Abstract
Chlamydia trachomatis infection can cause reactive arthritis that is associated with the persistence of chlamydial organisms in the joint. Fibroblasts of the synovial membrane represent host cells for Chlamydia during articular infection. In this study we investigated the expression of HLA class I molecules in synovial fibroblasts following infection with C. trachomatis D. The expression of HLA class I heavy chain (HLA-I) was up-regulated in infected cultures as shown by reverse transcription-PCR and immunoblotting. The increase in cell surface expression of HLA-I and β2 microglobulin on infected fibroblasts was demonstrated by flow cytometric analysis. Suppression of enhanced production of interferon-stimulated gene factor 3γ (ISGF3γ) in infected cell cultures by antisense oligonucleotide treatment reduced the level of HLA-I. Blocking antibodies to beta interferon (IFN-β) inhibited the Chlamydia-induced enhancement of both ISGF3γ and HLA-I. These findings show that the up-regulation of HLA-I in synovial fibroblasts infected with C. trachomatis is caused by the induction of IFN-β, which in turn stimulates the synthesis of ISGF3γ, a transcription factor participating in the regulation of the HLA-I gene. The IFN-β-mediated expression of HLA-I on Chlamydia-infected cells may be a regulatory factor in the immune response in chlamydial infections.
Chlamydia trachomatis, an obligate intracellular bacterium, is a major cause of urogenital infections that tend to chronicity. Chronic infections in women can lead to salpingitis and tubal occlusion. Moreover, Chlamydia is a triggering agent of reactive arthritis which is associated with the persistence of bacteria in the joint (9). Since primary target cells of urogenital C. trachomatis serovars (serovars D through K) represent nonprofessional phagocytes which constitutively express HLA class I but only weakly produce HLA class II molecules, it is conceivable that CD8+-T-cell-mediated immune mechanisms are important for the control of these chlamydial infections. Although a Th1 CD4+ response seems to be dominant in protective immunity during Chlamydia infection, the involvement of cytotoxic CD8+ T lymphocytes has been described previously (7, 17). A macaque model of salpingitis demonstrated that repeated chlamydial infections of the female upper genital tract lead to Th1-like cytokine milieus and dominant CD8+-T-cell infiltrates that are associated with progression to fibrosis and infertility (28). Chlamydia-induced reactive arthritis is associated with HLA-B27, which obviously influences the severity of disease (reviewed in reference 5). The arthritogenic peptide theory is one of the most-favored hypotheses of this association: some HLA-B27 alleles bind a specific arthritogenic peptide which is recognized by cytotoxic T lymphocytes. HLA-B27 binding peptides derived from different C. trachomatis proteins have been identified previously (14).
In the HLA class I antigen presentation pathway cytoplasmically located antigens are degraded to peptides which are transported into the endoplasmic reticulum, assemble with the HLA class I heavy chain (HLA-I) and β2 microglobulin (β2M) to form the HLA class I complex. This complex is transported to the cell surface and can be recognized by cytotoxic T lymphocytes. Although chlamydiae replicate within an endosome, there is increasing evidence that chlamydial proteins gain access to the host cell cytoplasm and enter the HLA class I pathway (4).
Inhibition of HLA molecule synthesis by intracellular pathogens is considered as a strategy to establish persisting infections (2). A recent study has demonstrated that C. trachomatis serovar L2, the causative agent of lymphogranuloma venereum, suppresses the expression of both HLA class I and HLA class II molecules in HeLa cells (30, 32). This effect was due to a chlamydial protease-like activity factor which is secreted into the host cell cytoplasm and degrades regulatory factor X-5 and upstream factor-1, two transcription factors participating in class I and class II gene expression, respectively (30, 31).
Immunoelectron microscopy on synovial membrane samples from patients with Chlamydia-induced reactive arthritis has revealed that synovial fibroblasts are suitable host cells for C. trachomatis during articular infection (18). Furthermore, Chlamydia-infected synovial fibroblasts from Lewis rats caused an arthritis with intense synovitis in rats after intra-articular injection into the knee joints (8). In previous studies we have demonstrated that the infection of synovial fibroblasts with C. trachomatis serovar D stimulates the production of beta interferon (IFN-β) and interferon-stimulated gene factor 3γ (ISGF3γ), the DNA-binding component of the ISGF3 complex (21, 22). The expression of HLA class I molecules can be up-regulated by type I IFN signaling through the induction of ISGF3 binding to the interferon-stimulated response element within the major histocompatibility complex class I promoter (19). Therefore, this study was designed to examine the effect of C. trachomatis D on HLA class I expression in human synovial fibroblasts and to investigate the involvement of IFN-β and ISGF3γ in the modulation of HLA class I production by chlamydiae.
MATERIALS AND METHODS
Chlamydia strains and fibroblast cultures.
C. trachomatis serovar D strain IC Cal 8 (obtained from the Institute of Ophthalmology, London, United Kingdom) was propagated in BGM cells as previously described (21). Infectivity titers of chlamydial stocks were quantified by titrating the number of inclusion-forming units (IFU) per milliliter in BGM cells. These titers were used to determine the infectious doses for fibroblasts.
Cultures of human synovial fibroblasts were established from synovial biopsy specimens obtained during meniscusectomies and arthroscopies of traumatic joint injury patients, using previously described methods (21). The fibroblasts used in this work were HLA-B27 negative as examined by a PCR assay according to the protocol of Bon et al. (1).
Chlamydia infection of synovial fibroblasts.
Prior to infection, cells were seeded into 35-mm-diameter culture wells. Confluent monolayers (7 × 105 to 106 cells/well) were infected with C. trachomatis by centrifugation at 4,000 × g at 37°C for 45 min. After the inoculum was decanted, the cells were washed in medium and further incubated with Dulbecco's modified Eagle's medium (Biochrom, Berlin, Germany) containing 10% fetal calf serum (Biochrom). For mock-infected cultures, cells were centrifuged without a chlamydial inoculum. For some experiments, infected fibroblasts were stimulated with tumor necrosis factor alpha (TNF-α) (200 U/ml; Biochrom) after infection. For heat inactivation, chlamydial suspensions were held at 75°C for 10 min prior to inoculation onto cell monolayers. For UV inactivation, chlamydial suspensions were placed under a UV lamp (15 W at 30 cm) for 15 min. Sheep polyclonal antibody to human IFN-β (Chemicon, Hofheim, Germany) was used to neutralize IFN-β activity in infected cultures.
Inhibition of ISGF3γ expression by antisense oligodeoxynucleotide (ODN) treatment.
Antisense (5′-TGCCCTGCCTGATGCCAT-3′) and sense (5′-ATGGCATCAGGCAGGGCA-3′) phosphorothioate ODNs corresponding to the human ISGF3γ cDNA sequence extending from the initiation codon to 18 nucleotides downstream were synthesized and high-performance liquid chromatography-purified by E. Birch-Hirschfeld (Institute of Virology, University of Jena, Jena, Germany). Transfection of fibroblasts with ODNs was performed using the Effectene transfection reagent kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
Reverse transcription (RT)-PCR assay.
Total RNA was prepared from mock-infected and Chlamydia-infected fibroblasts using the RNeasy Mini Kit (Qiagen). For each sample, first-strand cDNA was reverse transcribed from 1 μg of RNA in a total reaction volume of 20 μl, using the Promega (Mannheim, Germany) reverse transcription system. Each 25 μl of PCR mixture contained 1 μl of cDNA (corresponding to 50 ng of RNA), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 0.4 μM sense and antisense primers, and 1.25 U of Taq polymerase (Promega). The sequences of specific primers used in this work are given in Table 1. As an internal control, cDNA was amplified for pyruvate dehydrogenase (PDH). Twenty-five cycles of amplification were carried out in a TRIO-Thermoblock (Biometra, Göttingen, Germany). Reactions consisted of an initial incubation at 95°C for 7 min and then cycling at 95°C for 30 s, 60°C (ISGF3γ and PDH primers) or 54°C (IFN-β, HLA-I, and β2M primers) for 30 s, and 72°C for 1 min, with a final incubation at 72°C for 10 min. Negative controls were performed by omitting RNA from cDNA synthesis and PCR amplification (data not shown). Products were electrophoresed on 1% agarose gels and visualized with SYBR green staining. All HLA-I (A, B, C), β2M, ISGF3γ, and IFN-β band volumes (optical density × area [square millimeters]) were normalized against the PDH signal from the same sample. All results shown are representative of two separate experiments.
TABLE 1.
Primers used for RT-PCR
| Specificity | Primer paira | Product size (bp) | Refer- ence |
|---|---|---|---|
| ISGF3γ | 5′-GATACAGCTAAGACCATGTTCCGG-3′, | 1,182 | 26 |
| 5′-ACAAAGAGGACAGGTCAATCG-3′ | |||
| IFN-β | 5′-GATTCATCTAGCACTGGCTGG-3′, | 186 | 16 |
| 5′-CTTCAGGTAATGCAGAATCC-3′ | |||
| HLA-I | 5′-GTGGGCTACGTGGACGAC-3′, | 449 | 30 |
| 5′-TTCTCCAGGTATCTGCGG-3′ | |||
| β2M | 5′-TCTCGCTCCGTGGCCTTAG-3′, | 353 | 30 |
| 5′-ATGTCTCGATCCCACTTAACT-3′ | |||
| PDH | 5′-GGTATGGATGAGGACCTGGA-3′, | 105 | 24 |
| 5′-CTTCCACAGCCCTCGACTAA-3′ |
Top row, sense strand; bottom row, antisense strand.
The specificity of the PCR products was confirmed by the presence of a DNA band of the expected size (Table 1) and by sequencing in an automated ABI Prism 310 genetic analyzer (Perkin-Elmer, Applied Biosystems, Weiterstadt, Germany) as previously described (22).
Immunoblotting.
Immunoblot assays were carried out as we described in a previous paper (22). HLA-I protein was detected with a mouse monoclonal antibody purchased from DPC Biermann (Bad Nauheim, Germany) (Anogen clone LY5.1). ISGF3γ protein was detected with a mouse monoclonal antibody purchased from BD Transduction Laboratories (Heidelberg, Germany) (clone 6). Alkaline phosphatase-conjugated goat anti-mouse IgG (Dianova, Hamburg, Germany) was used as secondary antibody. The bands were visualized with 5-bromo-4-chloro-3-indolylphosphate toluidine salt-p-nitroblue tetrazolium chloride (BCIP/NBT) (Sigma, Deisenhofen, Germany). All results shown are representative of two separate experiments.
Flow cytometry.
Cells were detached from the culture wells with 0.2 mM EDTA, washed in phosphate-buffered saline, and incubated with fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal antibody to HLA-I (clone R-phycoery-thrin-2.6; BD PharMingen, Heidelberg, Germany) or FITC-conjugated rabbit polyclonal antibody to β2M (Dako, Hamburg, Germany) at room temperature for 20 min. For the dual-color assay, cell suspensions were incubated with RPE-conjugated monoclonal antibody to HLA-I (clone G46-2.6; BD Pharmingen). After washing, the cells were permeabilized with FACS permeabilizing solution (BD Immunocytometry Systems) and stained with FITC-conjugated antibody to C. trachomatis major outer membrane protein (Trinity Biotech, Frankfurt, Germany). Two additional washes were performed, and labeled cells were analyzed with a FACScan (BD Immunocytometry Systems) flow cytometer and CELL Quest software. Ten thousand cells were analyzed for each sample.
RESULTS
Up-regulation of HLA class I expression by C. trachomatis D in synovial fibroblasts.
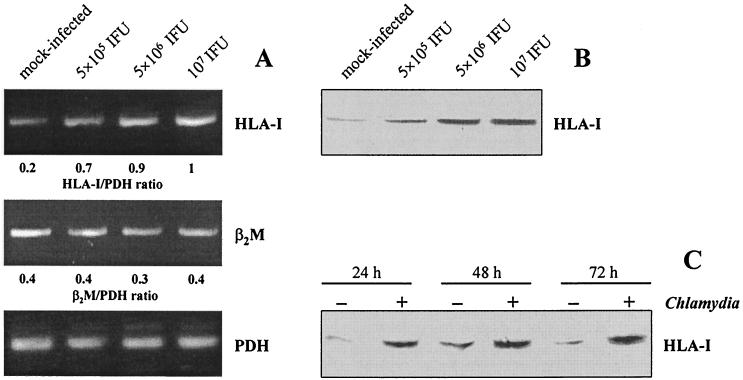
To study the effect of C. trachomatis infection on HLA class I expression in synovial fibroblasts, we first examined the levels of HLA-I and β2M mRNA using a semiquantitative RT-PCR assay. Mock-infected fibroblasts produced a basal level of HLA-I and β2M. Infection with C. trachomatis D caused an increase in the mRNA level of HLA-I in a dose-dependent manner at 24 h after infection, whereas the level of β2M mRNA remained unchanged (Fig. 1A). Expression of the housekeeping gene PDH was not affected in response to the infection.
FIG. 1.
HLA class I expression in synovial fibroblasts infected with C. trachomatis D. (A) HLA-I and β2M mRNA levels in cells infected with various doses of chlamydiae. RT-PCR analysis was conducted on total RNA extracted at 24 h after infection. (B) Immunoblot analysis of HLA-I protein in cells infected with various doses of chlamydiae. Total cell lysates were prepared at 48 h after infection. (C) Time course of HLA-I production determined by immunoblotting. Fibroblasts were infected with 5 × 106 IFU/well.
Further experiments were performed to examine whether enhanced HLA-I mRNA levels in infected cultures were associated with increased protein synthesis. Immunoblot analyses of total cell lysates showed a specific band of HLA-I of approximately 46 kDa. The amount of HLA-I protein increased with larger infectious doses (Fig. 1B). Chlamydia-exposed cells showed enhanced levels of HLA-I protein at 24, 48, and 72 h after infection compared to mock-infected cells (Fig. 1C).
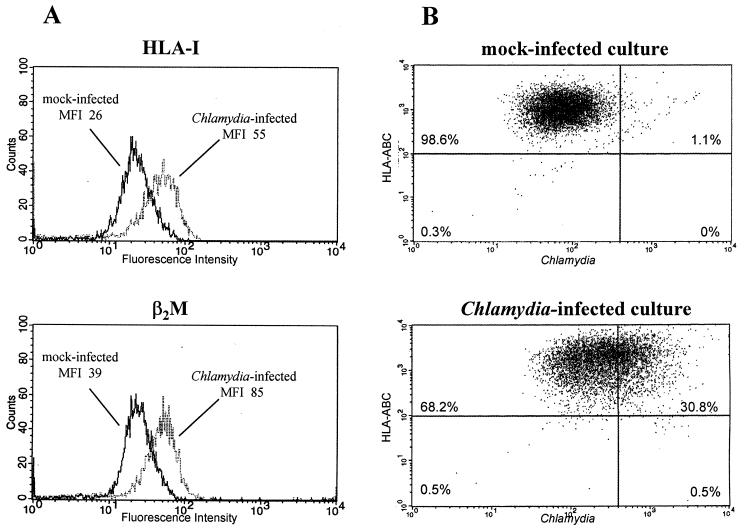
Cell surface expression of HLA class I was measured by flow cytometry. These experiments revealed an increase in the surface expression of both HLA-I and β2M following chlamydial infection (Fig. 2A). In infected cultures about 98% of the cells containing Chlamydia were HLA-I positive, indicating that there was no selective inhibition of HLA-I expression in Chlamydia-positive cells (Fig. 2B).
FIG. 2.
Flow cytometric analysis of HLA class I expression on fibroblasts with and without C. trachomatis infection. (A) HLA-I and β2M surface expression on cells in mock-infected and infected cultures. MFI, mean fluorescence intensity. (B) Dual-color assay for HLA-I and Chlamydia antigen expression. HLA-I-positive cells are shown in the top quadrants. Chlamydia-positive cells are shown in the right-hand quadrants. (A and B) The cells were infected with 5 × 106 IFU/well and harvested 48 h after infection. Results are representative of three experiments.
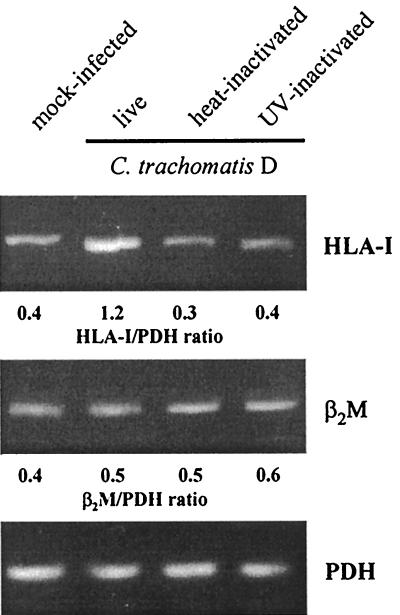
The stimulation of HLA-I expression required infectious chlamydiae, since fibroblasts exposed to heat or UV inactivated bacteria produced HLA-I mRNA levels similar to that of mock-infected cells (Fig. 3).
FIG. 3.
Effects of heat and UV inactivation of Chlamydia on HLA-I and β2M mRNA levels in synovial fibroblasts. Cells were infected with 5 × 106 IFU/well or centrifuged with an equivalent inoculum of inactivated chlamydiae. RT-PCR analysis was conducted on total RNA isolated at 24 h after infection.
Contribution of ISGF3γ and IFN-β to the HLA class I enhancement in C. trachomatis-infected fibroblasts.
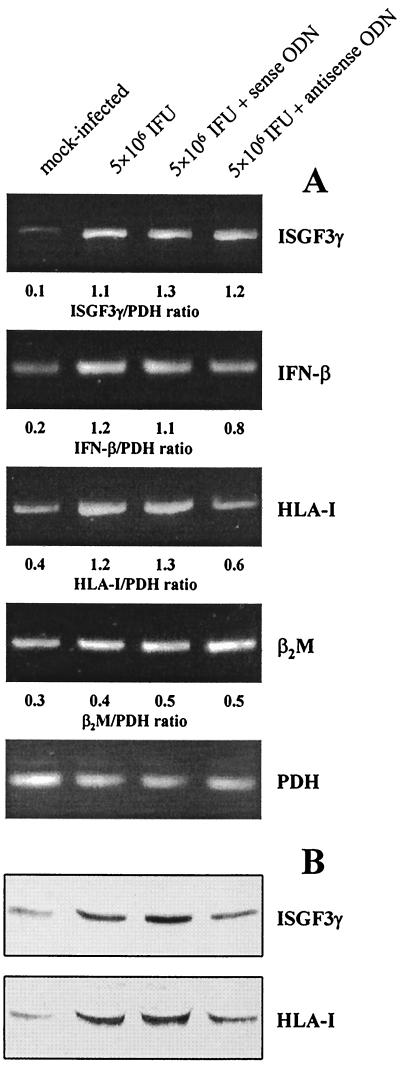
In previous studies we have shown that C. trachomatis infection up-regulates ISGF3γ and IFN-β synthesis in fibroblasts (21, 22). It is known that the expression of HLA-I is regulated by ISGF3 (19). Therefore, an antisense ODN technique was employed to investigate the role of ISGF3γ in the HLA-I enhancement in Chlamydia-infected synovial fibroblasts. We first examined the effect of different amounts of antisense and sense ODN (0.5, 1, and 2 μg per 35-mm-diameter culture well) on ISGF3γ protein levels in infected cultures (data not shown). Inhibition of ISGF3γ protein synthesis was observed at an amount of 2 μg of antisense ODN per culture well, whereas the level of mRNA remained unchanged (Fig. 4). In all further transfection experiments we used 2 μg of ODN per well and harvested the cells for RT-PCR and immunoblot assays at 24 h after transfection. As demonstrated in Fig. 4, inhibition of ISGF3γ production by antisense ODN was associated with a decrease in mRNA and protein levels of HLA-I. The induction of IFN-β mRNA was only slightly suppressed by antisense ODN (Fig. 4A). Treatment of Chlamydia-infected fibroblasts with ISGF3γ sense ODN had no inhibitory effect on the expression of any gene analyzed in this study (Fig. 4).
FIG. 4.
Effects of ISGF3γ antisense ODN on HLA class I expression in synovial fibroblasts infected with C. trachomatis D. The cells were incubated with medium alone or with ODN (2 μg/well) and collected 24 h after infection. (A) mRNA levels of ISGF3γ, IFN-β, HLA-I, and β2M were analyzed by RT-PCR. (B) ISGF3γ and HLA-I proteins were detected by immunoblotting.
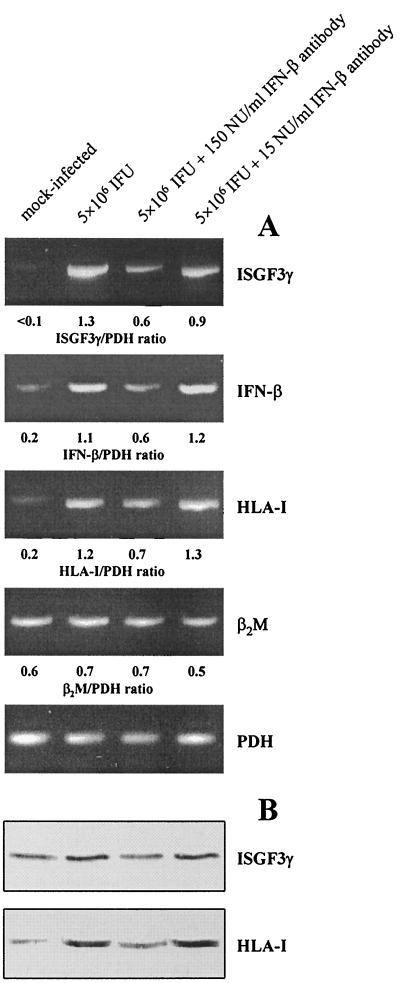
The up-regulation of ISGF3γ in synovial fibroblasts following chlamydial infection may be caused by the production of IFN-β. In the presence of neutralizing antibodies against IFN-β the production of ISGF3γ mRNA and protein was clearly inhibited when sufficient amounts of antibody were added (150 neutralizing units/ml) (Fig. 5). This effect was paralleled by a decrease in the levels of HLA-I mRNA and protein (Fig. 5). Furthermore, IFN-β neutralization results in partial inhibition of IFN-β gene induction, confirming that IFN-β regulates its own synthesis (Fig. 5A) (6, 29).
FIG. 5.
Blocking of Chlamydia-mediated HLA-I enhancement by IFN-β antibodies. Fibroblasts were infected with C. trachomatis D and incubated with medium alone or with anti-IFN-β. The cells were collected 24 h after infection. (A) mRNA levels of ISGF3γ, IFN-β, HLA-I, and β2M were analyzed by RT-PCR. (B) ISGF3γ and HLA-I proteins were determined by immunoblotting.
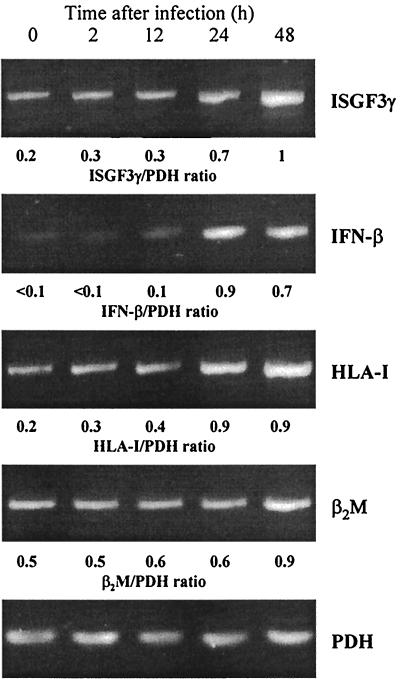
The time course of mRNA levels demonstrated that the accumulation of ISGF3γ mRNA did not precede the induction of IFN-β mRNA; stimulation of IFN-β, ISGF3γ, and HLA-I expression was observed 24 and 48 h after infection (Fig. 6).
FIG. 6.
Time course of ISGF3γ, IFN-β, HLA-I, and β2M mRNA expression in fibroblasts infected with C. trachomatis D. mRNA levels were determined by RT-PCR. The cells were infected with 5 × 106 IFU/well.
Effect of TNF-α on Chlamydia-stimulated HLA class I expression.
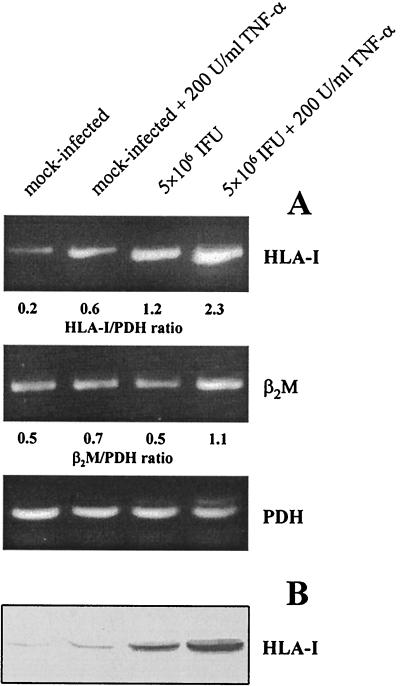
Since TNF-α interacts with chlamydial infection in the induction of the host cell type I IFN response, we examined the effect of TNF-α on HLA class I expression in C. trachomatis-infected fibroblasts (22, 23). Treatment of infected cells with TNF-α at 200 U/ml clearly enhanced the synthesis of HLA-I mRNA and protein, compared to mock-infected cells incubated with TNF-α or infected cells cultured in medium alone (Fig. 7). Moreover, β2M levels were found to be elevated in infected fibroblasts after TNF-α stimulation (Fig. 7A).
FIG. 7.
Effect of TNF-α treatment on HLA class I expression in C. trachomatis-infected fibroblasts. (A) Detection of HLA-I and β2M mRNA by RT-PCR. (B) Immunoblotting of HLA-I protein. Cells were collected 24 h after infection.
DISCUSSION
In this work the effect of C. trachomatis infection of synovial fibroblasts on HLA class I expression was investigated. We used this in vitro model because fibroblasts of the synovial membrane represent host cells for chlamydiae during articular infection in reactive arthritis (18). Whether fibroblasts are infected by Chlamydia during infections of the urogenital tract remains unclear. In situ hybridization of chlamydial DNA and detection of bacterial antigen in tubal biopsy specimens from women with postinfectious sterility suggested that C. trachomatis persists in submucosal tissues (20). These observations correspond to studies of a macaque model of Chlamydia-induced salpingitis, in which the pathogen was found not only in the mucosa but also in the submucosa and deep tissues, including connective tissue and smooth muscle layers (3).
We have shown that the infection of synovial fibroblasts with C. trachomatis D stimulates the expression of HLA class I molecules. This effect could be reduced by IFN-β neutralization in infected cultures, demonstrating that a mechanism of the HLA class I enhancement is a secondary effect of IFN-β induced by Chlamydia. The expression of HLA-I is regulated at the transcriptional level (27). The interferon-stimulated response element which is bound by interferon regulatory family (IRF) transcription factors such as IRF-1 and ISGF3γ, is one of the main control elements within the HLA-I promoter (27). Using an antisense ODN technique we could demonstrate that the increased production of ISGF3γ is involved in the up-regulation of HLA-I in C. trachomatis-infected cultures. Type I IFN can induce the transcription of IFN-stimulated genes by activating the function of the ISGF3 complex and by enhancing the level of ISGF3γ (12, 13). Therefore, the increased production of ISGF3γ is a critical regulator of the IFN response. Neutralization of IFN-β activity in infected cell cultures clearly reduced the level of ISGF3γ, whereas ISGF3γ antisense ODN treatment had only little effects on induction of the IFN-β gene. These findings indicate that the IFN-β initially produced in response to the chlamydial infection stimulates ISGF3γ production, resulting in enhanced expression of HLA-I.
The treatment of infected cells with TNF-α caused an additional increase in the synthesis of HLA-I, whereas TNF-α stimulation of mock-infected cells had only little effect. It is known that TNF-α alone inefficiently induces the IFN-β gene but acts in synergism with IFNs in the activation of IFN-inducible genes (10). Our observations correspond to previous studies in which an interaction of TNF-α and chlamydial infection in the induction of IFN-β and ISGF3γ was found (22, 23).
Recently, Zhong et al. reported that C. trachomatis suppresses the expression of HLA class I molecules in HeLa cells and other tumor cell lines (30). This phenomenon was due to a Chlamydia-specific factor (chlamydial protease-like activity factor) which is secreted into the host cell cytoplasm and degrades regulatory factor X-5, a transcription factor participating in HLA-I gene expression (27, 30, 31). However, the activation of specific CD8+ T cells in chlamydial infections provides an argument against the hypothesis of a general inhibition of HLA-I in all Chlamydia-infected cells. Our data indicate that the induction of the type I IFN response of the host cell plays an important role in the expression of HLA class I molecules after chlamydial infection.
In contrast to the stimulatory effect on HLA class I synthesis, C. trachomatis D inhibits the IFN-γ-induced expression of HLA class II molecules in synovial fibroblasts (21). Therefore, it is possible that an HLA class I-restricted CD8+-T-cell response participates in the recognition of Chlamydia-infected nonprofessional phagocytes. The involvement of cytotoxic T lymphocytes in the immune response to C. trachomatis infections has been demonstrated in several studies (7, 11, 15, 17, 25). Besides the ability to lyse infected cells, the production of IFN-γ is another potential mechanism for CD8+ T cells to limit the spread of infection (15).
In conclusion, we have shown that the infection of synovial fibroblasts with C. trachomatis D results in an up-regulation of HLA-I which is caused by the induction of IFN-β and ISGF3γ. Further studies are needed to elucidate whether the IFN-β-mediated expression of HLA class I molecules on Chlamydia-infected cells has any impact on the activation of CD8+ T lymphocytes and influences the control of chlamydial infection.
Acknowledgments
This work was supported by grant BMBF 01ZZ9602 from the Bundesministerium für Bildung und Forschung, Berlin, Germany.
We thank the collaborators of the Bundesinstitut für Gesundheitlichen Verbraucherschutz und Veterinärmedizin Jena for performing Mycoplasma testing, W. Lungershausen (Clinic of Surgery, University of Jena) for providing synovial tissue, and E. Birch-Hirschfeld (Institute of Virology, University of Jena) for providing PCR primers and phosphorothioate ODN.
Editor: J. D. Clements
REFERENCES
- 1.Bon, M. A. M., A. van Oeveren-Dybicz, and F. A. J. T. M. van den Bergh. 2000. Genotyping of HLA-B27 by real-time PCR without hybridization probes. Clin. Chem. 46:1000-1002. [PubMed] [Google Scholar]
- 2.Brodsky, F. M., L. Lem, A. Solache, and E. M. Bennett. 1999. Human pathogen subversion of antigen presentation. Immunol. Rev. 168:199-215. [DOI] [PubMed] [Google Scholar]
- 3.Cappuccio, A. L., D. L. Patton, C. C. Kuo, and L. A. Campbell. 1994. Detection of Chlamydia trachomatis deoxyribonucleic acid in monkey models (Macaca nemestrina) of salpingitis by in situ hybridization: implications for pathogenesis. Am. J. Obstet. Gynecol. 171:102-110. [DOI] [PubMed] [Google Scholar]
- 4.Fling, S. P., R. A. Sutherland, L. N. Steele, B. Hess, S. E. F. D' Orazio, J. F. Maisonneuve, M. F. Lampe, P. Probst, and M. N. Starnbach. 2001. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 98:1160-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaston, J. S. H. 2000. Immunological basis of Chlamydia-induced reactive arthritis. Sex. Transm. Infect. 76:156-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harada, H., Matsumoto, M. Sato, Y. Kashiwazaki, T. Kimura, M. Kitagawa, T. Yokochi, R. Sok-Pin Tan, T. Takasugi, Y. Kadokawa, C. Schindler, R. D. Schreiber, S. Noguchi, and T. Taniguchi. 1996. Regulation of IFN-α/β genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-α/β. Genes Cells 1:995-1005. [DOI] [PubMed] [Google Scholar]
- 7.Igietseme, J. U., D. M. Magee, D. M. Williams, R. G. Rank. 1994. Role for CD8+ T cells in antichlamydial immunity defined by Chlamydia-specific T-lymphocyte clones. Infect. Immun. 62:5195-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inman, R. D., and B. Chiu. 1998. Synoviocyte-packaged Chlamydia trachomatis induces a chronic aseptic arthritis. J. Clin. Investig. 102:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inman, R. D., J. A. Whittum-Hudson, H. R. Schumacher, and A. P. Hudson. 2000. Chlamydia and associated arthritis. Curr. Opin. Rheumatol. 12:254-262. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, D. R., and J. S. Pober. 1994. HLA class I heavy-chain gene promoter elements mediating synergy between tumor necrosis factor and interferons. Mol. Cell. Biol. 14:1322-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, S.-K., M. Angevine, K. Demick, L. Ortiz, R. Rudersdorf, D. Watkins, and R. DeMars. 1999. Induction of HLA class I-restricted CD8+ CTLs specific for the major outer membrane protein of Chlamydia trachomatis in human genital tract infections. J. Immunol. 162:6855-6866. [PubMed] [Google Scholar]
- 12.Kimura, T., Y. Kadokawa, H. Harada, M. Matsumoto, M. Sato, Y. Kashiwazaki, M. Tarutani, R. Sok-Pin Tan, T. Takasugi, T. Matsuyama, T. W. Mak, S. Noguchi, and T. Taniguchi. 1996. Essential and non-redundant roles of p48 (ISGF3γ) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells 1:115-124. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, R., and L. Korutla. 1995. Induction of expression of interferon-stimulated gene factor-3 (ISGF-3) proteins by interferons. Exp. Cell Res. 216:143-148. [DOI] [PubMed] [Google Scholar]
- 14.Kuon, W., H. G. Holzhütter, H. Appel, M. Grolms, S. Kollnberger, A. Traeder, P. Henklein, E. Weiss, A. Thiel, R. Lauster, P. Bowness, A. Radbruch, P. M. Kloetzel, and J. Sieper. 2001. Identification of HLA-B27-restricted peptides from the Chlamydia trachomatis proteome with possible relevance to HLA-B27-associated diseases. J. Immunol. 167:4738-4746. [DOI] [PubMed] [Google Scholar]
- 15.Lampe, M. F., C. B. Wilson, M. J. Bevan, and M. Starnbach. 1998. Gamma interferon production by cytotoxic T lymphocytes is required for resolution of Chlamydia trachomatis infection. Infect. Immun. 66:5457-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X. L., M. Boyanapalli, X. Weihua, D. V. Kalvakolanu, and B. Hassel. 1998. Induction of interferon synthesis and activation of interferon-stimulated genes by liposomal transfection reagents. J. Interferon Cytokine Res. 18:947-952. [DOI] [PubMed] [Google Scholar]
- 17.Magee, D. M., D. M. Williams, J. G. Smith, C. A. Bleicker, B. G. Grubbs, J. Schachter, and R. G. Rank. 1995. Role of CD8 T cells in primary Chlamydia infection. Infect. Immun. 63:516-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nanagara, R., F. Li, A. M. Beutler, A. P. Hudson, and H. R. Schumacher. 1995. Alteration of Chlamydia trachomatis biologic behaviour in synovial membranes: suppression of surface antigen production in reactive arthritis and Reiter's syndrome. Arthritis Rheum. 38:1410-1417. [DOI] [PubMed] [Google Scholar]
- 19.Nielsch, U., S. G. Zimmer, and L. E. Babiss. 1991. Changes in NF-κB and ISGF3 DNA binding activities are responsible for differences in MHC and β-IFN gene expression in Ad5- versus Ad12-transformed cells. EMBO J. 10:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patton, D. L., M. Askimazy-Elbhar, J. Henry-Suchet, L. A. Campbell, A. Cappuccio, W. Tannous, S. P. Wang, and C. C. Kuo. 1994. Detection of Chlamydia trachomatis in fallopian tube tissue in women with postinfectious tubal infertility. Am. J. Obstet. Gynecol. 171:95-101. [DOI] [PubMed] [Google Scholar]
- 21.Rödel, J., A. Groh, H. Vogelsang, M. Lehmann, M. Hartmann, and E. Straube. 1998. Beta interferon is produced by Chlamydia trachomatis-infected fibroblast-like synoviocytes and inhibits gamma interferon-induced HLA-DR expression. Infect. Immun. 66:4491-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rödel, J., A. Groh, M. Hartmann, K.-H. Schmidt, M. Lehmann, W. Lungershausen, and E. Straube. 1999. Expression of interferon regulatory factors and indoleamine 2,3-dioxygenase in Chlamydia trachomatis-infected synovial fibroblasts. Med. Microbiol. Immunol. 187:205-212. [DOI] [PubMed] [Google Scholar]
- 23.Rödel, J., S. Assefa, D. Prochnau, M. Woytas, M. Hartmann, A. Groh, and E. Straube. 2001. Interferon-β induction by Chlamydia pneumoniae in human smooth muscle cells. FEMS Immunol. Med. Microbiol. 32:9-15. [DOI] [PubMed] [Google Scholar]
- 24.Rolfs, A., I. Schuller, U. Finckh, and I. Weber-Rolfs. 1992. PCR: clinical diagnostics and research, p. 102-107. Springer-Verlag, Berlin, Germany.
- 25.Thoma-Uszinski, S., U. Simnacher, R. Marre, A. Essig. 1998. Clearance of Chlamydia trachomatis-induced polyserositis in SCID mice requires both CD4+ and CD8+ cells. Med. Microbiol. Immunol. 187:71-78. [DOI] [PubMed] [Google Scholar]
- 26.Tnani, M., and B. Bayard. 1999. Evidence for IRF-1-dependent gene expression deficiency in interferon unresponsive HepG2 cells. Biochim. Biophys. Acta 1451:59-72. [DOI] [PubMed] [Google Scholar]
- 27.van den Elsen, P. J., S. J. P. Gobin, M. C. A. J. van Eggermond, and A. Peijnenburg. 1998. Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics 48:208-221. [DOI] [PubMed] [Google Scholar]
- 28.van Voorhis, W. C., L. K. Barrett, Y. T. Cosgrove Sweeney, C. C. Kuo, and D. L. Patton. 1997. Repeated Chlamydia trachomatis infection of Macaca nemestrina fallopain tubes produces a Th1-like cytokine response associated with fibrosis and scarring. Infect. Immun. 65:2175-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Sato, K. Ozato, and T. Fujita. 1996. Autocrine amplification of type I interferon gene expression mediated by interferon-stimulated gene factor 3 (ISGF3). J. Biochem. 120:160-169. [DOI] [PubMed] [Google Scholar]
- 30.Zhong, G., L. Liu, T. Fan, P. Fan, and H. Ji. 2000. Degradation of transcription factor RFX-5 during the inhibition of both constitutive and interferon-γ-inducible major histocompatibility complex class I expression in Chlamydia-infected cells. J. Exp. Med. 191:1525-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong, G., P. Fan, H. Ji, F. Dong, Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong, G., T. Fan, and L. Liu. 1999. Chlamydia inhibits interferon-γ-inducible major histocompatibility complex class II expression by degradation of upstream regulatory factor 1. J. Exp. Med. 189:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]