Abstract
In a previous communication (Kindt et al., 2001) we reported preliminary results of Brownian dynamics simulation and analytical theory which address the packaging and ejection forces involving DNA in bacteriophage capsids. In the present work we provide a systematic formulation of the underlying theory, featuring the energetic and structural aspects of the strongly confined DNA. The free energy of the DNA chain is expressed as a sum of contributions from its encapsidated and released portions, each expressed as a sum of bending and interstrand energies but subjected to different boundary conditions. The equilibrium structure and energy of the capsid-confined and free chain portions are determined, for each ejected length, by variational minimization of the free energy with respect to their shape profiles and interaxial spacings. Numerical results are derived for a model system mimicking the λ-phage. We find that the fully encapsidated genome is highly compressed and strongly bent, forming a spool-like condensate, storing enormous elastic energy. The elastic stress is rapidly released during the first stage of DNA injection, indicating the large force (tens of pico Newtons) needed to complete the (inverse) loading process. The second injection stage sets in when ∼1/3 of the genome has been released, and the interaxial distance has nearly reached its equilibrium value (corresponding to that of a relaxed torus in solution); concomitantly the encapsidated genome begins a gradual morphological transformation from a spool to a torus. We also calculate the loading force, the average pressure on the capsid's walls, and the anisotropic pressure profile within the capsid. The results are interpreted in terms of the (competing) bending and interaction components of the packing energy, and are shown to be in good agreement with available experimental data.
INTRODUCTION
Virtually all viruses, whether they infect bacteria, plants or animals, have in common a fundamental structure that involves the viral genome (RNA or DNA) being encapsidated by a rigid protein shell (capsid). In almost all cases of plant and animal viral infections, the entire virus particle, capsid and all, enters the cell cytoplasm; the genome ends up being de-encapsidated—and thereby made available for integration into the host cell machinery—through a variety of scenarios (Lodish et al., 2000; Levy et al., 1994). Bacterial viruses (bacteriophages) on the other hand, are unique in that, with few exceptions, it is only the genome that enters the host cell, with the capsid remaining outside. This fact suggests that the genome must be strongly stressed inside the capsid, with the associated pressure sufficient to inject the genome into the host cell and thereby initiate the infection process.
Because of the early and central role that bacterial viruses have played in the development of molecular biology (Cairns et al., 1992), the structure of their encapsidated genomes has been the object of long and concerted studies (Black, 1989). A major theme running throughout all of this work is the challenge of accounting for how the genome can be confined in a capsid whose dimension is hundreds of times smaller than the genome length. In particular, the available volume in the capsid is barely big enough to accommodate the close-packed genome. Furthermore, because of the charge associated with the high density of phosphate groups, the nucleic acid chains are strongly self-repelling at the small separation distances characteristic of their packing in the viral capsids. Finally, in the case of double-stranded (ds) DNA, the capsid size (approximately hundreds of Ångstroms) is as small as the persistence length (ξ = 500 Å) of the genome, so that large elastic (bending) deformation energies are necessarily involved.
An especially well-studied and characteristic example of DNA packaging structure is provided by bacteriophage T7 (Cerritelli et al., 1997). Here it has been explicitly demonstrated that the double-stranded DNA chain is organized in a spool-like configuration, with average interchain separation as small as 25 Å (compared with the hard-core double-helix diameter of 20 Å and the interstrand distance in a relaxed toroidal DNA condensate of ∼28 Å), implying strong repulsive interactions between neighboring chain segments throughout the capsid. In addition, the average radius of curvature of the circumferentially-wound chain is as small as 100 Å near the hollow core of the packaged genome. The curvature stress associated with this strong bending force drives the chain to crowd on itself, resulting in smaller DNA-DNA spacing (Odijk, 1998; Kindt et al., 2001). The balance between the bending and interstrand repulsion forces dictates the structural characteristics of the encapsidated chain, and the pressure exerted by this chain on the capsid wall, as discussed theoretically and demonstrated numerically in the following sections.
Structural measurements on various viruses indicate that the symmetry of the DNA condensate inside the phage capsid is uniaxial rather than spherical (Earnshaw and Harrison, 1977; Cerritelli et al., 1997), presumably because the stiff DNA chain is packaged through a unique entry hole (the portal). Furthermore, in many cases, such as T7, this capsid portal is joined by a hollow cylindrical tail (the connector), whose connection reinforces the special radial direction of the packaged genome. In a recent, preliminary, communication (Kindt et al., 2001), we considered the force acting along this direction, resisting the entry of the DNA during packaging and driving its release during ejection. We found that this force is a strong function of the length of encapsidated chain. Specifically, it remains small (on the order of a few pN) throughout most of the packaging process, increasing sharply (to tens of pN) only after ∼3/4 of the genome has been loaded; conversely, during ejection, it drops quickly as soon as a small fraction of the DNA chain has been released. This behavior was found independently by experimental measurements of the loading force as a function of packaged chain length (Smith et al., 2001).
DNA packaging is an active process that requires the working of a motor protein (Lodish et al., 2000; Jülicher et al., 1997) that attaches itself to the portal of the empty capsid, grabs the replicated viral DNA by its end, and forces it into the capsid. This motor protein appears in the infected bacterial cell as one of the viral gene products; similarly, the empty pro-capsid and cylindrical tail arise from self-assembly of structural proteins that correspond to other gene products of the phage. The ejection process, on the other hand, can take place to a large extent spontaneously, because of its being driven by the high elastic energy stored in the capsid from the work of packaging.
In the present paper we formulate a model for the above packaging process via a phenomenological Hamiltonian for the encapsidation of a semiflexible linear polymer in a volume whose dimension is comparable to its persistence length and small compared to its overall (contour) length. We write the energy of an encapsidated chain as a sum of bending and interaction contributions. The bending elasticity is specified by a one-dimensional (1D) modulus, or persistence length, in the usual way (Grosberg and Khokhlov, 1994). The interactions between neighboring chains take into account the fact that dsDNA repels itself strongly at small interaxial spacings, even in the presence of polyvalent ions. These short-range repulsions are modeled using a simple functional form which has been derived from the osmotic stress measurements of Rau and Parsegian (1992). In a previous theoretical study Odijk (1998), focusing on DNA packaging within the capsid of the bacteriophage T7, has modeled the encapsidated chain as a perfect spool, with the hexagonally packed DNA circumferentially wound around the main (tail's) axis. Balancing the bending and interstrand interaction components to the packing energy, he determined the interaxial distance, finding good agreement with the experimental results of Cerritelli et al. (1997). Our model is similar to Odijk's in treating the bending and interstrand repulsion as the major contributions to the DNA packing energy. However, our analysis focuses on the structural and energetic changes, as well as the forces and pressures associated with the injection (or, conversely—loading) process. Also, our DNA-DNA interaction potential is quite different, involving an attractive minimum and exponential repulsion. Furthermore, unlike Odijk (1998) we do not assume that the encapsidated aggregate is a perfect spool (which is, however, an excellent approximation for highly loaded nucleocapsids, as well as for a purely repulsive interstrand potential). Rather, we allow for the continuous evolution in shape of an arbitrary uniaxial profile.
Theoretical models for bacteriophage DNA packaging and ejection have been proposed earlier by Reimer and Bloomfield (1978) and by Gabashvili et al. (1991a,b; 1992). The primary contribution of Reimer and Bloomfield was to provide the first systematic estimates of the magnitudes of the several free-energy components of DNA packaging, notably: the bending energy of the double-stranded genome; the electrostatic interactions between neighboring portions of chain (as functions of monovalent and multivalent salt concentrations); and the free energy due to configurational entropy loss of the confined chain. Gabashvili et al. (1991b) contributed a discussion of the effects of interaction between the packaged chain and the inner wall of the capsid. In particular, they argued that the ejection of DNA should in general be incomplete, rather than all-or-none, due not only to the possibility of chain-wall attractions, but also to changes in capsid size and/or the poor quality of solvent. Later work (Gabashvili and Grosberg, 1991a) focused on the effects of friction in determining the rate of ejection of the phage genome from its capsid. Specifically, various kinetic scenarios were related to alternate sources of friction, e.g., reptation of the chain along its length within its spool structure, rotation of the overall spool with respect to the inner capsid walls, and translational motion of the dsDNA through the hollow cylindrical tail of the phage.
We focus here on predicting the structural evolution and statistical thermodynamics (energies, forces, and pressures) of dsDNA as it is packaged in, and released from, bacteriophage capsids. To determine the configuration of the encapsidated chain, we posit local hexagonal ordering and a uniaxial symmetry of the packaged DNA. The cross-sectional profile and the interstrand distance of the encapsidated chain are then determined by functional minimization of its free energy with respect to these variables, and subject to the boundary conditions imposed by the presence of the impenetrable capsid wall. The force required for DNA loading into the capsid and the average pressure exerted by the condensate on the capsid walls are derived from the derivatives of the condensate's free energy with respect to the genome length and capsid volume, respectively. Allowing for small elastic deformations of the capsid shell we can also derive the anisotropic distribution of the local pressure along the capsid walls.
Our key finding is that the average pressure of DNA is a strongly increasing function of its encapsidated length. More explicitly, for a typical phage such as λ, whose capsid size is comparable to the genome's (dsDNA) persistence length (ξ ≈ 500 Å) and yet hundreds of times smaller than the DNA contour length (∼15 μm), we find that the pressure increases sharply from just a few atmospheres when half the genome is packaged to ∼50 atm when the capsid is fully loaded. The largest rate of increase occurs at a loading fraction of ∼3/4, at which point the interaxial spacing begins to drop sharply. We show in particular that this is the point where the packaged DNA is no longer able to fill in the capsid with smaller radii of curvature because of the prohibitive elastic energy cost that would have to be paid by bending on these small length scales. Beyond this point, additional length of DNA is accommodated by the chain becoming crowded on itself (i.e., by a decrease in separations between neighboring chain segments), and thereby experiencing the energy increase associated with short-range repulsions.
THEORETICAL FORMULATION
In this section we present a theoretical model describing the energetics and structure of the DNA chain in the course of its spontaneous injection from (or, inversely, its forced loading into) the viral capsid. We shall use L = Lin + Lout to denote the total length of the dsDNA chain, with Lin denoting the length of DNA remaining in the capsid after a length Lout has been ejected. Assuming that at any stage during the injection both the internal (Lin) and external (Lout) chains have had sufficient time to relax, the free energy of the system can be expressed as a sum,
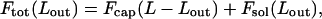 |
(1) |
of the free energies of the DNA within the capsid, and that in solution (i.e., released into the external environment—simple aqueous solution in the in vitro case, or cell cytoplasm in vivo).
In general, the DNA within the capsid is highly stressed, implying that Fcap and hence Ftot decrease rapidly with Lout, at least in the early stages of the injection process. The decrease of Ftot may proceed monotonically, all the way to Lout = L, but may also stop at some 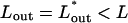 , corresponding to a minimum of Ftot,
, corresponding to a minimum of Ftot,
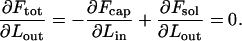 |
(2) |
The derivatives 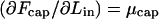 and
and 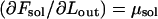 are the chemical potentials (per unit length, or base pair) of DNA within the capsid and solution, respectively. Injection proceeds as long as the driving force,
are the chemical potentials (per unit length, or base pair) of DNA within the capsid and solution, respectively. Injection proceeds as long as the driving force, 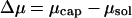 is positive, and stops if the internal and external chemical potentials become equal at some
is positive, and stops if the internal and external chemical potentials become equal at some 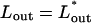 . Conversely, the chemical potential difference
. Conversely, the chemical potential difference
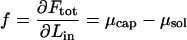 |
(3) |
is the minimum mechanical force that must be supplied by the motor protein to load the DNA into the capsid.
The free energy of a DNA condensate, either within the capsid (Fcap) or in solution (Fsol), can be written as a sum of two contributions,
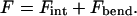 |
(4) |
The first term here, Fint, accounts for the interaction free-energy between different parts of the long DNA chain. Of particular relevance to our model is the interaction energy between (spatially, not chemically) neighboring segments of a DNA chain arranged as a compact, hexagonally packed, aggregate. In this case Fint = Fbulk + Fsurf = −Lɛ(d) + αLsurf ɛ(d), with L denoting the total length of DNA, d the distance between neighboring strands, and ɛ(d) the cohesive energy per unit length of DNA in the bulk of the aggregate. (If the interaction energy were due only to nearest-neighbor interstrand potentials, say ω(d) per unit length, then ɛ(d) = −3ω(d); ɛ > 0, corresponding to attractive interactions.) The second term, Fsurf, is relevant for finite-size aggregates such as the toroidal or spool-like condensates of interest here, and represents the surface correction to the interaction free energy; Lsurf is the total DNA length at the aggregate's surface, and α is a geometric factor expressing the fraction of nearest neighbor DNA-DNA contacts lost upon creating a surface. In all calculations we shall use α = 1/2. (Choosing a somewhat smaller α, e.g., one-third, does not affect our results.)
The second term in Eq. 4, Fbend, is the elastic bending energy associated with the semiflexible DNA chain. It is an integral of local contributions, 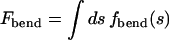 , with fbend(s) = (1/2)κ/R(s)2 the 1D bending energy per unit length at point s along the DNA contour, and R(s) the local radius of curvature at s; κ = ξkBT is the 1D bending rigidity, with ξ = 500 Å the persistence length (Grosberg and Khokhlov, 1994) of dsDNA.
, with fbend(s) = (1/2)κ/R(s)2 the 1D bending energy per unit length at point s along the DNA contour, and R(s) the local radius of curvature at s; κ = ξkBT is the 1D bending rigidity, with ξ = 500 Å the persistence length (Grosberg and Khokhlov, 1994) of dsDNA.
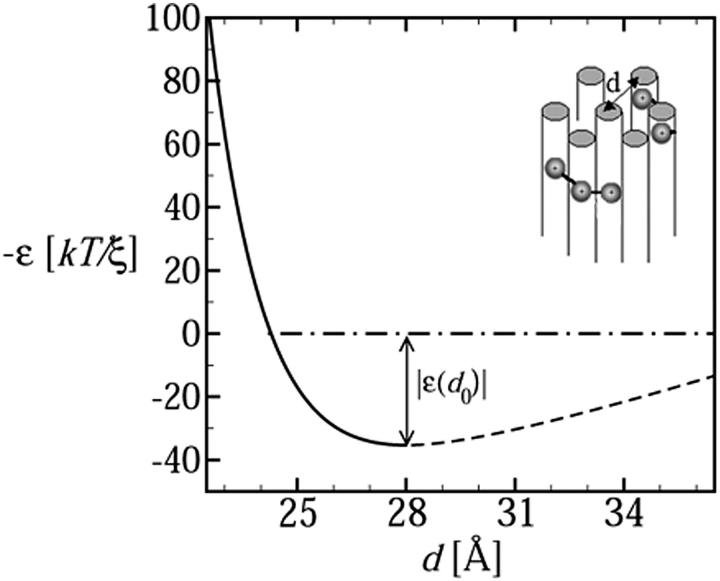
The minimal hard-core distance between neighboring dsDNA molecules is dmin ≈ 20 Å. However, strong interstrand repulsion generally sets in at larger distances, ∼d ≈ 25–30 Å, reflecting electrostatic (counterion confinement) and hydration forces. The van der Waals attraction between DNA strands is generally weak compared to these forces. On the other hand, as is well known, the presence of polyvalent cations, acting as condensing agents, can induce an attractive minimum in ɛ(d), as shown in Fig. 1. The attractive interaction mediated by the polyvalent counterions is responsible for the condensation of dsDNA into compact, typically toroidal aggregates (Bloomfield, 1996; Golan et al., 1999; Hud and Downing, 2001). The specific function ɛ(d) shown in Fig. 1 will be used later in the free-energy minimization procedure for determining the shape of the encapsidated and free chain portions. It has been derived from experimental data as outlined next.
FIGURE 1.
The cohesive energy per unit length of DNA packed in a hexagonal array, as a function of the interstrand distance. The inset illustrates hexagonal packing of dsDNA rods.
Consistent with experiment and our variational approach below, suppose (just for the determination of ɛ(d)) that the DNA condensate in solution is a perfect torus, composed of hexagonally packed dsDNA. Using R to denote the major radius of the torus and L the length of DNA packed in this torus, the free energy of the condensate is, to a very good approximation, given by
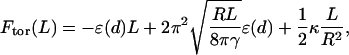 |
(5) |
where 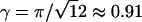 is the volume fraction of hexagonally close-packed cylinders. The first two terms in this expression represent the bulk and surface contributions to Fint, and the third is the average bending energy of the torus. In this last equation and throughout the paper, unless specified otherwise, energies are measured in units of kBT and length in units of ξ.
is the volume fraction of hexagonally close-packed cylinders. The first two terms in this expression represent the bulk and surface contributions to Fint, and the third is the average bending energy of the torus. In this last equation and throughout the paper, unless specified otherwise, energies are measured in units of kBT and length in units of ξ.
Minimizing Ftor with respect to R, we find that the equilibrium radius of the torus is given by
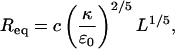 |
(6) |
with ɛ0 = ɛ(d0) denoting the cohesive energy per strand in an optimally packed (free-energy-minimized) torus, where, by definition, d = d0 (c = 0.99 is a numerical constant). Substituting Req from the last equation into Eq. 5, we obtain
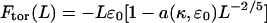 |
(7) |
as an approximate expression for the torus energy. The second term in the square brackets is the sum of the surface and bending energies (both scaling with 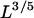 ), with a(κ,ɛ0) = 4.46 (κ/ɛ0)1/5. (For κ = ξkBT and ɛ0 = 35.3 kBT/ξ, as used in most of our calculations (see below), we have a = 2.18 with ξ as our unit of length.)
), with a(κ,ɛ0) = 4.46 (κ/ɛ0)1/5. (For κ = ξkBT and ɛ0 = 35.3 kBT/ξ, as used in most of our calculations (see below), we have a = 2.18 with ξ as our unit of length.)
Equation 6 agrees very well with the exact relationship between Req and L, as obtained from the full (numerical) minimization of F with respect to the bundle's shape and density. Here, however, we use it only to derive a numerical value for ɛ0. Using κ = ξkBT and the experimentally measured values for the dimensions of the torus formed in solution by λ-phage DNA, namely L = 330ξ, Req = 300 Å = 0.6ξ (Golan et al., 1999), we obtain ɛ0 ≈ kBT / ξ.
We shall see below that the DNA condensate in solution is always packed at d = d0 and hence only ɛ0 is necessary to evaluate its structural characteristics. On the other hand, the encapsidated DNA is highly stressed, and d (in the fully loaded capsid) is substantially smaller than d0. Knowledge of the function ɛ(d), especially in the d ≤ d0 regime, is thus crucial for predicting the DNA structure and pressure within the capsid. We determined the repulsive part of ɛ(d) by integrating the measured osmotic force between hexagonally packed DNAs, Π(d), as reported by Rau and Parsegian (1992). That is, 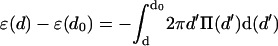 , thus obtaining ɛ(d) − ɛ(d0) in the repulsive regime, d ≤ d0; see Fig. 1. (Using the form Π(d) = Π0{exp[−(d − d0)/c] − 1}, we find Π0 = 1.2 × 10−4 kBT/Å3 c = 1.4 Å and d0 = 28 Å.) The minimum, at d = d0 ≈ 28 Å, signifies the interstrand distance at which the (extrapolated) osmotic force vanishes. This numerical value is characteristic of DNA in solutions containing polyvalent counterions of the kind found in many viruses. (Interestingly, in a recent cryoelectron microscopy study, Hud and Downing (2001) found that, in solution containing polyvalent cations, the dsDNA of the λ-phage condenses into a torus, with equilibrium interaxial distance d0 = 28 Å.) The depth of the minimum in ɛ(d) is set equal to ɛ0 = 35.3 kBT/ξ, based on Eq. 6 and the measured value of Req for λ-DNA in solution (Golan et al., 1999), as discussed above. The attractive part of the potential, i.e., ɛ(d) for d > d0, turns out to play no role in our analysis or calculations.
, thus obtaining ɛ(d) − ɛ(d0) in the repulsive regime, d ≤ d0; see Fig. 1. (Using the form Π(d) = Π0{exp[−(d − d0)/c] − 1}, we find Π0 = 1.2 × 10−4 kBT/Å3 c = 1.4 Å and d0 = 28 Å.) The minimum, at d = d0 ≈ 28 Å, signifies the interstrand distance at which the (extrapolated) osmotic force vanishes. This numerical value is characteristic of DNA in solutions containing polyvalent counterions of the kind found in many viruses. (Interestingly, in a recent cryoelectron microscopy study, Hud and Downing (2001) found that, in solution containing polyvalent cations, the dsDNA of the λ-phage condenses into a torus, with equilibrium interaxial distance d0 = 28 Å.) The depth of the minimum in ɛ(d) is set equal to ɛ0 = 35.3 kBT/ξ, based on Eq. 6 and the measured value of Req for λ-DNA in solution (Golan et al., 1999), as discussed above. The attractive part of the potential, i.e., ɛ(d) for d > d0, turns out to play no role in our analysis or calculations.
To illustrate the dramatic crowding of DNA within a fully loaded viral capsid consider, for instance, the λ-phage. The total length of its genome is L = 330ξ and the radius of its (nearly spherical) capsid is Rc = 0.55ξ, implying a capsid volume of Vc = 4πRc3/3 ≈ 0.697ξ3. Let us momentarily ignore end and curvature effects and suppose that the entire amount of the λ-DNA is packed uniformly and hexagonally within the capsid interior. This implies an interstrand distance d dictated by π(d/2)2L = 0.91 × 0.697ξ3 (recall that γ = 0.91 is the maximal packing fraction). Thus d = 0.0495ξ = 24.75 Å, well within the repulsive range of the interstrand potential function; see Fig. 1.
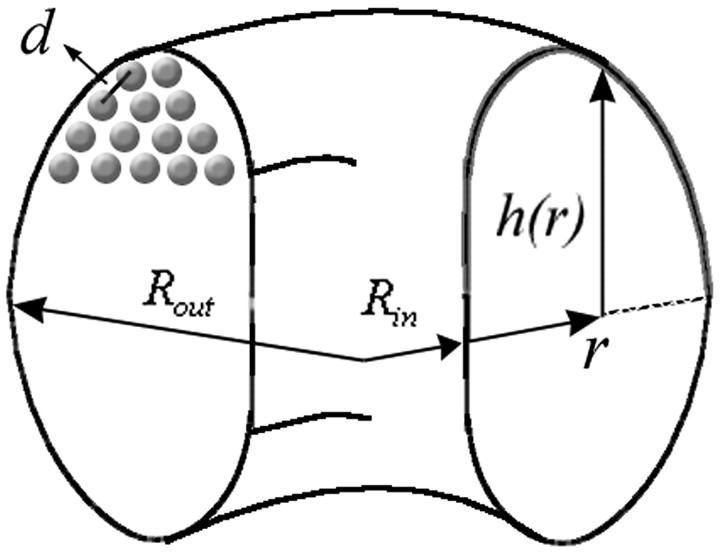
We assume that the DNA condensate in solution as well as in the capsid posses cylindrical symmetry, as shown schematically in Fig. 2. Following Ubbink and Odijk (1996) we describe the shape of the condensate in terms of the profile function, h(r), shown in Fig. 2. Using this function, the free energy of a uniaxial DNA condensate is given by
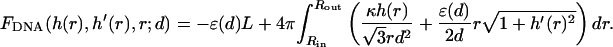 |
(8) |
The first term here, −ɛ(d)L, is the bulk cohesive energy of the condensate, as if it were a portion of an infinite hexagonal array of DNA rods with interaxial distance d. The surface correction, accounting for the finite size of the (curved) condensate appears in the second term of the integral. The first term in the integrand is the local bending energy associated with all circumferentially wound DNA at a distance r from the center of the aggregate. (Note that the curvature energy per single DNA loop is 2πr(κ/2)/r2. The number of loops between r and r + dr, both above and below the condensate's midplane, is 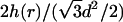 .) The integral in Eq. 8 extends from the inner to the outer radius of the bundle (see Fig. 2).
.) The integral in Eq. 8 extends from the inner to the outer radius of the bundle (see Fig. 2).
FIGURE 2.
The profile function of the DNA condensate.
For any given d, the profile function must satisfy the volume conservation constraint,
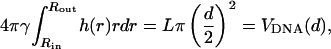 |
(9) |
with VDNA(d) denoting the volume of the condensate.
The equilibrium profile, corresponding to a given d, is obtained by minimizing F subject to Eq. 9 or, equivalently, by minimizing the (Legendre transformed) free-energy functional 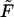 (h(r), h′(r), r; d, λ) = F + λVDNA, with λ denoting the Lagrange multiplier conjugate to the volume conservation condition, Eq. 9.
(h(r), h′(r), r; d, λ) = F + λVDNA, with λ denoting the Lagrange multiplier conjugate to the volume conservation condition, Eq. 9.
For the condensate in solution, minimization of 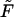 results in Euler-Lagrange equations (Ubbink and Odijk, 1996), whose solution indicates (as we shall see in the next section) an essentially perfect toroidal shape for the outside condensate. Furthermore, the condensate in solution is relaxed, i.e., d = d0. Minimizing
results in Euler-Lagrange equations (Ubbink and Odijk, 1996), whose solution indicates (as we shall see in the next section) an essentially perfect toroidal shape for the outside condensate. Furthermore, the condensate in solution is relaxed, i.e., d = d0. Minimizing 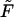 for the DNA condensate within the capsid is more complicated owing to the additional boundary conditions imposed by the presence of the rigid, impenetrable, capsid wall. This restriction (implying inequality constraints on h(r)) prohibits analytical evaluation of h(r). Consequently, F (h(r), h′(r), r; d({h(r)}, r) has been minimized numerically, with d({h(r)}, r) substituted from the packing constraint, Eq. 9, subject to the condition that h(r) cannot exceed the capsid limits.
for the DNA condensate within the capsid is more complicated owing to the additional boundary conditions imposed by the presence of the rigid, impenetrable, capsid wall. This restriction (implying inequality constraints on h(r)) prohibits analytical evaluation of h(r). Consequently, F (h(r), h′(r), r; d({h(r)}, r) has been minimized numerically, with d({h(r)}, r) substituted from the packing constraint, Eq. 9, subject to the condition that h(r) cannot exceed the capsid limits.
The average pressure exerted on the capsid walls, for a given length of loaded DNA, Lin, may be defined in analogy to the thermodynamic pressure:
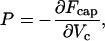 |
(10) |
with Vc denoting the capsid's volume. It must be noted, however, that the DNA condensate inside the capsid is not isotropically packed. Thus, in principle, the local pressure on the capsid wall may vary both in magnitude and direction. In other words, the derivative in Eq. 10 may depend on how the volume is changed. In the following we shall use P, as defined in Eq. 10, for the free-energy change corresponding to a change in Vc preserving the spherical shape of the capsid; i.e., dVc = 4πRc2dRc, with Rc denoting the radius of the (supposedly) spherical capsid shell. We shall refer to P as the average isotropic pressure.
An alternative procedure for calculating the pressure inside the capsid, as well as its anisotropic distribution, is to replace the rigid walls by a repulsive potential resisting the expansion of the (highly compressed and strongly bent) DNA condensate. Similar methods are often used in simulation studies to determine the pressure exerted by a fluid on the walls of a container. The version of this procedure employed here is to represent the empty protein capsid as an elastic spherical shell of radius Rc, consisting of a continuous distribution of radial springs. More explicitly, let R,θ,φ denote a system of polar coordinates whose origin coincides with the center of the empty capsid and whose z-axis coincides with the axis of the condensate. Consider now a small area element, dA = Rc2 dΩ = Rc2 sinθdθdφ, on the capsid's envelope at θ,φ. If the direction Ω = θ,φ corresponds to a point where the outside of the packaged-chain profile h(r) lies at a distance RΩ from the center that exceeds Rc, then we associate a local harmonic restoring force −k [RΩ − Rc] dA with this area element. Because the DNA condensate is uniaxial we are only concerned here with changes in capsid shape δRΩ ≡ RΩ − Rc = Rθ − Rc ≡ δRθ which are independent of the azimuthal angle φ. The total elastic energy penalty associated with a small (Rθ − Rc ≪ Rc, for all θ) but arbitrary (uniaxial) deformation of the capsid wall is then
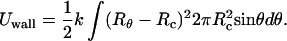 |
(11) |
The total free energy of the loaded virus, (nucleocapsid), is now given by
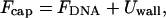 |
(12) |
with FDNA denoting the packing energy of the DNA condensate, as given by Eq. 8. Inasmuch as 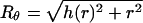 , (h(r) = r/tanθ), Uwall becomes a functional of the profile function h(r). The equilibrium profile, describing both the shape of the DNA condensate and the capsid's envelope, is dictated by the minimum of Fcap = FDNA + Uwall with respect to {h(r)} or, equivalently, {Rθ}; that is, δFcap = δFDNA+δUwall = 0 for all possible variations in capsid's shape, {δRΩ}, ({δRθ} for the uniaxial variations of interest here). Expressing δFDNA in the form
, (h(r) = r/tanθ), Uwall becomes a functional of the profile function h(r). The equilibrium profile, describing both the shape of the DNA condensate and the capsid's envelope, is dictated by the minimum of Fcap = FDNA + Uwall with respect to {h(r)} or, equivalently, {Rθ}; that is, δFcap = δFDNA+δUwall = 0 for all possible variations in capsid's shape, {δRΩ}, ({δRθ} for the uniaxial variations of interest here). Expressing δFDNA in the form 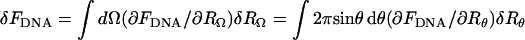 , the equilibrium condition reads:
, the equilibrium condition reads: 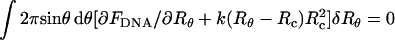 for all {δRθ} around the equilibrium configuration. Thus, at equilibrium, the differential, (or local) pressure, p(Ω) = p(θ), exerted by the condensate on the capsid's wall along the direction Ω = θ,φ is given by
for all {δRθ} around the equilibrium configuration. Thus, at equilibrium, the differential, (or local) pressure, p(Ω) = p(θ), exerted by the condensate on the capsid's wall along the direction Ω = θ,φ is given by
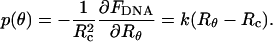 |
(13a) |
The equilibrium shape of the nucleocapsid is given by {Rθ}, which slightly deviates from the sphere of radius Rc, characterizing the empty capsid. Clearly, as k increases, the difference |Rθ − Rc| decreases, for all θ. However, the product k(Rθ − Rc) = p(θ) approaches a constant value, corresponding to (1/Rc2)(∂FDNA/∂Rθ) with the derivative evaluated at Rθ→Rc. In this hard-capsid limit we also have Uwall→0, so that all the energy of the loaded phage is stored within the DNA condensate, Fcap = FDNA. It may be noted, however, that the procedure just outlined for evaluating Rθ and p(θ) is also applicable to DNA (or other) condensates trapped within compartments bounded by softer walls such as lipid vesicles (Lambert et al., 1998, 2000) or viral pro-capsids. It can also be applied to capsids characterized by nonuniform k's or nonspherical equilibrium shapes.
The change in the free energy of the condensate, corresponding to an arbitrary (yet small) deviation of the capsid's shape ({δRθ}) from its initial, equilibrium, configuration is given by 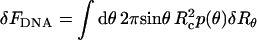 . The corresponding change in volume is
. The corresponding change in volume is 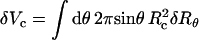 . The ratio −δFDNA/δVc ≡
. The ratio −δFDNA/δVc ≡ 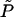 depends, of course, on the shape of the deformation.
depends, of course, on the shape of the deformation.
For a uniform expansion of the capsid from Rc to Rc + δRc, (i.e., δRθ = δRc for all θ), we find 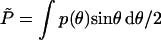 . Recall that, by definition, the average isotropic pressure in Eq. 10 is the change in Fcap upon a uniform spherical expansion of the hard capsid walls; i.e., in this case
. Recall that, by definition, the average isotropic pressure in Eq. 10 is the change in Fcap upon a uniform spherical expansion of the hard capsid walls; i.e., in this case 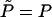 , or
, or
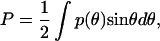 |
(13b) |
with p(θ) = k(Rθ − Rc) evaluated at the large k limit.
RESULTS AND DISCUSSION
The numerical results presented and analyzed in this section are concerned with the structure, energy, force, and pressure characteristics of DNA packaging in viral capsids and its injection into solution. Most of the calculations were carried out for a model system, resembling the λ-phage. Thus, the protein capsid is modeled as a spherical shell of radius Rc = 275 Å = 0.55ξ. The total length of the viral genome is L = 330ξ =16.5 μm.
Energetics and structure
The free energy of the partially loaded capsid, Ftot(Lout) = Fcap(L − Lout) + Fsol(Lout), has been determined by minimizing both contributions to Ftot with respect to their DNA packing profiles, h(r). The minimization of Fsol(Lout) reveals that the DNA chain in solution organizes into a relaxed (d = d0 = 28 Å), perfectly shaped torus, for all values of Lout. This result is to be expected in view of the fact that the toroidal shape allows minimal surface-to-volume ratio (hence maximal cohesive energy (−Loutɛ(d0)) at a minimal bending energy cost. Eq. 6 turns out to provide an excellent approximation for the radius of the torus derived from the full minimization procedure, and Eq. 7 for the packing energy of the bundle in solution.
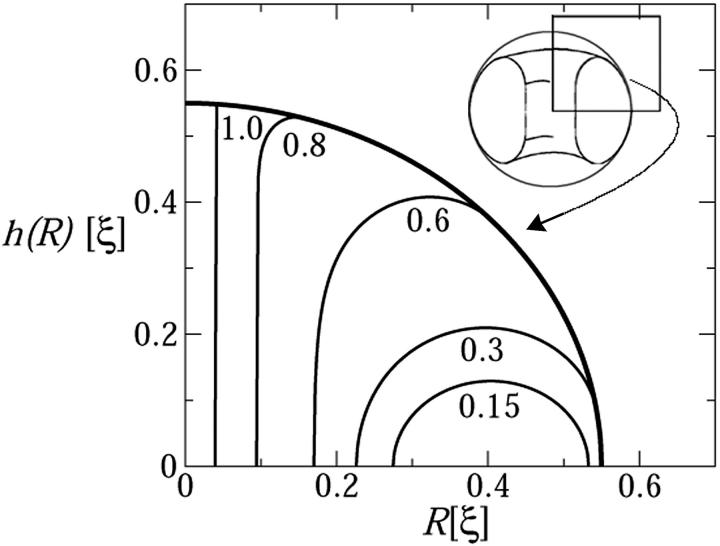
More interesting behavior is exhibited by the encapsidated chain. For small values of Lin we find, as anticipated, that the encapsidated chain is a perfect and relaxed (d = d0) torus. This behavior prevails as long as the loaded length of DNA, Lin, is small enough to ensure that the relaxed torus can comfortably be accommodated within the capsid. Namely, the encapsidated chain forms a torus for DNA length satisfying Req(Lin) < Rc − (Rout − Rin)/2. Fig. 3, which shows DNA packing profiles obtained via full minimization (see Eq. 8) of Fcap(Lin), confirms this behavior for small loading fractions; Lin/L ≤ 1/4 or so, in good agreement with the value predicted by Eq. 6.
FIGURE 3.
DNA packing profiles within the viral capsid. The figure shows the contour lines corresponding to the top-right quarter of the condensate's cross section, for different values of the loading fraction, Lin/L, as labeled.
It should be noted that the profiles shown in Fig. 3 were derived for DNA chains which are not attracted to, nor repelled by, the capsid walls; rather, they are just constrained to lie inside them. For certain viruses, however, e.g., the HK97-phage (Wikoff et al., 2000), the inner capsid wall is known to be positively charged, suggesting that the leading parts of the loaded genome are attracted to the walls. The inclusion of such interactions in our model is straightforward, and later in this section we briefly address their (qualitatively minor) effects on the genome injection process.
As soon as the outer radius of the relaxed torus exceeds Rc, the shape of the encapsidated condensate must deviate from that of a perfect torus. In our treatment of the λ-phage this happens when Lin ≈ L/4. The deformation involving the lowest energetic cost corresponds to a shape transformation into a spool-like structure, as shown in Fig. 3. In the course of this continuous, torus-to-spool transition, the inner face of the condensate becomes increasingly flatter whereas its outer face adopts the shape of the capsid's wall. This transformation evolves gradually with Lin and is driven by the tendency of the DNA bundle to minimize the bending energy penalty associated with lowering its inner radius, Rin, in the course of DNA loading. The surface energy penalty associated with the concomitant increase in surface to volume ratio is negligible.
In addition to the change in condensate's shape, there is another degree of freedom for accommodating the increasing amount of DNA loaded into the capsid, namely, compressing the bundle to higher densities than those corresponding to the optimal interaxial spacing d = d0. Note, however, that this option involves a strong (exponential) increase in interstrand repulsion, and does not become operative until the (rate of increase of) bending energy penalty exceeds the increase in interstrand compression energy. Before this crossover point, DNA packing into the growing spool enables an increase in the condensate's volume at a small change in Rin and thus tolerable bending energy cost. In Fig. 3 we see that the torus-to-spool transition begins at Lin/L ≈ 0.3 and is essentially completed when Lin/L ≈ 0.7, with d = d0 throughout this range; see Fig. 4. Thus, in this range the increase in bending energy is, indeed, still small compared to that of compressing the bundle into the repulsive interaction regime (d < d0). DNA compression begins immediately and steeply afterwards, as further analyzed in the next section.
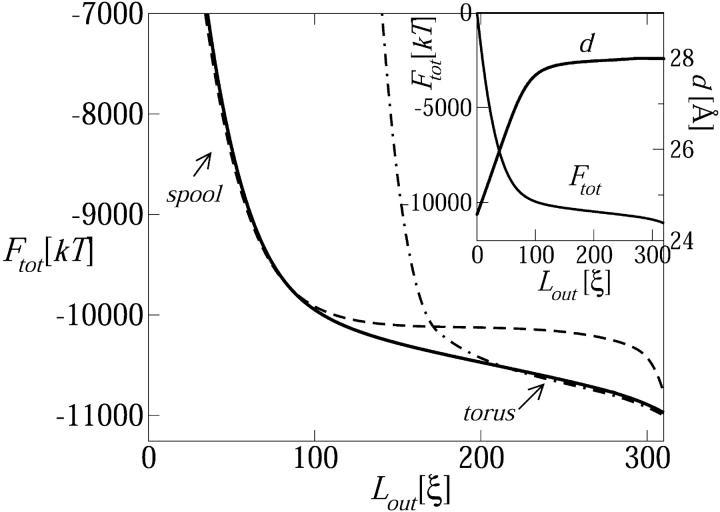
FIGURE 4.
The total free energy of the DNA chain, Ftot = Fcap + Fsol, as a function of the ejected length, Lout (Ftot ≡ 0 at Lout = 0). The dashed and dashed-dotted curves describe Ftot for a DNA chain whose encapsidated part, Fcap, is treated as a perfect spool or a perfect torus (but adjustable d), respectively. The inset shows Ftot for the entire range of possible Lout values and the corresponding variation in the interaxial spacing, d.
Ignoring momentarily all energetic aspects, simple geometric considerations reveal that the maximal length of hexagonally packed DNA which can be loaded into the λ-capsid, with d = d0 = 28 Å, is Lin ≈ 0.8L, significantly less than the total genome length. Thus, to complete the loading process, d must be compressed to values lower than d0. From the calculation of d shown in Fig. 4 (see inset), the interstrand distance begins falling below d0 already at Lin/L ≈ 0.7 which, as noted above, marks the completion of the spool structure. Clearly then, above this loading fraction, the bending energy penalty increases so steeply with the decreasing (already small) values of Rin that the system begins opting for lower values of d, despite the substantial energetic cost. The competition between the bending and interstrand repulsion forces is further discussed below.
Fig. 4 displays the total free energy of the DNA chain, Ftot = Fcap + Fsol, as a function of the ejected length Lout = L − Lin. Viewed in the opposite direction, i.e., as a function of Lin, the figure describes the loading energy. The dramatic increase in Ftot at the late stages of the loading process, namely Lin/L ≥ 2/3, is due entirely to the repulsive energy, Fcap(d < d0), stored in the highly compressed DNA spool. Eventually, this stored energy will be released in the course of the spontaneous injection of the viral genome, once its portal is opened. The inset to Fig. 4 demonstrates clearly the direct correlation between the rapid decrease in Ftot and the increase in d toward its relaxed value (d0 = 28 Å) as Lout increases. Note that d increases approximately linearly to a constant value (d0). Purohit and Phillips (unpublished) have shown that this simplified dependence allows one to obtain closed-form analytical results for the packaging free energy, force, and pressure, as a function of Lin.
Also shown in Fig. 4, are two total free-energy curves obtained by modeling the encapsidated chain (i.e., the Fcap contribution to Ftot) as being either a perfect torus or a perfect spool for all 0 ≤ Lin ≤ L. As in the full calculation, the interstrand distance d (of either the torus or the spool within the capsid) is allowed to adjust so as to minimize Fcap for any given Lin. As expected, the torus curve provides a good model for the early stages of loading (large Lout = L − Lin) but fails badly as soon as its outer radius exceeds the capsid's radius, which happens at Lin/L ≈ 0.3. The spool curve provides an excellent approximation to the late stages of loading (early stages of injection). Note that the spool description is applicable for all values of Lin in cases of negligible cohesive energy (ɛ0 ≪ kBT), because under these conditions the surface energy term, which favors the torus over the spool, becomes completely irrelevant.
As mentioned in the Introduction, the spool shape of the loaded DNA chain has been most convincingly established by Cerritelli et al. (1997) for bacteriophage T7. Based on electron microscopy measurements these authors have also determined the interhelix spacing, d, for several (large) loading fractions Lin/L, as given in Table 1. The T7 capsid, similar to that of λ, is nearly spherical, with a radius Rc = 275 Å = 0.55ξ, but it contains a protein connector of height Rc and radius b = 105 Å, restricting the inner spool radius within one hemisphere of the capsid to Rin ≥ b. Its full genome length is L = 13.6 μm = 272ξ. Taking these special structural characteristics into account, we applied our free-energy minimization procedure to calculate the interhelical spacing d in T7, for the three loading fractions (Lin/L's) reported experimentally (Cerritelli et al., 1997). The results, revealing excellent agreement with experiment, are given in Table 1. Similarly good agreement between experiment and theory had previously been obtained by Odijk (1998), whose model has been discussed briefly in the Introduction. Our larger values for d indicate the steeper short range repulsion of our interstrand interaction potential.
TABLE 1.
Interhelical spacing in the T7-phage for three (high) values of the loading fraction, Lin/L, as measured by Cerritelli et al. (1997), and calculated theoretically by Odijk (1998) and by the present work
| Interhelical distance [Å]
|
|||
|---|---|---|---|
| Lin/L | Experiment | Odijk | This work |
| 100% | 25.4 | 24.9 | 25.6 |
| 92.1% | 26.4 | 25.9 | 26.5 |
| 84.4% | 27.5 | 27.0 | 27.4 |
Twenty-five years ago, Earnshaw and Harrison (1977) reported the interhelical spacing in λ-phage derived from diffraction measurements. Their results are given in Table 2, including one value for an overloaded capsid, (the experimental error is on the order of ±2 Å (Earnshaw and Harrison, 1977). The results predicted by our theory, corresponding to the capsid radius (Rc = 275 Å = 0.55ξ) used in all our calculations in this paper, are given in the second column. The agreement between theory and experiment, especially with respect to the rate of change, is quite reasonable. Better agreement can be obtained by using a slightly smaller capsid volume, Rc = 270 Å = 0.54ξ, as shown in the third column. It should be stressed, however, that our main goal in showing this alternative calculation is to demonstrate how sensitive the interstrand distance (and hence the compression energy) is to very small variations in capsid volume, or more precisely to the ratio between capsid and DNA volumes (as well as to other factors such as d0 and the slope of the repulsive potential wall).
TABLE 2.
Interhelical spacing in the λ-phage for five (high) values of the loading fraction, Lin/L; as measured by diffraction measurements (Earnshaw and Harrison, 1977), and calculated theoretically for two different capsid radii
| Interhelical distance [Å]
|
|||
|---|---|---|---|
| Theory
|
|||
| Lin/L | Experiment | Rc = 275 Å | Rc = 270 Å |
| 105% | 23.2 | 24.0 | 23.4 |
| 100% | 23.6 | 24.6 | 24.0 |
| 89% | 24.6 | 25.9 | 25.3 |
| 88% | 24.7 | 26.0 | 25.5 |
| 78% | 25.8 | 27.1 | 26.7 |
Loading force
The overall picture emerging from the structural-energetic analysis of the previous section is that the injection process consists of two stages. The first stage involves a fast relief of elastic strain, with Ftot decreasing rapidly with Lout and a corresponding relaxation of d toward d0. The second stage is slower and driven by an interplay between the bending and surface energies. Viewed in the reverse direction, only a small force is needed for DNA loading as long as d = d0, i.e., as long as the loading fraction is less than Lin/L ≈ 0.7. At somewhat higher loading fractions the loading force, Eq. 3, begins its steep increase, as shown in Fig. 5. This increase is dominated by the strong DNA-DNA repulsion at d < d0, but reflects a nontrivial interplay between interstrand repulsion and DNA bending.
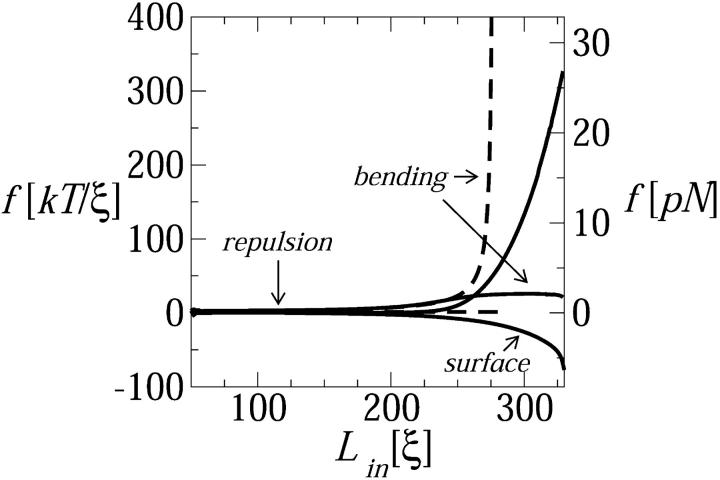
FIGURE 5.
The surface, bending, and DNA-DNA repulsion components of the loading force, as a function of the loaded genome length. The total force curve overlaps the repulsive component. The dashed curves describe the repulsion and bending forces corresponding to a model calculation in which d is not allowed to fall below 27 Å: the effect of this constraint on the surface term is negligible and therefore not shown.
The individual—surface, bending, and interstrand repulsion—contributions to the loading force are also shown in Fig. 5. Note that the bending and surface contributions are relatively small throughout the loading process. Thus, the total force curve essentially overlaps the DNA-DNA interaction contribution. In Fig. 5 we see that the force begins its steep increase at Lin ≈ 0.8L, with the major contribution arising from interstrand compression. That is, d falls below d0 and essentially all of the work done by the loading force is stored as an interstrand repulsive energy. It should be noted, however, that the DNA bending energy plays a crucial role in this process—albeit indirectly—by forcing the chain to crowd upon itself as it avoids bending on too small a length scale.
Careful inspection of Fig. 5 reveals that when the loaded length exceeds Lin ≈ 200ξ, it is the bending force rather than interstrand repulsion which begins increasing with Lin, reflecting the decrease in the inner radius of the loaded chain. These two forces change roles at Lin ≈ 250ξ, at which point the inner spool radius is so small that further increase in the rate of change of bending energy becomes excessively large, whereas interstrand compression involves a large yet more tolerable force. To illustrate this interpretation, we show in Fig. 5 the results of a calculation corresponding to a loading process where d is not allowed to decrease below a certain interstrand distance, say, d = 27 Å. (That is, an infinite repulsive wall has been superimposed on our ɛ(d) at d = 27 Å.) For this system, once the interstrand spacing reaches this limiting value the repulsive force becomes constant, whereas the bending force increases extremely rapidly. More significantly, the increase in the bending force is steeper than the increase in the repulsive force in the unconstrained system. Thus, although energetically very costly, reducing d is the only way to pack the entire genome into the capsid.
Using optical tweezers to pull on the dsDNA genome of the bacteriophage φ29, Smith et al. (2001) have recently measured the force necessary to stop (stall) the loading of DNA by the portal motor protein of this phage. The capsid of this virus is, roughly, a 420 Å × 540 Å prolate ellipsoid and its available volume is ∼1/2 the volume of the λ-capsid; correspondingly, its genome length (19.3 kilobases ≈ 65ξ) is less than half that of λ. Notwithstanding these differences, the loading mechanism in φ29 appears to involve two regimes: a fast stage (i.e., small stalling force) followed by a slow loading stage, indicating that the action of the motor protein is progressively resisted by an opposite force exerted by the packaged genome portion. From their stalling force measurements, Smith et al. (2001) have concluded that the internal, opposing force starts increasing when ∼1/2 of the genome is packed, reaching ∼50 pN toward the end of the loading process. These values are of the same order of magnitude as those derived from our model; see Fig. 5. A quantitative comparison between theory and experiment is not warranted here because the structural characteristics of the φ29 phage are quite different from those of our present model. Also, the force measurements were carried out in solutions containing counterions (sodium and magnesium; see Smith et al., 2001) which do not mediate DNA attraction, suggesting the need for a larger loading force. It may also be noted that the loading process may involve dissipative losses associated with the dynamical character of the experiment, suggesting that the measured force is an upper bound to the calculated, statistical-thermodynamic force.
Pressure
The average pressure exerted by the encapsidated DNA on the capsid walls (Eq. 10) is shown in Fig. 6, as a function of the loaded genome length. The onset of the increase in P correlates with the onset of the decrease in d to values lower than the optimal spacing d0. This happens at Lin/L ≈ 3/4 (see Fig. 4), where the encapsidated chain already forms a spool condensate; see Fig. 3. Qualitatively, the rapid increase of P with Lin is quite straightforward, because an increase in Lin implies a lower d and hence stronger DNA-DNA repulsion, resulting in larger pressure on the capsid wall. Further insight into the (essentially exponential) increase of P with Lin is provided by the following (approximate) considerations.
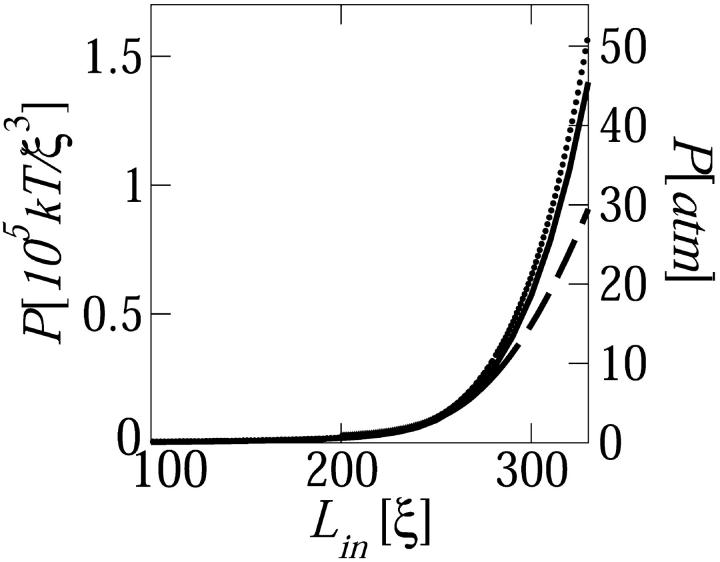
FIGURE 6.
(Solid line), The average (thermodynamic) pressure on the capsid wall, P = −∂Fcap/∂Vc, as a function of the length of DNA loaded into the capsid. (Dashed line), the average pressure calculated for a capsid wall represented by a harmonic restoring force with k = 107kBT/ξ4; for k ≥ 109kBT/ξ4 the calculated pressure is indistinguishable from the thermodynamic pressure (solid curve). (Dotted line), The osmotic pressure in a macroscopic phase of hexagonally packed DNA.
At the late stages of loading, the capsid's volume Vc is mostly occupied by the DNA spool, whose volume is VDNA ∼ Lind2. The volume of the small cylindrical core in the center of the capsid is Vc − VDNA = Vhole ≈ 2πRcRin2, with Rin and 2Rc denoting the radius and length of the hole, respectively. To a good approximation, a uniform increase in Vc, (dVc = 4πRc2dRc), implies a corresponding change in VDNA; namely, bundle expansion (dVc = dVDNA ∼ Linddd), resulting in the relief of DNA packing strain. This is because a change in Rin is strongly opposed by the enormous bending energy penalty (see Fig. 3), which means that dVhole/dVDNA ∼ (Rin/Rc)2 ≪ 1. Now, inasmuch as for large Lin/L most of the loading energy is stored as interstrand repulsion, Ftot ≈ Fcap(Lin) ∼ −Linɛ(d) and hence P = −∂Fcap/∂Vc = ∂Fcap/∂VDNA ∼ (1/d)∂ɛ/∂d = Π(d). In other words, the average pressure inside the (nearly fully loaded) capsid is, to a good approximation, given by the pressure Π(d) determined by osmotic stress measurements, Rau and Parsegian (1992). Indeed, Fig. 6 confirms that Π(d) ≈ P(d).
Also shown in Fig. 6 is a pressure curve calculated for a DNA condensate entrapped in a capsid bounded by elastic walls, represented by an harmonic restoring potential with a force constant, k = 107kBT/ξ4. Smaller values of k result in lower P curves, and vice versa. This is because a softer wall (lower k) allows the capsid to expand more easily beyond the equilibrium position of the empty capsid. This expansion implies a lower volume fraction of the encapsidated chain and hence a smaller pressure on the capsid's walls. Upon increasing the wall rigidity to k ≥ 109kBT/ξ4, the pressure calculated for the elastic capsid converges to the isotropically averaged pressure determined from Eq. 10. The average isotropic pressure corresponds to the hard-wall (infinite k) limit, as discussed in the previous section.
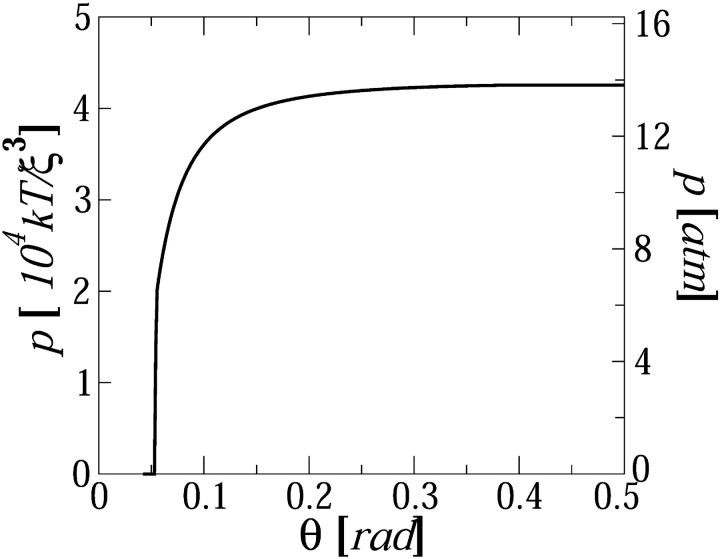
Finally, in Fig. 7 we show the pressure profile, p(θ), in the northern hemisphere of the capsid, as calculated from Eq. 13 (the pressure in the southern hemisphere is its mirror image). Recall that p(θ) is the local radial force per unit area acting at the point Rc,θ,φ on the wall of the capsid; its angular average (see Eq. 14) is the average isotropic pressure. Our calculation reveal that the force is nearly constant for a wide range of polar angles around the equator (θ = π/2), falling sharply to zero at some finite angle near the pole. For Lin = 290ξ this happens at θ* ≈ 0.05, as shown in Fig. 7. Not surprisingly, this is approximately the angle subtended by the cylindrical hole in the center of the spool. Indeed, larger Lin's correspond to smaller θ*, and to larger p(θ > θ*) values. Perhaps the most significant aspect of Fig. 7 is that the pressure distribution along the capsid walls appears to be quite uniform, providing a possible explanation for the high mechanical stability of viral capsids. Purohit and Phillips (unpublished) have shown how the elastostatic continuum mechanics of thick shells can be used to relate the critical lateral stress within a capsid—estimated from atomistic-level structure and energetics of capsid proteins—to the maximum uniform pressure that can be tolerated by a fully packaged phase. This limit of mechanical stability (of order 50 atm) is consistent with the pressures discussed above from our several routes.
FIGURE 7.
The pressure profile along one hemisphere of the viral capsid, for Lin = 290ξ.
Incomplete injection
The initial, very steep decrease of Ftot with Lout (see Fig. 4) is a direct consequence of the extreme compression of the genome within the capsid. This appears to be a general mechanism which viruses use to ensure irreversible spontaneous injection of their genome once the portal is opened. Our theory predicts that the steep initial decline of Ftot = Fcap + Fsol is followed by a more moderate yet monotonic decrease of Ftot with Lout; see Fig. 4. The change in slope occurs at Lout/L ≈ 1/3, when the interstrand distance has relaxed to d ≈ d0. The slower change in Ftot during this second stage of injection reflects a delicate balance between the bending, DNA compression, and surface energy contributions to Fcap and Fsol. Clearly, the predictions of our theory for this regime may be affected by details, such as the form of ɛ(d). Similarly, the rate of release during the second injection stage should be more sensitive to changes in experimental conditions, e.g., the ionic composition of the host solution. For instance, if a sufficient concentration of polyvalent counterions is not present, DNA condensation will not take place and the ejected DNA is better described as a self-avoiding semiflexible polymer rather than a compact torus. Given the specific conditions of the system in question, the theoretical model can be modified accordingly.
A question of more general interest is whether the injection process may involve an energetic barrier. Because of the large release of elastic energy immediately after injection begins, a barrier of this kind is more likely to appear in the second injection stage. Quite generally, an injection barrier may be due to a decrease in the packing energy of the encapsidated chain, or an increase in the energy of the released chain, or both.
A possible mechanism that lowers the entrapped chain energy is attraction to the capsid walls, which, for certain viruses, are known to be positively charged. In a recent study it was reported (Wikoff et al., 2000) that the HK97 phage has 3800 positive charges on the inner walls of its Rc = 330 Å capsid, corresponding to a surface charge density of ∼0.3 charges per nm2. (The surface charge density on one of the flat faces of a monolayer array composed of parallel dsDNA chains at distance d = 28 Å from each other is roughly the same, indicating good charge-matching.) Assuming an attractive electrostatic energy of ≈ kBT per DNA-wall ion-pair, one estimates that a net energetic barrier of ∼1000 kBT may resist the release of the last layer (corresponding to Lin ≈ 50ξ) of DNA. This non-negligible, yet small binding energy could probably assist DNA compactization in the early stages of loading, but does not appear as a significant barrier for the release process.
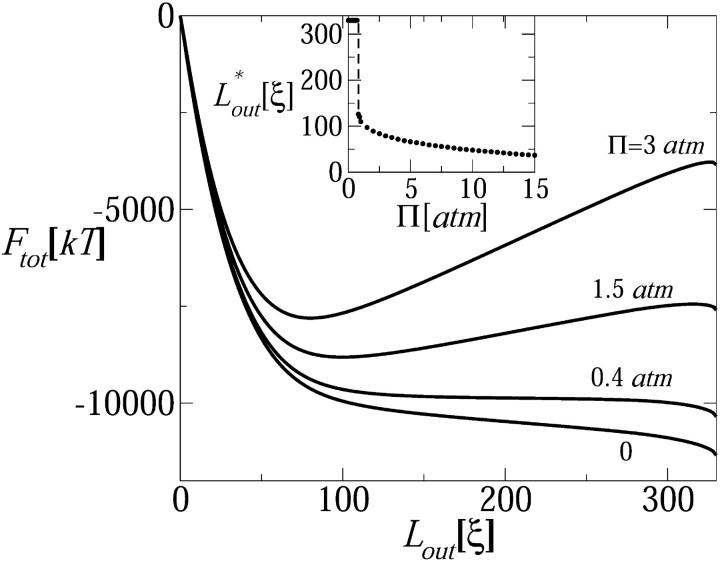
In principle, DNA injection can be opposed, even arrested, by increasing the osmotic pressure in solution, thus adding an unfavorable contribution to Fsol. More specifically, suppose that a neutral polymer (e.g., PEG) has been added to the external solution, resulting in an osmotic pressure Π. Upon DNA injection, solvent enters the capsid to fill up the released volume, whereas the stressing polymer is too large to enter the capsid and must remain in solution. This results in a larger volume fraction of polymer in solution and a corresponding free-energy increase in the system of ΠVsol(Lout), with Vsol(Lout) ≅ d02Lout the volume in solution occupied by the released DNA. More precisely, Vsol = αLout, with α as a geometric factor depending on the shape of the ejected chain. E.g., if the chain is organized as a relaxed (d = d0) torus, then α = (π/0.91)(d0/2)2 ≈ d02. Adding this osmotic pressure term to the total free energy, Eq. 4 is replaced by
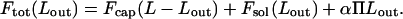 |
The osmotic pressure term in Eq. 15 opposes the injection, suggesting that for large enough Π one should expect a barrier to injection at some intermediate Lout. In Fig. 8 we show Ftot as a function of Lout for several values of Π. For Π larger than a certain threshold value Π*, a minimum in Ftot (implying a barrier to injection) appears at Lout ≈ 100ξ, shifting toward lower values of Lout as Π increases. The threshold osmotic pressure is Π* ≈ 0.4 atm, and can be rationalized as follows. From the Ftot curve in Fig. 8 corresponding to Π = 0 (see also Fig. 4), we note that, during the second stage of DNA release,  decreases nearly linearly with Lout, so that ∂Ftot0/∂Lout ≈ Δμ ≈ 3.6kBT/ξ ≈ constant, Δμ denoting the nearly constant chemical potential difference during the second injection stage. Thus a minimum in Ftot is expected for all
decreases nearly linearly with Lout, so that ∂Ftot0/∂Lout ≈ Δμ ≈ 3.6kBT/ξ ≈ constant, Δμ denoting the nearly constant chemical potential difference during the second injection stage. Thus a minimum in Ftot is expected for all 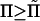 ≈ Δμ/d02 ≈ 0.4 atm, consistent with the results shown in Fig. 8.
≈ Δμ/d02 ≈ 0.4 atm, consistent with the results shown in Fig. 8.
FIGURE 8.
The effect of osmotic pressure in solution on the total chain free energy, as a function of the ejected genome length. A minimum in Ftot appears for Π ≥ 0.5 atm. Assuming that DNA ejection stops (or at least delayed) at 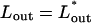 corresponding to the minimum in Ftot, the inset shows
corresponding to the minimum in Ftot, the inset shows 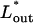 as a function of the external osmotic pressure.
as a function of the external osmotic pressure.
CONCLUDING REMARKS
The basic result of our present work is a demonstration of the interplay between chain bending and repulsion energies in determining the structural and energetic properties of packaged DNA in phage capsids. This problem is related intimately to the more general one of a self-repelling, semiflexible chain confined to a volume whose dimensions are comparable to the chain persistence length but small compared to its contour length. Our key conclusion—based on the bending and repulsion energies of double-stranded DNA—is that the bending energy is dominant, in the sense that it ultimately prohibits the chain from filling in the core of the capsid, forcing it instead to be crowded on itself in a spool-like structure. This situation arises only after a large fraction (of order two-thirds to three-quarters) of the genome is packaged. Up until this point, increasing lengths of the chain are accommodated by its bending with smaller radii of curvature and thereby filling up progressively more of the capsid volume. Beyond this point, however, additional length is accommodated largely by the chain crowding onto its nearest neighbors, filling in a spool-like volume with decreasing interaxial spacing. Because of the strong short-range repulsions acting between neighboring chains, this latter process is associated with a dramatic increase in packaging stress. We show in particular that the force required of the viral motor protein—to load the chain at the capsid portal—increases from a few pN to tens of pN as the final 20% of the genome is packaged; (in good agreement with the recent experimental determination of motor packaging force as a function of loading fraction (Smith et al., 2001). Similarly, we calculate the angular distribution of pressure acting on the inner wall of the capsid and demonstrate that its average is a strongly increasing function of the fraction of chain loaded, rising steeply from a few atmospheres to tens of atmospheres in the final 20% of packaging.
We have also treated the inverse process of DNA ejection, relevant to the first step of the viral infection cycle in which the phage ejects its genome into a host bacterial cell. This process is driven initially by precisely the stored stress established in the packaging step of phage replication, i.e., by the force f = −∂Ftot/∂Lout discussed at length above. As long as the chain free-energy Ftot is a monotonically decreasing function of ejected length Lout, this force will remain repulsive and drive the chain completely out of the capsid. In the presence of an attraction between the chain and the inside of the viral capsid, however, the ejection force will vanish whereas some of the chain remains inside. This is the case first treated phenomenologically by Gabashvili et al. (1991a,b).
More generally, we expect that the ejection of phage DNA into its host bacterial cell will be incomplete because of the osmotic pressure in the cell. More explicitly, the high concentration of cytoplasmic proteins gives rise to an effective force (work of insertion per unit length) which resists the ejection force associated with stored packaging stress. Indeed, we find that as soon as this osmotic pressure exceeds half an atm (and realistic estimates for macromolecular crowding in bacterial cells suggest that it does), at most, one-third of the genome is ejected. Accordingly, it becomes important to investigate physical mechanisms that make possible the delivery of the rest of the genome to the infected cell. One scenario for pulling in the remaining DNA involves transcription of the genes that have been delivered, i.e., translocation is driven by motor protein action of the host cell's RNA polymerase (see, for example, the case of T7 (Garcia and Molineux, 1996). Another, alternative, scenario involves the adsorption of DNA-binding proteins on the ejected (cytoplasmic) portion of the viral genome; the adsorption here gives rise to an effective force (binding energy per unit length) which pulls the rest of the chain into the cell.
Acknowledgments
We thank Dr. Phillips and Dr. Purohit for informing us about their work before publication.
The financial support of the US-Israel Binational Science Foundation; the Israel Science Foundation (to A.B.S.); and the National Science Foundation (to W.M.G.), is gratefully acknowledged. The Fritz Haber Center is supported by the Minerva Foundation, Munich, Germany.
References
- Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267–292. [DOI] [PubMed] [Google Scholar]
- Bloomfield, V. A. 1996. DNA condensation. Curr. Opin. Struct. Biol. 6:334–341. [DOI] [PubMed] [Google Scholar]
- Cairns, J., G. S. Stent, and J. D. Watson, editors. 1992. Phage and the Origins of Molecular Biology. Cold Spring Harbor Laboratory Press, New York.
- Cerritelli, M. E., N. Cheng, A. H. Rosenberg, C. E. McPherson, F. P. Booy, and A. C. Steven. 1997. Encapsidated conformation of bacteriophage T7 DNA. Cell. 91:271–280. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W. C., and S. C. Harrison. 1977. DNA arrangement in isometric phage heads. Nature. 268:598–602. [DOI] [PubMed] [Google Scholar]
- Gabashvili, I. S., and A. Y. Grosberg. 1991a. Reptation of DNA from bacteriophage. Biophysics. 36:790–796. [Google Scholar]
- Gabashvili, I. S., A. Y. Grosberg, D. V. Kuznetson, and G. M. Mrevlishvili. 1991b. Theoretical model of DNA packing in the phage head. Biophysics. 36:782–789. [PubMed] [Google Scholar]
- Gabashvili, I. S., and A. Y. Grosberg. 1992. Dynamics of double stranded DNA reptation from bacteriophage. J. Biomol. Struct. Dyn. 9:911–920. [DOI] [PubMed] [Google Scholar]
- Garcia, L. R., and I. J. Molineux. 1996. Transcription independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J. Bacteriol. 178:6921–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan, R., L. I. Pietrasanta, W. Hsieh, and H. G. Hansma. 1999. DNA toroids: stages in condensation. Biochemistry. 38:14069–14076. [DOI] [PubMed] [Google Scholar]
- Grosberg, A. Y., and A. R. Khokhlov. 1994. Statistical Physics of Macromolecules. AIP Press, New York.
- Hud, N. V., and K. H. Downing. 2001. Cryoelectron microscopy of λ-phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc. Natl. Acad. Sci. USA. 98:14925–14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jülicher, F., A. Ajdari, and J. Prost. 1997. Modeling molecular motors. Rev. Mod. Phys. 69:1269–1281. [Google Scholar]
- Kindt, J., S. Tzlil, A. Ben-Shaul, and W. M. Gelbart. 2001. DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. USA. 98:13671–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, O., L. Plançon, J. L. Rigaud, and L. Letellier. 1998. Protein-mediated DNA transfer into liposomes. J. Mol. Biol. 30:761–765. [DOI] [PubMed] [Google Scholar]
- Lambert, O., L. Letellier, W. M. Gelbart, and J. L. Rigaud. 2000. DNA delivery by phage as a strategy for encapsulating toroidal condensates of arbitrary size into liposomes. Proc. Natl. Acad. Sci. USA. 97:7248–7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, J. A., H. F. Fraenkel-Conrat, and R. A. Owens. 1994. Virology, 3rd Ed. Prentice Hall, New York.
- Lodish, H., A. Berk, S. L. Zipursky, P. Matsudaria, D. Baltimore, and J. Darnell. 2000. Molecular Cell Biology, 4th Ed. Freeman, New York.
- Odijk, T. 1998. Hexagonally packed DNA within bacteriophage T7 stabilized by curvature stress. Biophys. J. 75:1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, D. C., and V. A. Parsegian. 1992. Direct measurements of the intermolecular forces between counterion-condensed DNA double helices. Biophys. J. 61:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer, S. C., and V. A. Bloomfield. 1978. Packaging of DNA in bacteriophage heads: some considerations on energetics. Biopolymers. 17:785–792. [DOI] [PubMed] [Google Scholar]
- Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. E. Anderson, and C. Bustamante. 2001. The bacteriophage φ29 portal motor can package DNA against a large internal pressure. Nature. 413:748–752. [DOI] [PubMed] [Google Scholar]
- Ubbink, J., and T. Odijk. 1996. Deformation of toroidal DNA condensates under surface stress. Europhys. Lett. 33:353–358. [Google Scholar]
- Wikoff, W. R., L. Liljas, R. L. Duda, H. Tsuruta, R. W. Hendrix, and J. E. Johnson. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 289:2129–2133. [DOI] [PubMed] [Google Scholar]