosson and Moffat, 2001) and the LOV1 domain from C. reinhardtii (Fedorov et al., 2003) have been solved by x-ray crystallography to 2.7 Å and 1.9 Å, respectively. The overall protein fold of both LOV domains is nearly identical and characteristic for the large family of Per-Arnt-Sim (PAS) domains. At the polar end, the isoalloxazine ring of FMN is hydrogen-bonded to asparagine and glutamine residues whereas the apolar end of the ring is coordinated via hydrophobic interactions. The terminal phosphate group of the ribityl side chain interacts with two nearby arginine residues. A reactive cysteine residue (C57) is ∼4 Å away from the C4a of FMN, the site where it is covalently attached after illumination. The same structural principles apply to the single LOV domain of the Ytva protein from B. subtilis (Losi et al., 2002).
FIGURE 1.
The chromophore FMN and the light-induced thiol adduct where the sulfur of C57 is covalently linked to the carbon at position 4a of the isoalloxazine ring.
The LOV domains absorb blue light with the absorbance maximum at around 450 nm and vibronic side bands at 425 and 475 nm. The band of the higher electronic transition (S0–S2) is observed at 360 nm. The fluorescence emission peaks at 495 nm with a shoulder at 520 nm. The fluorescence quantum yield for LOV1 has been determined to 0.17 and the associated fluorescence lifetime is 2.9 ns (Holzer et al., 2002).
After light excitation, the LOV domains exert a self-contained photocycle (Salomon et al., 2001; Swartz et al., 2001; Kasahara et al., 2002). The first intermediate is strongly red-shifted (λmax = 710 nm in LOV1 from C. reinhardtii and 660 nm in LOV2 from oat) and corresponds most probably to the triplet state of FMN. This state decays in the early microsecond domain to a strongly blue-shifted intermediate (λmax = 390 nm), which has been suggested to represent the signaling state (Swartz et al., 2001). In LOV-390, the cysteine residue in the chromophore binding pocket is covalently linked to FMN via the C4a to form the thio-photoproduct (Fig. 1) (Salomon et al., 2001; Crosson and Moffat, 2002; Fedorov et al., 2003).
The time-constant for the transition of LOV1-390 back to the initial dark state LOV1-447 is in the range of minutes. For LOV1 from C. reinhardtii this reaction is strongly accelerated in the alkaline pH region suggesting a rate-limiting base-catalyzed reaction during the breakage of the thio-adduct (Kottke et al., 2003). The involvement of a proton transfer limited reaction in the recovery of the dark state of LOV2 from oat has been inferred from the kinetic isotope effect (KIE = 3) upon H2O/D2O exchange (Swartz et al., 2001).
The described chemical transitions during the light-activated turnover of the LOV domain can be investigated in detail by vibrational spectroscopy. Resonance-Raman spectroscopy selectively probes the vibrational modes of those molecules whose electronic transition is excited by the laser. The excitation far from the resonance still leads to the enhancement of chromophore bands, though with much lesser efficiency. The major advantage of the so-called preresonance approach is the fact that strongly fluorescent molecules are accessible by Raman spectroscopy. Whereas resonance Raman spectroscopy selects for the chromophore modes, vibrations of the whole protein are detected by infrared spectroscopy. To resolve single vibrations from the manifold of vibrations of a protein, reaction-induced FTIR difference spectroscopy is employed (see Vogel and Siebert, 2000, for a recent review). In this approach, the protein is activated to form a reaction intermediate. The recorded infrared spectrum is ratioed against the spectrum of the dark state. Both vibrational methods (Raman scattering and infrared absorption) were extremely helpful in the elucidation of the reaction mechanism of a prototypic chromophoric protein, bacteriorhodopsin, where conformational changes of the cofactor, the protein backbone, and amino acid side chains, as well as proton transfer steps and changes in hydrogen-bonding, have been detected (Mathies, 1995;Heberle et al., 2000).
Vibrational spectroscopy on flavoproteins has been mainly performed by resonance-Raman spectroscopy. It was used to study the chemical state of the flavin, i.e., protonation and redox state, and how it is modulated by the immediate protein environment (McFarland, 1987). By these means, details of the catalytic mechanism of flavin-binding enzymes could be resolved (Murgida et al., 2001; Zheng et al., 2001; Zheng et al., 1999).
In this report, we measured the vibrational spectrum of the FMN chromophore of LOV1 by preresonance Raman spectroscopy. The FMN bands have been assigned on the basis of previous vibrational analysis of free FMN. By these means, we were able to identify the FMN modes in the light-induced infrared difference spectrum of LOV1. The remainder of the bands in the FTIR difference spectrum arises from the apoprotein. The band at 2567 cm−1 could be unequivocally assigned to the thiol S-H stretching vibration of C57, the residue that forms the thio-adduct with FMN. The fact that C57 is protonated before blue-light excitation is of central importance for the reaction mechanism of LOV1 and challenges earlier interpretations of the pH-dependence of the fluorescence emission of LOV2 that the thiolate of C57 is the reactive species (Swartz et al., 2001). Other amino acid side-chain vibrations were tentatively assigned on the basis of their frequency and their spatial location in the three-dimensional structure. It is noted that similar work on the LOV2 domain of Phot1 from oat has been published during the review process of this manuscript (Swartz et al., 2002).
MATERIAL AND METHODS
LOV1 from C. reinhardtii (NCBI entry: AJ416556) is expressed in Escherichia coli in high yield (60–70 mg purified protein per 1 l medium). (For further details on expression, purification, and characterization by UV/Vis spectroscopy, see Kottke et al., 2002). The protein was used in a final concentration of ∼700 μM. The solution is buffered with 10 mM phosphate to pH 8. For isotopic replacement, the sample was concentrated (Centricon YM-10, Amicon, Beverly, MA) and diluted with phosphate buffered D2O at least three times.
FTIR spectroscopy
FTIR difference spectroscopy was performed on a Bruker IFS 66v spectrometer. In the conventional transmission technique, a droplet of the sample solution was applied to a BaF2 cuvette and sealed by a cover window of the same material (see Heberle and Zscherp, 1996, for more experimental details). The temperature of the sample was adjusted to 20°C by a circulating water bath in all experiments. Spectra were recorded across the whole midinfrared range (7000–800 cm−1) with 2 cm−1 resolution. Stray light from the exciting blue-light source (Schott KL 1500 equipped with BG 12 filter) was blocked by a germanium filter placed in front of the mercury cadmium telluride detector.
For ATR spectroscopy, where sample conditions are easier to control, an out-of-compartment accessory was used that employs a diamond disk as the internal reflection element (Nyquist et al., 2001). A droplet of 20 μl of the protein solution was applied to the diamond surface and concentrated by a stream of nitrogen until the ratio of the band heights around 1650 cm−1 (amide I overlapped by H2O bending mode) and 1550 cm−1 (amide II) was ∼2:1. Finally, the sample container was sealed by a Perspex lid on which an optical fiber is mounted for illumination from the blue-light source. A broadband interference filter was inserted in front of the detector to protect from stray light in light-induced difference spectroscopy. As a photometric benefit, the free spectral range is limited to the range from 1900 cm−1 to 950 cm−1. Spectral resolution was 4 cm−1 in the ATR experiments (Heberle and Zscherp, 1996). It is stressed that the protein used for infrared spectroscopy was always dissolved in aqueous medium. A semidry or rehydrated film has never been employed.
Raman spectroscopy
Raman spectra were acquired under preresonant conditions with near-infrared excitation from the 752 nm line from a Krypton laser (Innova 90K, Coherent, Dieburg, Germany). The laser radiation is fed into a LabRam spectrometer (Jobin Yvon, Bensheim, Germany), where the laser emission was focused by a microscope objective (Olympus) to a spot size of 20 μm with a typical power of 10 mW at the sample. The sample was prepared by gently drying the protein on the inner surface of a quartz cuvette. The cuvette was half filled with distilled water to rehydrate the protein without dissolving it. Photons from the sample are collected in back-scattering configuration. Rayleigh scattering is rejected by a holographic notch filter. The inelastically scattered photons are dispersed by a grating (1800 lines/mm) and detected by a Peltier-cooled CCD camera. The resulting data points of the Raman spectrum are spaced by 0.5 cm−1, and the spectral resolution was 2 cm−1.
RESULTS AND DISCUSSION
FTIR difference spectroscopy is used to study the vibrational changes associated with the transition from the dark state of the LOV1 domain to the long-lived photointermediate LOV1-390 upon excitation with blue light. The very slow decay time (τ = 200 s under the applied conditions; Kottke et al., 2003) leads to the accumulation of sufficient molecules in the intermediate state to be detectable by infrared difference spectroscopy. Of primary interest is the protonation state of C57 before it reacts with FMN to form a thio-adduct (Fig. 1).
Light-induced FTIR difference spectroscopy on LOV1
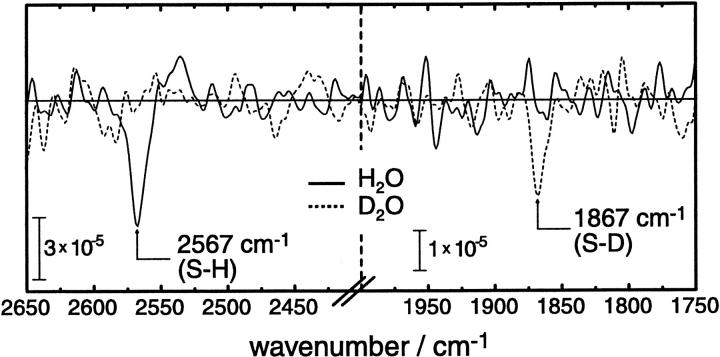
Fig. 2 shows the frequency range where S-H stretching vibrations of thiols are known to absorb (Bare et al., 1975). As evidenced for the LOV2 domain of Avena sativa (oat) by recent NMR experiments (Salomon et al., 2001), a cysteine residue is covalently linked to the FMN chromophore after blue-light excitation. It was argued from fluorescence spectroscopy that it is the (deprotonated) thiolate that is the reactive species in Phot1 LOV2 from oat (Swartz et al., 2001). In contrast, the infrared difference spectra presented in Fig. 2 provide strong evidence that the thiol is protonated before the formation of the covalent bond with FMN in LOV1. The assignment of the negative band at 2567 cm−1 to the S-H stretching vibration is based on the following line of reasoning:
In this frequency range, only a limited number of vibrations are known to absorb, of which the S-H of the cysteine thiol is the most abundant in proteins.
The band is negative, which represents the depletion of this bond upon photoexcitation. This agrees well with the fact that the cysteine binds to FMN via the sulfur atom (Salomon et al., 2001).
The band is downshifted by 700 cm−1 in D2O (Fig. 2, dashed line). Such a strong isotope shift is characteristic for S-H stretching vibrations (Bare et al., 1975; Rath et al., 1994).
The extinction coefficient is very small as can be qualitatively deduced from the comparison of the absorbance scales in Figs. 2 and 3. This is also typical for S-H stretching vibrations (Bare et al., 1975).
FIGURE 2.
Infrared difference spectra of LOV1 in the S-H stretching region obtained after continuous illumination with blue light. The data shown are a zoom-out from the original spectra that have been recorded across the whole midinfrared range (7000–800 cm−1). The negative band shifts from 2567 cm−1 in H2O (continuous line) to 1867 cm−1 in D2O (dashed line) accompanied by a decrease in extinction. Note the different absorbance scale in the two panels.
FIGURE 3.
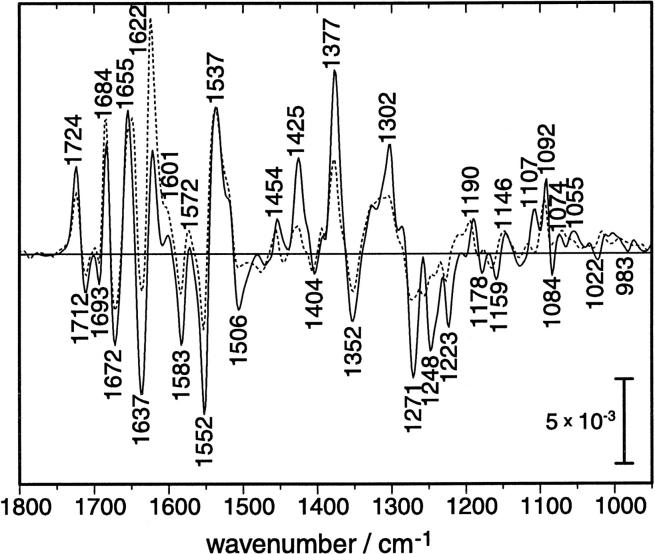
Light-induced FTIR difference spectra of LOV1. The spectrum of a highly concentrated protein solution in H2O (continuous line) is overlaid by the spectrum of the protein dissolved in D2O (dashed line). The indicated frequencies correspond to the measurement in H2O and are summarized in Table 1.
It is obvious from the crystal structure of LOV1 (Fedorov et al., 2003) that C57 is the only residue in the binding pocket of FMN with a thiol side chain. The two other cysteines, C32 and C83, are facing away from the isoalloxazine ring and undergo no change upon illumination of LOV1 (Fedorov et al., 2003). Therefore, the band at 2567 cm−1 is assigned to the S-H stretching vibration of C57 (Table 1). The band has negative amplitude because the proton of the thiol is lost upon bond formation with the isoalloxazine ring. The frequency of 2567 cm−1 indicates that the S-H group is surrounded by an environment that is intermediate in its hydrogen-bonding capacity. A strongly hydrogen-bonded S-H gives rise to the stretching frequency of around 2550 cm−1, whereas in the absence of any hydrogen bonding this frequency upshifts to ∼2600 cm−1 (Bare et al., 1975; Rath et al., 1994). The band at 2567 cm−1 is due to a single molecular vibration, as the halfwidth of 12 cm−1 suggests (Bare et al., 1975), and there are no indications for other bands that might overlap with the S-H vibration of C57.
TABLE 1.
Vibrational bands of the LOV1 domain of phototropin detected by FTIR difference spectroscopy and by preresonance-Raman spectroscopy
| Infrared difference bands of the LOV1-390 intermediate (pos) and the dark state (neg) | Raman bands of the dark state of LOV1 | Raman bands of FMN in H2O | Vibrational assignment | |
|---|---|---|---|---|
| pos | neg | |||
| 2567 | S-H of C57 | |||
| 1724 | ||||
| 1712 | 1710 | symmetric C=O stretch (mainly ν(C4=O))† | ||
| 1693 | ||||
| 1684 | ||||
| 1672 | n.d.* | n.d. | asymmetric C=O stretch (mainly ν(C2=O)) | |
| 1655 | ||||
| 1637 | n.d. | 1632 | band I: ν(C=C) of ring I | |
| 1622 | ||||
| 1601 | ||||
| 1583 | 1580 | 1585 | band II: ν(C4a=N5), ν(C4-C4a), ν(C10a=N1) | |
| 1572 | ||||
| 1552 | 1548 | 1551 | band III: ν(C5a=C6), ν(C8-C9), ν(C5a=N5) | |
| 1537 | ||||
| 1506 | 1506 | 1504 | band IV: ν(C6=C7), ν(C9-C9a), ν(C10a=N1) | |
| 1459 | 1468 | band V: ν(C10a=N1), ν(C4a=N5), ν(C10a-N10), ν(C9a-N10) | ||
| 1454 | ||||
| 1425 | ||||
| 1404 | 1404 | 1411 | band VI: ν(C4-N3), ν(C4-C4a), ν(C2-N3), δ(C9-H), δ(C4=O)‡ | |
| 1377 | ||||
| 1352 | 1352 | 1355 | band VII: ν(C10a-N10), ν(C1′-N10), ν(C5a=C9a), ν(C10a=C4a) | |
| 1302 | ||||
| 1282 | band IX: ν(C4-N3), ν(C8-CCH3), ν(C9a-N10), ν(C7-CCH3) | |||
| 1271 | ||||
| 1248 | 1245 | 1259 | band X: ν(C4-N3), ν(C2-N3), ν(C4-C4a), ν(C2-N1), δ(C2=O) | |
| 1223 | 1228 | 1229 | band XI: ν(C7-CCH3), ν(C10a-C4a), ν(C1′-N10), ν(C9a-N10), ν(C2-N3) | |
| 1190 | ||||
| 1178 | 1184 | 1185 | band XII: ν(C9-C9a), ν(C5a-N5), ν(C4a-C10a) | |
| 1159 | 1163 | 1163 | band XIII: δC6-H, ν(C5a-N5), ν(C9a-N10), ν(C10a-N10) | |
| 1146 | ||||
| 1125 | 1132 | |||
| 1107 | ||||
| 1092 | ||||
| 1084 | 1083 | |||
| 1074 | ||||
| 1068 | ||||
| 1055 | ||||
| 1022 | 1035 | |||
| 992 | 993 | |||
| 983 | 983 | |||
| 790 | 790 | ring I deformation | ||
| 762 | ring I deformation | |||
| 744 | 739 | ring I deformation | ||
See Figs. 2–4 for the spectra. The assignment to molecular vibrations has been done on the basis of the published data and calculations (Dutta et al., 1980; Nishina et al., 1998; Altose et al., 2001; Abe and Kyogoku, 1987; Lively and McFarland, 1990; Bowman and Spiro, 1981; Hazekawa et al., 1997; Kim and Carey, 1993; McFarland, 1987; Zheng et al., 1999; Rath et al., 1994).
n.d.: not determined.
ν: stretching mode.
δ: bending mode.
The formation of the covalent bond between C57 and FMN results in vibrational changes of the chromophore. Therefore, the light-induced difference spectrum in the frequency range of 1800–950 cm−1 shows a manifold of difference bands (Fig. 3, continuous line). Most of the difference bands arise from the chromophore because FMN is the strongest dipole of the protein. However, vibrational contributions from the surrounding protein moiety can also be expected.
Bands at 1724, 1684, 1655, 1622, 1601, 1572, 1537, 1454, 1425, 1377, 1302, 1190, 1146, 1107, 1092, 1074, and 1055 cm−1 appear after blue-light excitation at the expense of bands at 1712, 1693, 1672, 1637, 1583, 1552, 1506, 1404, 1352, 1271, 1248, 1223, 1178, 1159, 1084, 1022, and 983 cm−1 of the dark state (Table 1, left panel). We note the close similarity of the IR difference spectrum of LOV1 from C. reinhardtii to that of LOV2 from oat (Swartz et al., 2002). The exchange of H2O to D2O leads to only minor changes in the light-induced difference spectrum (Fig. 3, dashed line). Apart from amplitude changes, the majority of difference bands above 1350 cm−1 are insensitive to the isotopic replacement, which is common for flavin and flavoproteins (Visser et al., 1983). Only the positive band at 1655 cm−1 downshifts by 5 cm−1 in the presence of D2O. The strongest influence of D2O is exerted on the three negative bands in the region between 1280 cm−1 and 1220 cm−1. Therefore, the vibrations underlying these bands must involve exchangeable protons. The only dissociable proton of the isoalloxazine ring is bound to N3. Therefore, these bands might represent potent indicators for hydrogen-bonding interaction between N3-H and its respective acceptor. However, the asymmetric P-O stretching vibration of the phosphate moiety of the ribityl side chain also absorbs in the frequency region between 1240 and 1220 cm−1. The asymmetric P-O stretch is a very sensitive indicator for the hydrogen-bonding strength of the phosphate moiety (Pohle, 1990). R58 and R74 hold the phosphate group by hydrogen-bonding from the guanidyl nitrogens. The exchange of either of these residues by mutagenesis will enable the band assignment.
The two positive bands at 1425 cm−1 and 1302 cm−1 disappear almost completely upon H/D exchange. Thus, these modes most probably involve the newly created N5-H vibration of the thio-adduct, which is not present in the dark state. However, the change in the force constant of the N3-H bond cannot be excluded. After formation of the thio-adduct, the planarity of ring III is removed and N3-H moves to a different spatial position with altered distance to the mutual hydrogen-bond acceptor.
These tentative assignments are supported by resonance Raman spectroscopy to selectively probe the chromophore modes. The acquired Raman spectrum allows determination of the vibrational contributions of FMN to the infrared spectrum.
The chromophore vibrations of the LOV1 dark state
The Raman spectrum (Fig. 4, top spectrum) has been acquired under preresonant conditions with far-red excitation at 752 nm. Two reasons favor this approach. First, the strong fluorescence emission of LOV1 renders it extremely difficult to observe the Raman bands under resonance conditions. To circumvent this problem, high concentrations of KI can be applied that quench the fluorescence (McFarland, 1987). However, the fluorescence emission is negligible in the range >752 nm, where the preresonance Raman experiment is performed. Second, excitation of the chromophore under fully resonant conditions inevitably leads to the formation of the long-lived photoproduct LOV1-390. The corresponding vibrational bands will overlap with those of the dark state, rendering a clear-cut assignment of the resonance Raman spectrum of the dark state of LOV1 extremely difficult.
FIGURE 4.
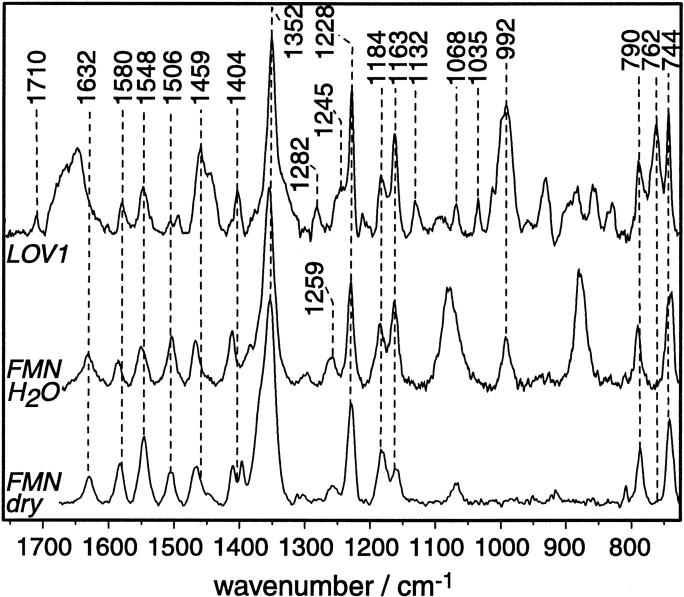
Resonance-Raman spectrum of LOV1 in the unilluminated dark state (top trace). The spectrum has been obtained under preresonant conditions with excitation at 752 nm. For comparison, the Raman spectrum of FMN dissolved in H2O (middle trace) and of FMN in the solid state (bottom trace) are depicted. Dashed vertical lines correspond to the frequency of those LOV1 bands that are discussed in the text.
The preresonance Raman spectrum of the dark state of LOV1 is characteristic for the FMN chromophore (Fig. 4, top trace) as can be deduced from the comparison with free FMN (Fig. 4, middle and bottom trace, for FMN dissolved in H2O and in the solid state, respectively). Fortunately, other investigators have already assigned the FMN bands by isotope labeling and normal mode analysis (Table 1). The preresonance Raman spectrum of FMN in H2O is identical to the spectrum obtained with excitation at 647 nm (Kim and Carey, 1993). The band intensities change dramatically, however, when the excitation wavelength is tuned into the electronic transition and fully resonant conditions are established (see e.g., (Swartz et al., 2002)). The frequencies of the FMN modes are not drastically altered upon insertion into the protein binding pocket (Visser et al., 1983), which is also observed for LOV1 (Fig. 4). Therefore, the assignment of the Raman bands to molecular vibrations is straightforward. As a general rule, bands in the high-frequency region arise from fairly isolated vibrations that are not as strongly coupled then those in the low-frequency region.
The band at 1710 cm−1 is due to the symmetric stretching vibration of the two carbonyls (mainly C4=O with minor contribution from the C2=O stretch, (Abe and Kyogoku, 1987)). The asymmetric carbonyl stretch is supposed to absorb at around 1670 cm−1 albeit with weak intensity (Zheng et al., 1999). In this spectral region, a broad band is seen that arises most probably from the amide I vibration of the protein backbone (predominantly C=O stretching vibration) that hides the clear identification of the asymmetric carbonyl stretch (mainly C2=O stretch). The same is true for the benzene C=C stretching mode at around 1632 cm−1 (band I, according to the notation introduced by Bowman and Spiro, 1981). which is seen only as a shoulder on the low-frequency tail of the amide I band. The band is, however, obvious in the Raman spectrum of FMN dissolved in H2O (Fig. 4, middle trace) and in dry FMN (bottom trace) measured under otherwise identical conditions.
The band at 1580 cm−1 (band II) involves contributions from the C4a=N5 stretching mode (Table 1). Consequently, it represents a marker bands for monitoring the light-induced reaction with the thiol group of C57. The 5 cm−1 lower frequency of band II in the dark state of LOV1 as compared to the frequency of FMN in H2O indicates stronger hydrogen bonding environment of the protein binding pocket than in water. This agrees well with the x-ray structure where the three amides, Q61, N89, N99, and Q120, graft the isoalloxazine ring (Fedorov et al., 2003). Band III and band IV may also represent marker bands for possible hydrogen bonding interaction because both involve C = N stretching vibrations, with N(5) and N(1), respectively, as vibrating atoms. However, the correlation of the H-bonding strength with the Raman frequency of these two bands is not straightforward because it depends also on other factors (see Tegoni et al., 1997, for a discussion).
Band V, band VI, and the very strong band VII appear at similar frequencies for FMN dissolved in water and FMN in the binding pocket, respectively. Band VIII, which is expected in the region around 1300–1305 cm−1 (Bowman and Spiro, 1981), could not be clearly identified in the preresonance Raman spectrum of FMN and LOV1. The band at 1282 cm−1 is tentatively assigned to band IX due to the similar frequency in other FMN binding proteins (Tegoni et al., 1997). However, the band is not detectable in the preresonance Raman spectra of free FMN. The reason for this is unclear.
Band X is considered to represent another marker band for hydrogen-bonding interaction of FMN with its immediate environment. The band appears in free FMN at 1259 cm−1 in accordance with literature data (see e.g., McFarland, 1987). In the Raman spectrum of LOV1, however, such a band is not detectable at this frequency. Apparently, it is downshifted as much as 14 cm−1 to appear as a shoulder at 1245 cm−1 on the strong peak at 1228 cm−1. It has been elaborated by Tegoni et al. (1997), that the frequency of band X correlates with the number of H-bonds at the isoalloxazine moiety. We conclude from the low frequency of band X in LOV1 that the isoalloxazine ring is held by only five H-bonds in the protein binding pocket. This is in agreement with the x-ray structures of LOV1 (Fedorov et al., 2003) and LOV2 (Crosson and Moffat, 2001) where the carbonyl oxygen of C(2) forms H-bonds with the amide N-H of Q61 and of N89, respectively. Likewise, the oxygen at C4 is H-bonded to the amide N-H of N99 and of Q120. The fifth H-bond is the proton of N3 interacting with the amide carbonyl of N89. Interestingly, neither N1 nor N5 is in hydrogen-bonding distance to any residue.
The bands at 1228 cm−1 (band XI), at 1184 cm−1 (band XII), and at 1163 cm−1 (band XIII) are due to stretching vibrations of all three rings of the isoalloxazine moiety. Neither of these involves vibrations of exchangeable protons. These bands appear at the same frequency in dry FMN, when dissolved in water and when incorporated in LOV1 (Fig. 4). Thus, the vibrational assignment is taken over from that of free FMN (Table 1).
Further low-frequency vibrational bands are found for FMN bound to LOV1, and some of them also appear in free FMN. Very strong are those from deformation vibrations of ring I in the 700 cm−1 range. Vibrations in the low-frequency region are usually very sensitive to the conformation of the vibrator. However, there is not sufficient consensus on the vibrational assignment of these modes. Therefore, we will refrain from the detailed discussion of these bands. Moreover, the Raman data serve the primary goal to disentangle the vibrational contributions of FMN to the FTIR difference spectrum. The far-infrared range is currently not accessible by our FTIR spectrometer. Thus, we will compare in the following only the midinfrared Raman bands with those in the light-induced infrared difference spectrum.
Resonance Raman versus infrared difference spectroscopy of LOV1
The comparison of the Raman spectrum of the dark state of LOV1 with the light-induced infrared difference spectrum provides a means to assign the chromophore bands. The selection rules usually exclude the observation of a particular vibration by Raman scattering if it is infrared active, and vice versa. While this principle holds true for small molecules it is violated when coupled vibrations are studied such as occur in large molecules. In the latter case, the same vibration is observable by both techniques, albeit with different intensity.
The spectra depicted in Fig. 5 reveal that most of the negative bands in the infrared difference spectrum (top spectrum) coincide with the Raman bands (bottom spectrum; see also Table 1). Consequently, they can be assigned to chromophore vibrations of the dark state of LOV1. The assignment of the respective modes in the LOV1-390 intermediate requires Raman experiments on this state with the concomitant mode analysis, which are currently under way. However, some of the infrared bands of the LOV1-390 intermediate can already be assigned with some confidence. Of particular importance are those at high frequency because they are due to fairly isolated vibrations and, hence, are more easily interpreted on a molecular basis.
FIGURE 5.
Comparison of the preresonance-Raman spectrum of the dark state of LOV1 (bottom trace) with the blue-light-induced FTIR difference spectrum. Spectra have been replotted from Figs. 3 and 4. Vertical dashed lines indicate bands of the chromophore of the dark (unilluminated) state of LOV1 that are detected by both vibrational techniques.
The negative band at 1712 cm−1 in the FTIR difference spectrum (1710 cm−1 in the Raman spectrum) is shifted to 1724 cm−1 in the LOV1-390 intermediate. The light-induced shift of the C4=O band is readily explained by the formation of the thio-adduct. The resulting sp3-hybridization of C4a isolates the carbonyl C4=O from the conjugated π-system of the isoalloxazine ring. This leads to the stronger bond as observed by the frequency upshift in the infrared spectrum. Less hydrogen bonding to the amide nitrogen of N99 may also account for the upshift. Spectral overlap by the C=O stretching vibration of the asparagine (N89, N99) or the glutamine residues (Q61) that line the chromophore binding pocket, is unlikely because these vibrations are usually observed in the range below 1700 cm−1 (Rahmelow et al., 1998).
The stretching mode of the C2=O bond is assigned to the negative band at 1672 cm−1 in the FTIR difference spectrum. The frequency is lower then that of the C4=O stretch by 40 cm−1, which is typical for FMN (Abe and Kyogoku, 1987; Zheng et al., 1999). The corresponding band could not be clearly detected by preresonance Raman spectroscopy because of the broad amide I band (see above). The intensity of the negative band at 1672 cm−1 is much stronger than that at 1712 cm−1, although both arise from carbonyl stretches. The former might be overlapped by amide I vibrations arising from backbone changes in the helical part of the protein structure.
As a consequence of the assignment of the C2=O stretch, the negative band at 1693 cm−1 in the FTIR difference spectrum is not due to an FMN mode but might arise from the C=O stretch of one of the two Asn or the Gln residue of the FMN binding pocket. However, the change in the amide I vibration of a β-turn motif can also be responsible for the negative band at 1693 cm−1. Point mutants will help to elucidate this assignment in future studies.
Of particular interest is band II at 1580 cm−1 of the dark state of LOV1 because it involves the C4a=N5 stretching mode (Table 1). It is evident that the nucleophilic attack of the thiol of C57 at the C4a position of the isoalloxazine ring will lead to strong alterations of modes that involve this atom. Indeed, this band can be identified in the infrared difference spectrum as a negative band. Band III (1548 cm−1) is a coupled vibration of C = C stretches of ring I of the isoalloxazine ring system with the C5a=N5 stretch of ring II. The composition of band IV at 1506 cm−1 is very similar. It is a coupled vibration of C = C stretches of ring I with an admixture from the C10a-N1 stretch. Although neither of these atoms directly participates in the thio-linkage of the photoproduct, the energy and coupling of these vibrators are influenced by the π-electron redistribution in the ring system upon the formation of the thioether bond.
All of those vibrations where the carbon atom at position 4a is involved should be changed upon light excitation. Consequently, band VI at 1404 cm−1 and band VII at 1352 cm−1 as assigned by preresonance Raman spectroscopy show negative bands in the IR difference spectrum. Band V at 1459 cm−1, however, also involves the C4a=N5 stretching vibration but a corresponding band is not seen in the IR difference spectrum. Either the assignment is not correct in this respect or the negative IR band is compensated by an overlapping positive feature. The negative band at 1444 cm−1 of the IR difference spectrum corresponds in frequency to the shoulder of band V in the Raman spectrum. This shoulder is not seen in the Raman spectrum of free FMN (Fig. 4) and, therefore, the origin is as yet unclear.
The IR difference spectrum displays three characteristic negative bands at 1271, 1248, and 1223 cm−1. The latter two bands appear at similar frequencies in the preresonance Raman spectrum (Fig. 5) and have been assigned to band X and XI of FMN (Table 1). The sensitivity of the three negative IR difference bands to H/D-exchange (Fig. 3) suggests that these vibrations include contributions from the only exchangeable proton at N3 of the isoalloxazine ring system. The band at 1271 cm−1 has no obvious counterpart in the Raman spectrum. Consequently, this band does not arise from isoalloxazine vibrations but either from the ribityl or from some amino acid side chain. The Raman bands at 1184 cm−1 and at 1163 cm−1 correspond to the IR bands at 1178 cm−1 and 1159 cm−1. These bands have been assigned to band XII and band XIII, respectively (Table 1).
Finally, the assignment of the chromophore bands in the IR difference spectrum is far from being comprehensive. Some of the assignments are still tentative and await confirmation by the use of isotopomers. Moreover, those positive bands in the FTIR difference spectrum that indicate vibrational bands of the chromophore in the intermediate state will be disentangled from the vibrational changes of the apo-protein by resonance Raman spectroscopy on the photoproduct. The band assignment is facilitated by normal mode analysis similarly to that performed on a thio-adduct model compound of FMN (Swartz et al., 2002). The remaining difference bands of the apo-protein that represent structural rearrangements of particular amino acid side chains will be elaborated by employing point mutants. Such studies are currently under way.
CONCLUSIONS
The FMN modes of the dark state of LOV1 were identified by preresonance Raman spectroscopy and assigned to molecular vibrations. We conclude from the comparison with FTIR spectroscopy that most of the bands are due to FMN modes in the light-induced difference spectrum. The participation of amide modes of residues (Q61, N89, N99, and Q120) that interact with the polar part of FMN was discussed.
Large backbone changes of the protein can be excluded during the phototransformation of the dark state into the putative signaling state. It is important to note in this respect that the infrared difference spectra of LOV1 have been acquired on dissolved samples. This rules out effects of drying on the conformational changes of the protein backbone as has been observed for the structurally similar photoactive yellow protein PYP (Hoff et al., 1999). The IR difference bands in the amide region, which are indicative for backbone changes, are almost completely assignable to chromophore modes. Still, smaller amide difference bands might occur that are masked by overlapping FMN modes. The absence of large conformational changes agrees well with the x-ray crystallographic model of this state (Fedorov et al., 2003).
The band at 2567 cm−1 is assigned to the S-H stretching vibration of C57, the residue that forms the transient thio-adduct with the chromophore FMN. The occurrence of this band is evidence that C57 is protonated in the dark state of LOV1. The consequence on the reaction mechanism of LOV1 with an uncharged thiol in the vicinity of the FMN chromophore is elaborated in the companion paper (Kottke et al., 2003). Future infrared experiments on the LOV2 domain of C. reinhardtii will clarify whether the protonation state of this critical residue represents the molecular difference between LOV1 and LOV2. It has been shown during the review process of this paper (Iwata et al., 2002) that the reactive cysteine residue of LOV2 from Adiantum, C966, is also protonated before reacting with the FMN moiety.
Acknowledgments
J.H. is grateful to Georg Büldt (Jülich) for fruitful discussions and continuous support. We are indebted to Tina Schireis for excellent technical assistance and Rebecca Nyquist for discussions and improving the English style.
K.A. thanks the fellowship provided by the Alexander von Humboldt foundation. The work was supported by the Deutsche Forschungsgemeinschaft (GK640 and SFB 521 to P.H., and SFB 189 to J.H.).
Abbreviations used: ATR/FTIR, attenuated total reflection Fourier transform infrared spectroscopy; LOV, light, oxygen, voltage; FMN, flavin mononucleotide.
References
- Abe, M., and Y. Kyogoku. 1987. Vibrational analysis of flavin derivatives: normal coordinate treatments of lumiflavin. Spectrochimica Acta. A. 43:1027–1037. [Google Scholar]
- Altose, M. D., Y. Zheng, J. Dong, B. A. Palfey, and P. R. Carey. 2001. Comparing protein-ligand interactions in solution and single crystals by Raman spectroscopy. Proc. Natl. Acad. Sci. USA. 98:3006–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bare, G. H., J. O. Alben, and P. A. Bromberg. 1975. Sulfhydryl groups in hemoglobin. A new molecular probe at the alpha1 beta 1 interface studied by Fourier transform infrared spectroscopy. Biochemistry. 14:1578–1583. [DOI] [PubMed] [Google Scholar]
- Bowman, W. D., and T. G. Spiro. 1981. Normal mode analysis of lumiflavin and interpretation of resonance Raman spectra of flavoproteins. Biochemistry. 20:3313–3318. [DOI] [PubMed] [Google Scholar]
- Briggs, W. R., and J. M. Christie. 2002. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7:204–210. [DOI] [PubMed] [Google Scholar]
- Christie, J. M., M. Salomon, K. Nozue, M. Wada, and W. R. Briggs. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA. 96:8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson, S., and K. Moffat. 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA. 98:2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson, S., and K. Moffat. 2002. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 14:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, P. K., R. Spencer, C. Walsh, and T. G. Spiro. 1980. Resonance Raman and coherent anti-stokes Raman scattering spectra of flavin derivatives. Vibrational assignments and the zwitterionic structure of 8-methylamino-riboflavin. Biochim. Biophys. Acta. 623:77–83. [DOI] [PubMed] [Google Scholar]
- Fedorov, R., I. Schlichting, E. Hartmann, T. Domratcheva, M. Fuhrmann, and P. Hegemann. 2003. Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from. Chlamydomonas reinhardtii. Biophys. J. In press. [DOI] [PMC free article] [PubMed]
- Hazekawa, I., Y. Nishina, K. Sato, M. Shichiri, R. Miura, and K. Shiga. 1997. A Raman study on the C4=O stretching mode of flavins in flavoenzymes: hydrogen bonding at the C4=O moiety. J. Biochem. (Tokyo). 121:1147–1154. [DOI] [PubMed] [Google Scholar]
- Heberle, J., J. Fitter, H. J. Sass, and G. Büldt. 2000. Bacteriorhodopsin: the functional details of a molecular machine are being resolved. Biophys. Chem. 85:229–248. [DOI] [PubMed] [Google Scholar]
- Heberle, J., and C. Zscherp. 1996. ATR/FT-IR difference spectroscopy of biological matter with microsecond time resolution. Appl. Spectrosc. 50:588–596. [Google Scholar]
- Hoff, W. D., A. Xie, I. H. Van Stokkum, X. J. Tang, J. Gural, A. R. Kroon, and K. J. Hellingwerf. 1999. Global conformational changes upon receptor stimulation in photoactive yellow protein. Biochemistry. 38:1009–1017. [DOI] [PubMed] [Google Scholar]
- Holzer, W., A. Penzkofer, M. Fuhrmann, and P. Hegemann. 2002. Spectroscopic characterization of flavin mononucleotide bound to the LOV1 domain of Phot1 from Chlamydomonas reinhardtii. Photochem. Photobiol. 75:479–487. [DOI] [PubMed] [Google Scholar]
- Huang, K., T. Merkle, and C. F. Beck. 2002. Isolation and characterization of a Chlamydomonas gene that encodes a putative blue-light photoreceptor of the phototropin family. Physiol Plant. 115:613–622. [DOI] [PubMed] [Google Scholar]
- Iwata, T., S. Tokutomi, and H. Kandori. 2002. Photoreaction of the Cysteine S-H Group in the LOV2 Domain of Adiantum Phytochrome3. J. Am. Chem. Soc. 124:11840–11841. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., T. E. Swartz, M. A. Olney, A. Onodera, N. Mochizuki, H. Fukuzawa, E. Asamizu, S. Tabata, H. Kanegae, M. Takano, J. M. Christie, A. Nagatani, and W. R. Briggs. 2002. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 129:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., and P. R. Carey. 1993. Observation of a carbonyl feature for riboflavin bound to riboflavin-binding protein in the red-excited Raman spectrum. J. Am. Chem. Soc. 115:7015–7016. [Google Scholar]
- Kottke, T., J. Heberle, D. Hehn, B. Dick, and P. Hegemann. 2003. Phot LOV1: photocycle of a blue light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys J. 84: In press. [DOI] [PMC free article] [PubMed]
- Lively, C. R., and J. T. McFarland. 1990. Assignment and the effect of hydrogen bonding on the vibrational normal modes of flavins and flavoproteins. J. Phys. Chem. 94:3980–3994. [Google Scholar]
- Losi, A., E. Polverini, B. Quest, and W. Gärtner. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies, R. A. 1995. Biomolecular vibrational spectroscopy. Methods Enzymol. 246:377–389. [DOI] [PubMed] [Google Scholar]
- McFarland, J. T. 1987. Flavins. In Resonance Raman Spectra of Polyenes and Aromatics. T. G. Spiro, editor. John Wiley and Sons, New York. 211–302.
- Murgida, D. H., E. Schleicher, A. Bacher, G. Richter, and P. Hildebrandt. 2001. Resonance Raman spectroscopic study of the neutral flavin radical complex of DNA photolyase from Escherichia coli. J. Raman Spectrosc. 32:551–556. [Google Scholar]
- Nishina, Y., K. Sato, R. Miura, K. Matsui, and K. Shiga. 1998. Resonance Raman study on reduced flavin in purple intermediate of flavoenzyme: use of [4-carbonyl-18O]-enriched flavin. J. Biochem. (Tokyo). 124:200–208. [DOI] [PubMed] [Google Scholar]
- Nyquist, R. M., D. Heitbrink, C. Bolwien, T. A. Wells, R. B. Gennis, and J. Heberle. 2001. Perfusion-induced redox differences in cytochrome c oxidase: ATR/FT-IR spectroscopy. FEBS Lett. 505:63–67. [DOI] [PubMed] [Google Scholar]
- Pohle, W. 1990. Interpretation of the influence of hydrogen bonding on the stretching vibrations of the PO2- moiety. J. Mol. Struct. 242:333–342. [Google Scholar]
- Rahmelow, K., W. Hübner, and T. Ackermann. 1998. Infrared absorbances of protein side chains. Anal. Biochem. 257:1–11. [DOI] [PubMed] [Google Scholar]
- Rath, P., P. H. Bovee-Geurts, W. J. DeGrip, and K. J. Rothschild. 1994. Photoactivation of rhodopsin involves alterations in cysteine side chains: detection of an S-H band in the Meta I→Meta II FTIR difference spectrum. Biophys. J. 66:2085–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon, M., W. Eisenreich, H. Dürr, E. Schleicher, E. Knieb, V. Massey, W. Rüdiger, F. Müller, A. Bacher, and G. Richter. 2001. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc. Natl. Acad. Sci. USA. 98:12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz, T. E., S. B. Corchnoy, J. M. Christie, J. W. Lewis, I. Szundi, W. R. Briggs, and R. A. Bogomolni. 2001. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276:36493–36500. [DOI] [PubMed] [Google Scholar]
- Swartz, T. E., P. J. Wenzel, S. B. Corchnoy, W. R. Briggs, and R. A. Bogomolni. 2002. Vibration spectroscopy reveals light-induced chromophore and protein structural changes in the LOV2 domain of the plant blue-light receptor phototropin 1. Biochemistry. 41:7183–7189. [DOI] [PubMed] [Google Scholar]
- Tegoni, M., M. Gervais, and A. Desbois. 1997. Resonance Raman study on the oxidized and anionic semiquinone forms of flavocytochrome b2 and L-lactate monooxygenase. Influence of the structure and environment of the isoalloxazine ring on the flavin function. Biochemistry. 36:8932–8946. [DOI] [PubMed] [Google Scholar]
- Visser, A. J., J. Vervoort, D. J. O'Kane, J. Lee, and L. A. Carreira. 1983. Raman spectra of flavin bound in flavodoxins and in other flavoproteins. Evidence for structural variations in the flavin-binding region. Eur. J. Biochem. 131:639–645. [DOI] [PubMed] [Google Scholar]
- Vogel, R., and F. Siebert. 2000. Vibrational spectroscopy as a tool for probing protein function. Curr. Opin. Chem. Biol. 4:518–523. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., J. Dong, B. A. Palfey, and P. R. Carey. 1999. Using Raman spectroscopy to monitor the solvent-exposed and “buried” forms of flavin in p-hydroxybenzoate hydroxylase. Biochemistry. 38:16727–16732. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., M. A. Wagner, M. Schuman Jorns, and P. R. Carey. 2001. Selective enhancement of ligand and flavin Raman modes in charge-transfer complexes of sarcosine oxidase. J. Raman Spectrosc. 32:79–92. [Google Scholar]