Abstract
Fimbriae have been shown to play an essential role in the adhesion of pathogenic gram-negative bacteria to host cells. In the enteroinvasive bacterium Yersinia pseudotuberculosis, we characterized a previously unknown 11-kb chromosomal locus involved in the synthesis of type IV pili. The locus consists of 11 open reading frames forming a polycistronic unit and encoding putative Pil proteins, PilLMNOPQRSUVW. When introduced into Escherichia coli, the Y. pseudotuberculosis operon reconstituted bundles of filaments at a pole on the bacterial surface, demonstrating that the pil locus was functional in a heterogenous genetic background. Environmental factors regulated transcription of the Y. pseudotuberculosis operon; in particular, temperature, osmolarity, and oxygen tension were critical cues. Deletion of the type IV pilus gene cluster was associated with a reduction of Y. pseudotuberculosis pathogenicity for mice infected orally. Forty-one percent of Y. pseudotuberculosis strains isolated from human or animal sources harbored the type IV pilus locus. Therefore, the pil locus of Y. pseudotuberculosis might constitute an “adaptation island,” permitting the microorganism to colonize a vast reservoir.
Type IV pili are appendages emanating from the surfaces of several gram-negative bacteria, including species pathogenic for animals and plants (Actinobacillus pleuropneumoniae, Aeromonas veronii biovar sobria, Aeromonas caviae, Dichelobacter nodosus, Eikenella corrodens, enteropathogenic Escherichia coli [EPEC] and enterotoxigenic E. coli [ETEC], Legionella pneumophila, Moraxella bovis, Neisseria meningitidis, Neisseria gonorrhoeae, Pseudomonas aeruginosa, Salmonella enterica, and Vibrio cholerae) (9, 11, 12, 15, 16, 18, 19, 20, 21, 25, 27, 40, 45, 49). These structures, which may be peritrichous or polar on the cell surface, sometimes form bundles and have been implicated in a variety of bacterial functions, including cell adhesion (24), bacteriophage adsorption (17, 19), plasmid transfer (19), and twitching motility, a form of flagellum-independent locomotion (26, 44). Pili are composed of pilin subunits, the primary structures of which are conserved (24). All pilins are produced from prepilin molecules through cleavage of the leader sequence by a prepilin peptidase. There is a long hydrophobic segment (20 to 30 amino acid residues [aa]) at the N-terminal region of the mature pilin protein. Type IV pili are classified into two subclasses, IVA and IVB, on the basis of similarities in the deduced amino acid sequences of prepilins. Type IVA prepilins have a very short leader sequence (5 to 6 aa), and the N-terminal amino acid of the mature pilin is a methylated phenylalanine. Type IVB prepilins tend to have longer signal sequences (13 to 30 aa), and the N-terminal amino acid of the mature protein may be a methionine (methylated in some cases), leucine, tryptophan, or serine. The assembly machinery involved in the formation of fimbriae consists of a set of proteins encoded by genes either scattered throughout the bacterial genome (IVA subclass) or organized into operons consisting of 11 to 14 genes (IVB subclass). Many of the proteins involved in type IV pilus biogenesis are homologous to proteins involved in type II secretion and natural transformation (DNA uptake) systems in various bacteria (6, 7, 10, 14, 18, 28, 30, 34, 41).
Fimbriae of the IVB subclass have been described exclusively in enteric pathogens such as V. cholerae, E. coli (EPEC and ETEC), and S. enterica (1, 11, 12, 43, 48). The gram-negative bacterium Yersinia pseudotuberculosis is responsible for animal and human infections of the digestive tract (ileitis and mesenteric lymphadenitis) following ingestion of contaminated food or water (3). Here, we provide evidence that the enteropathogenic species Y. pseudotuberculosis also harbors a functional IVB subclass fimbrial gene cluster. We report the genetic organization of this cluster and its distribution among strains of this species. We also show that the type IV pilus gene cluster contributes to the pathogenicity of Y. pseudotuberculosis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Ninety-two human and animal strains of Y. pseudotuberculosis were used: 47 were of serotype I (367/89, 376/89, 892/97, 102/88, 442/93, 310/89, YPT2, YPT3, YPT4, YPT6, YPT7, YPT8, YPT9, YPT10, YPT11, YPT12, 347/89, 72/91, 2/91, 2775, 32777, 32953, 2783, 2790, 2853, WS53/94, WS37/95, WS27/93, WS31/96, WS34/94, 33036, 33034, 33052, 32845, 32849, 33053, 32982, 417/94, 40/92, 736/91, 3625/92, 1924/95, 9314/74, 58/87, 1937/88, 2063/88, and 145/89), 11 were of serotype II (165/89, 297/89, 300/89, MA, 1432/94, 2515, 2892, 2926, 2929, WS25/91, and 33054), 22 were of serotype III (304/89, 201/90, 97/88, 32829, 32842, 32984, 32826, 32861, 32945, 32977, 32975, 32992, YPIII, 2666, 2887, 2889, YPT1, YPT5, 32607, 33001, 766/95, and 1216/93), 5 were of serotype IV (ST, AH, NT, 3255/96, and 1830), 4 were of serotype V (199/90, 2821, 2843, and 99/91), and 3 were of serotype VI (682/90, 1553, and 1554). They were kindly provided by S. Aleksić (Hygiene Institut Hamburg, Hamburg, Germany), E. Falsen (University of Göteborg, Göteborg, Sweden), D. Caugant (National Institute of Public Health, Oslo, Norway), E. Carniel (Institut Pasteur, Paris, France), R. Van Noyen (Imelda Hospital, Bonheiden, Belgium), and N. Takeda (Kurashiki Central Hospital, Okayama, Japan). Strain MIV is a mutant engineered from the parent strain 32777 (serotype I). The E. coli DH5α, NM554 (32), and MC1061 strains (46) were used as hosts for cloning. E. coli SY327λpir and SM10λpir were recipients for replication of the suicide vector pCVD442 and its derivatives. The cloning vectors used in this work and the recombinant plasmids constructed in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Reference or origin |
|---|---|---|
| pUC18 | Cloning vector; Ap | Appligene |
| pACYC184 | Cloning vector; Tet | New England BioLabs |
| pHC79 | Cloning vector; Ap | Stratagene |
| pCVD442 | Suicide vector containing the counterselectable marker sacB; Ap | 5 |
| pUC4K | Km; source of aminoglycoside phosphotransferase aphA-Ia gene | Amersham Pharmacia |
| pLS92.1 | pUC18Ω; 3.4-kb EcoRV-digested DNA from Y. pseudotuberculosis 32777 | This work |
| pMM2.1 | pHC79Ω; 30- to 35-kb partially Sau3A-digested DNA from Y. pseudotuberculosis 32777 | This work |
| pMM2.A6 | pHC79Ω; 30- to 35-kb partially Sau3A-digested DNA from Y. pseudotuberculosis 32777 | This work |
| pMM3.D6 | pHC79Ω; 30- to 35-kb partially Sau3A-digested DNA from Y. pseudotuberculosis 32777 | This work |
| pMM5 | pACYC184Ω; 10.8-kb NcoI-digested fragment from pMM2.1 | This work |
| pMM1 | pUC18Ω; 1,693-bp SstI/BamHI PCR-generated fragment with primers 1 and 2 encompassing the upstream region of pilL from Y. pseudotuberculosis 32777 | This work |
| pMM2 | pMM1Ω; 1,162-bp BamHI/PstI PCR-generated fragment with primers 3 and 4 encompassing part of pilV and its 3′ flanking region from Y. pseudotuberculosis 32777 | This work |
| pMM3 | pMM2Ω; 1,264-bp BamHI-digested fragment from pUC4K containing aphA-Ia | This work |
| pMM4 | pCVD442Ω; 4,524-bp SstI-digested fragment from pMM3 | This work |
Ap, Km, and Tet, resistance to ampicillin, kanamycin, and tetracycline, respectively; Ω, in vitro insertion.
Bacterial growth conditions.
Y. pseudotuberculosis and E. coli strains were grown routinely in Luria-Bertani (LB) broth or agar medium. Ampicillin (100 μg ml−1), tetracycline (12 μg ml−1), kanamycin (50 μg ml−1), sucrose (10%), IPTG (isopropyl-β-d-thiogalactopyranoside), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were added to media for bacterial selection when necessary. For anaerobic bacterial growth, LB broth was first boiled to remove dissolved air, and liquid paraffin was added to the hot medium. For gene transcription studies, the osmolarity of the LB broth was adjusted by adding 100 mM KCl or 200 mM l-arabinose (final concentrations). The pH of the LB broth was adjusted to 5.5, 7.0, or 8.5 and maintained by supplementing the medium with sulfonate buffers with the appropriate pKa values: MOPS [3-(N-morpholino)propanesulfonic acid], pKa = 7.2; MES [2-(N-morpholino)ethanesulfonic acid], pKa = 6.1; and TAPS [tris(hydroxymethyl)methyl-aminopropanesulfonic acid], pKa = 8.4 (29). The pH of each medium was measured before and after bacterial growth to ensure it did not change (<0.1 U).
DNA and RNA preparation, analysis, and hybridization.
Genomic DNA extraction and small-scale plasmid DNA isolation were performed as previously described (8). Large-scale plasmid DNA preparations were purified on Qiagen columns according to the manufacturer's recommendations. DNA was digested with the appropriate restriction endonucleases purchased from Life Technologies or Roche Diagnostics, and the resulting fragments were separated by gel electrophoresis. Restriction fragments were eluted from agarose gels with the Qiaquick gel extraction kit (Qiagen GmbH). DNA fragments were transferred from gels to Hybond-N+ membranes (Amersham) by the Southern technique. Colony blotting was performed as previously described (36).
Total RNAs were extracted from growing Yersinia cells with the SV total RNA isolation kit (Promega) following the manufacturer's instructions. For slot blot analysis, RNA was spotted onto a Hybond-N+ membrane.
The DIG hybridization and detection kit (Roche Diagnostics) was used for hybridization. Hybridization was carried out under stringent conditions with a digoxigenin-labeled DNA probe.
Synthetic oligonucleotides.
Oligonucleotide primers were custom synthesized (Sigma and Genset) for PCR generation of DNA fragments used for cloning or probing or for reverse transcriptase (RT)-PCR assays. Their nucleotide sequences (5′-3′) were as follows: YopH1, CATCGTCAGGTATCTCGA; YopH2, CAATCAGTTGCGCAGTAC; 1f, TCCGTTCGACAGAATGC; 1r, TTGTGCTGGGCGGTGGTGT; 2f, GCCGAGGTAGGCTCAAC; 2r, GGCTGGCTTGCGCAAACA; 3f, GGCTGAAGGGAGTATTG; 3r, AAGGCTGAGTTTTAGCGC; 4f, GGTCATAACGGTCTGG; 4r, CGGTAATCTGTTGCGCT; 5f, GGTGTTAGCCAGCATAGA; 5r, GGATAGATTCCAGTTGGC; 6f, TGTCGTTGAGGTCACT; 6r, CGAACTATCAGCTATACG; 7f, TGTGCATTGCCAGCAA; 7r, ATCGGTATGAACCTCC; 8f, TGAGCGAGTCAACAAGCG; 8r, TCAAGGATACCCCAGCCA; 9f, AGGACACTGGCAGAATCG; 9r, GAGGTCATATTGCTGAAC; 10f, AGTCAAGGTCTATGATCC; 10r, AAGCGCGATAACTGCACG; 11f, CACGCTATTGGACACA; 11r, GCACGTCAGAATAGTAG; 12f, TATGTTGGCCAATGGC; 12r, GTTAGTCGATCAGCTAC; 1, AAAAGAGCTCAGCCACTATGCCATTGGTC; 2, CGCGGATCCCCAGCGATGATAGTTAAGC; 3, CGCGGATCCAGCTATTAGCCTCTGCTGG; 4, AAAACTGCAGGAGCTCTCAGTGTGAGCAATCACTC; pilrs1, TCCTTGCTGACCGAAGGTG; pilrs2, AGGTACGCCACCGACCTGA; pil7, AAAGTTTTAACTAATCGC; pil6, CGTTTTTTTATTTCCCCG; pil5, GCGTTGTCCACCCGTTCGCCG; pil3, ACCATTTCGGCGGGCATGCTGATA; 5′REG1, CTCTGTTGGCGATGGCTT; 5′REG2, AATGACATTGGCCAGC; 3′REG1, GATCGACGATCTGGTC; 3′REG2, GCAGTAGATGTCAGTG; RRNA1, GAGAGTTTGATCCTGGCTCAG; and RRNA2, AAGGAGGTGATCCAGCCG.
PCR.
PCR amplification was performed as described elsewhere (37) with AmpliTaq Gold polymerase purchased from Perkin-Elmer Applied Biosystems. Digoxigenin-labeled PCR products were generated with the PCR DIG labeling mix from Roche Diagnostics. PCR products were purified on Dye-ex spin columns or with a Qiaquick PCR purification kit purchased from Qiagen. RT-PCR assays were performed following the manufacturer's recommendations with a kit (Access RT-PCR system) purchased from Promega.
DNA cloning and sequencing.
DNA fragments were ligated to endonuclease-restricted vectors according to standard techniques with T4 DNA ligase (Life Technologies). Recombinant plasmid DNA was introduced into E. coli by transformation. Chromosomal DNA inserted into the cosmid pHC79 was packaged into bacteriophage lambda head particles with a packaging kit (Giga Pack II) purchased from Stratagene, and E. coli NM554 cells were transduced as described elsewhere (36).
DNA was sequenced by the dideoxy chain termination method using the ABI PRISM dichloRhodamine Dye Terminator sequencing kit with Amplitaq DNA polymerase FS (Perkin-Elmer) according to the manufacturer's instructions. Extension products were analyzed with the Applied Biosystems model ABI 377XL automated DNA sequencer (Perkin-Elmer). The nucleotide sequences obtained were analyzed with Perkin-Elmer software (Sequence Navigator).
Transmission electron microscopy (TEM).
Bacteria were scraped from 24-h cultures grown at 37°C on the surface of LB agar and suspended in 30 μl of 0.05 M Tris-buffered saline (pH 7.4). A carbon-coated Formvar copper grid (200 mesh; Electron Microscopy Sciences) was placed on the bacterial suspension for 2 min. The grid was then stained for 2 min with 4% uranyl acetate, washed twice for 15 s each time with distilled water, and finally air dried. The grid was examined in a Hitachi-7500 transmission electron microscope.
Mouse experimental infection.
Six week-old female inbred BALB/c mice (Iffa Credo) were challenged either by the intravenous route (a 0.3-ml bacterial suspension in sterile phosphate-buffered saline) or by the intragastric (i.g.) route (0.2 ml of bacterial suspension in sterile distilled water) using a gastric tube. For gastric inoculation, the mice were first starved for 18 h. Bacterial inoculums were prepared from overnight cultures in LB broth at 28°C. The cultures were centrifuged, and the bacterial pellets were washed once and resuspended in distilled water or phosphate-buffered saline. The animals were kept in positive-pressure cabinets during experimentation, and mortality was monitored daily for 21 days after challenge. For each experimental infection, the presence of the virulence plasmid pYV was confirmed by PCR on bacterial thermolysates using the YopH1 and YopH2 primers, which are internal to yopH, a gene located on pYV (4). Infected animals were monitored for 3 weeks, and the 50% lethal dose (LD50) was calculated from groups of 5 (intravenous model) or 10 (i.g. model) mice according to the method of Reed and Muench (33).
Nucleotide sequence accession number.
The nucleotide sequence data have been deposited in the GenBank nucleotide sequence database under accession number AY047316.
RESULTS
Y. pseudotuberculosis strain 32777 harbors a type IV pilus gene cluster.
While cloning for another research project, we isolated pLS91.2, a recombinant plasmid bearing a ∼3.4-kb EcoRV DNA fragment from Y. pseudotuberculosis 32777. Analysis of the sequence of the DNA insert revealed the presence of four open reading frames (ORFs) (orf1 to -4), only two of which (orf2 and orf3) were complete. Similarity searches with the deduced amino acid sequences of the proteins encoded by these ORFs showed that ORF1, ORF2, ORF3, and ORF4 displayed 55, 57, 59, and 55% similarity to PilL, PilM, PilN, and PilO, respectively, of S. enterica. These Pil proteins are thought to be involved in the biogenesis of type IV pili in this pathogen (19). To identify the complete type IV pilus gene cluster in Y. pseudotuberculosis, a cosmid library was constructed from strain 32777 DNA. Briefly, bacterial DNA partially digested with Sau3A was ligated to cosmid pHC79 at the BamHI site, and E. coli NM554 was transformed. Recombinants were further screened by colony blot hybridization under stringent conditions, using PCR-generated DNA probes at the 5′ and 3′ ends of the pLS91.2 insert. Three positive clones, NM554 (pMM2.1), NM554 (pMM2.A6), and NM554 (pMM3.D6), were isolated.
The type IV pilus gene cluster from Y. pseudotuberculosis forms a single polycistronic unit.
Cosmid pMM2.1 contained an 11-kb locus composed of 11 ORFs, the organization of which is shown in Fig. 1A. In terms of the arrangement of the first 10 genes and their products (see below), the Y. pseudotuberculosis gene cluster strongly resembles the pil locus recently discovered in S. enterica (19, 48). For this reason, we adopted, by analogy, the gene designation put forward for the Salmonella pil genes. The pil cluster was incomplete in cosmid pMM2.A6 and in pMM3.D6, which contained only pilLMN and pilNOPQRSUVW, respectively.
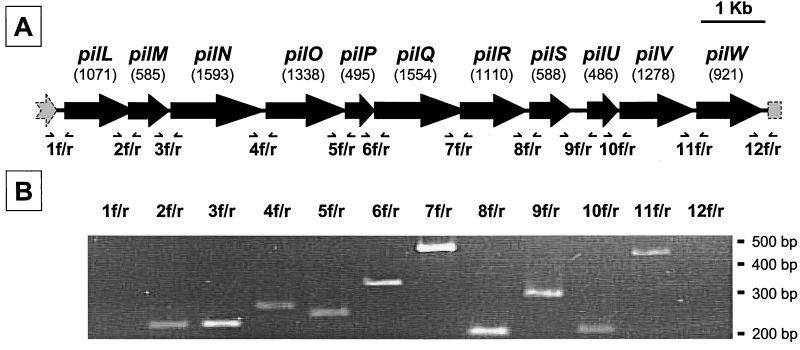
FIG. 1.
The Y. pseudotuberculosis pil operon. (A) Genetic organization of the pil gene cluster. The pil gene cluster consists of 11 genes (the sizes in base pairs are shown in parentheses) with the same polarity and apparently arranged in an operon-like manner (see the text for details). (B) Transcriptional analysis of the pil gene cluster. RNA extracted from exponentially growing Y. pseudotuberculosis cultures at 37°C were analyzed by RT-PCR using primer sets 1 to 12. Forward (f) and reverse (r) primers were designed within adjacent genes of the pil locus and flanking regions (panel A). The amplification products were analyzed by agarose gel electrophoresis.
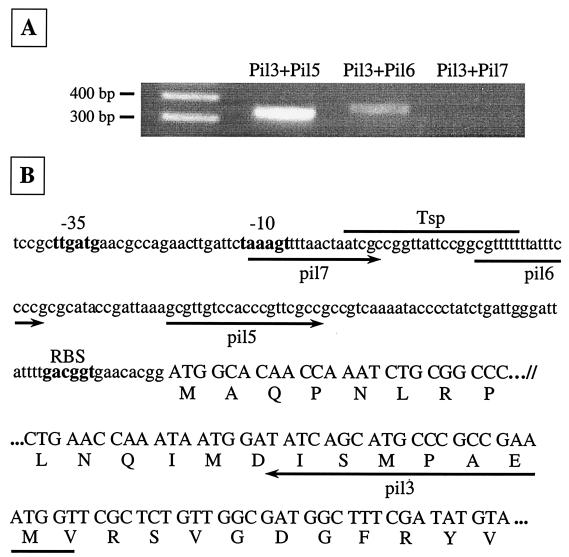
The mean G+C content of the pil cluster was 50.8%, and for each pil gene, the percentage of G+C residues was between 47.6 (pilW) and 53.3% (pilU). All pil ORFs were present on the same strand, and intergenic spaces ranged from −146 to 179 nucleotides. Some pil genes were preceded by a putative Shine-Dalgarno sequence. Taken together, these facts suggested that the pil gene cluster may be transcribed as a single unit. The instability of the large mRNA transcript made Northern blot analysis impossible. The polycistronic organization of the locus was confirmed by RT-PCR experiments on total RNA extracted from strain 32777 using 12 sets of primers (1 to 12) (Fig. 1 A). No amplification product was obtained with primer sets 1 and 12, whereas primer sets 2 to 11 yielded amplicons of the expected sizes (Fig. 1B). These results indicated that pilLMNOPQRSUVW are most likely cotranscribed as a single mRNA, since RT-PCR does not definitively preclude the existence of other nested transcripts. Given the instability of the mRNA, several attempts at primer extension analysis were unsuccessful. Thus, the pil operon promoter was further delineated by RT-PCR assays (Fig. 2). The primer pairs pil3-pil5 and pil3-pil6 yielded fragments of 334 and 364 bp, respectively. In contrast, no fragment was generated with the primers pil3 and pil7 by RT-PCR, even though PCR on total DNA amplified the expected fragment (397 bp; not shown). We thus concluded that the potential transcriptional start point (Tsp) is located between bp −97 and −120 upstream of the ATG codon. This is in agreement with the presence of putative −10 and −35 sequences identified upstream of the Tsp (Fig. 2B). Nucleotide sequence analysis of the intergenic space downstream of pilW revealed a perfect palindrome (5′-AGATAACCCTGATAAAGGGTTATCT-3′) sequence (underlined) forming a stem-loop transcriptional terminator (ΔG = −13.8 kcal/mol [50]) 48 bp after the stop codon of the last gene of the pil operon. Additionally, the putative transcriptional terminator was followed by an AT-rich region. The functionality of this terminator was confirmed by RT-PCR results as shown in Fig. 1A.
FIG. 2.
Promoter analysis of the pil operon. (A) RNA extracted from exponentially growing Y. pseudotuberculosis cultures at 37°C were analyzed by RT-PCR to identify the putative promoter region of the pil locus. Three forward oligonucleotides (pil5, pil6, and pil7) upstream of the ATG codon of pilL, in the promoter region, were used in combination with the reverse primer pil3 located 265 bp downstream of the start codon. The amplification products were analyzed by agarose gel electrophoresis. (B) The nucleotide (nt) sequence of the 163-bp DNA segment upstream of pilL and the 5′ end of pilL (nt 1 to 24 and 226 to 297) is shown in lowercase and uppercase letters, respectively. Forward and reverse oligonucleotides (described in Materials and Methods) are symbolized by arrows pointing right and left, respectively, below the nucleotide sequence. The positions of the presumed −35 and −10 sequences and ribosome binding sites (RBS) are indicated in boldface. The Tsp of the pil operon lies between oligonucleotides pil6 and pil7.
The type IV pilus gene clusters from Y. pseudotuberculosis and S. enterica are highly homologous.
The deduced amino acid sequences of the proteins encoded by the pilLMNOPQRSUV genes display 30 to 55% identity and 46 to 70% similarity to PilLMNOPQRSUV proteins involved in the biogenesis of type IV pili in Salmonella (19, 48). The main characteristics of the gene products are given in Table 2. On the basis of protein similarities, the putative products of pilS and pilV are thought to be structural prepilins. Table 2 shows a comparison of signal sequences of prepilins of the type IVB family. A putative signal sequence cleavage site has been identified between the 14th and 15th amino acids of prePilS and the 11th and 12th amino acids of prePilV. Interestingly, this putative cleavage site for both prePilS and prePilV is between a glycine and a tryptophan based on sequence alignments with all known prepilins from the type IVB family. The mature products of pilS and pilV also have a 20-aa hydrophobic domain in their N-terminal regions (Table 3), whereas in their C-terminal regions, several cysteine residues (two for PilS at positions 140 and 177; eight for PilV at positions 327, 344, 365, 388, 402, 405, 411, and 424) may form intramolecular disulfide bridges, a feature shared by almost all type IV pilins (24). Y. pseudotuberculosis PilU, the putative product of the gene pilU, is homologous to prepilin peptidases; sequence analysis of PilU showed the presence of a pair of aspartate residues at positions 33 and 95. These residues are completely conserved among type IV prepilin peptidases and have been shown to be essential for enzyme activity (23). Finally, the last gene of the Y. pseudotuberculosis pil cluster, pilW, encodes a product showing 61% identity with a Yersinia pestis putative transposase. It is noteworthy that this gene is not present within the type IV pilus gene cluster in Salmonella (19, 48).
TABLE 2.
Main characteristics of ORFs identified within the 11-kb locus of Y. pseudotuberculosis strain 32777
| ORF | Size (aa) | Molecular mass (Da) | pI | Homolog as determined by BLAST | % Identity/homology (amino acid overlap) | GenBank accession no. |
|---|---|---|---|---|---|---|
| PilL | 356 | 38,710 | 8.9 | PilL encoded by S. enterica serovar Typhi (unknown function) | 43/53 (356) | AF000001 |
| PilM | 194 | 21,620 | 10.7 | Lipoprotein PilM encoded by S. enterica serovar Typhi (unknown function) | 42/57 (125) | AF000001 |
| PilN | 530 | 57,189 | 8.7 | Lipoprotein PilN encoded by plasmid R64 from S. enterica serovar Typhimurium (unknown function) | 45/59 (528) | D88588 |
| PilO | 445 | 49,158 | 9.0 | PilO encoded by S. enterica serovar Typhi (unknown function) | 33/50 (445) | AF000001 |
| PilP | 164 | 17,560 | 9.2 | PilP encoded by S. enterica serovar Typhi (unknown function) | 36/54 (149) | AF000001 |
| PilQ | 517 | 57,826 | 7.3 | Nucleotide-binding protein PilQ encoded by S. enterica serovar Typhi (unknown function) | 55/70 (495) | AF000001 |
| PilR | 369 | 41,411 | 8.6 | Integral membrane protein PilR encoded by plasmid R64 from S. enterica serovar Typhimurium (unknown function) | 44/59 (362) | D88588 |
| PilS | 195 | 20,191 | 9.7 | Prepilin PilS encoded by plasmid R64 from S. enterica serovar Typhimurium | 49/62 (189) | D88588 |
| PilU | 161 | 17,461 | 5.9 | Prepilin peptidase PilU encoded by S. enterica serovar Typhi | 30/46 (158) | AF000001 |
| PilV | 425 | 45,880 | 8.8 | Minor pilin PilV encoded by plasmid R64 from S. enterica serovar Typhimurium | 46/58 (423) | D88588 |
| PilW | 306 | 35,089 | 6.7 | Putative transposase Y1072 from Y. pestis plasmid pMT1 | 61/78 (306) | AF074611 |
TABLE 3.
Comparison of signal sequences of prepilins of the type IVB family
| Prepilin | Microorganism | Amino acid sequence of the N terminus of prepilina |
|---|---|---|
| ▾ | ||
| prePilS | Y. pseudotuberculosis | MLSPVASRKQPHSG WGILESGGVALVVIVVIAVV |
| prePilS | S. enterica serovar Typhimurium | MLVENINTTLTGNNKKNEPHDKG WAILEQGTIALVVLFVIVVV |
| prePilS | S. enterica serovars Dublin and Typhi | MKNETEGKMMNEVSTLNPCNRPDRG MSADAGATALFILVIIGVIA |
| prePilV | Y. pseudotuberculosis | MTFKTRALHRG WAMMSTGIALLILVIVVIWA |
| prePilV | S. enterica serovar Typhimurium | MKKYDRG WASLETGAALLIVMLLIAWG |
| prePilV | S. enterica serovars Dublin and Typhi | MKKQKHDGG FVAMSVGAGLLIVLVMASLA |
| preBfpA | E. coli (EPEC) | MVSKIMNKKYEKG LSLIESAMVLALAATVTAGV |
| preLngA | E. coli (ETEC) | MLSVYNRTQKFKAEARKKIAKYHELRKQRG MSLLEVIIVLGIIGTIAAGV |
| preTcpA | V. cholerae | MQLLKQLFKKKFVKEEHDKKTGQEG MTLLEVIIVLGIMGVVSAGV |
Deduced from prepilin genes (accession numbers in GenBank, AF004308, AF304486, X74730, AF000001, D88588, and AF247502). The pilin monomers are synthesized as precursor proteins with a hydrophilic leader peptide of variable length that is processed at a consensus cleavage site (arrowhead) by a prepilin peptidase usually recognizing the consensus motif GXXXXE. The signal sequences of prepilins and the first 20 aa (hydrophobic domain) of the mature proteins are shown.
Expression of the Y. pseudotuberculosis pil operon in E. coli K-12 reconstitutes bundle-forming pili.
Since sequence comparisons showed high homologies between the Y. pseudotuberculosis and S. enterica pil gene clusters, we investigated whether the Y. pseudotuberculosis pil cluster is functional and capable of forming type IV pili. We thus initially analyzed E. coli NM554 containing the recombinant plasmids pMM2.1, pMM2.A6, and pMM3.D6; the last two encompass an incomplete pil operon (see above). Two hundred and fifty bacterial cells from each strain obtained after 24-h culture on LB agar at 37°C were observed by TEM. Microscopy was performed without prior knowledge of the identities of the cloned inserts in recombinant plasmids. Bundle-forming pili protruded from the surface in the polar regions of 147 (59%) NM554(pMM2.1) cells (Fig. 3). In contrast, these appendages were absent from all 250 of the NM554(pMM2.A6) and NM554(pMM3.D6) cells examined (data not shown). These results demonstrated that plasmid pMM2.1, which harbors the entire pil gene cluster, was capable of inducing pilus biogenesis in E. coli, in contrast to the truncated pil gene clusters (plasmids pMM2.A6 and pMM3.D6).
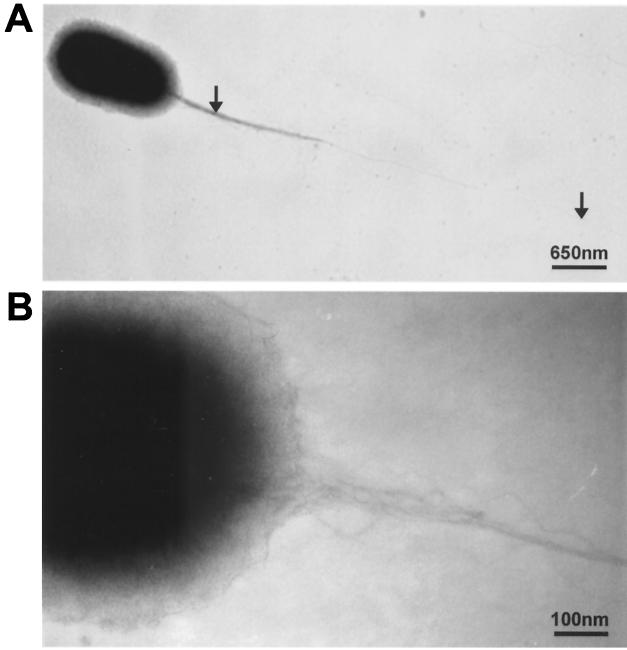
FIG. 3.
Electron micrographs of E. coli containing the recombinant plasmid pMM2.1, which harbors the Y. pseudotuberculosis pil gene cluster. The strain was examined by TEM after uranyl acetate negative staining. A long pilus (6 μm) emanates from one polar position (A, arrows) and is constituted of bundles of fibers with a diameter of 5 nm (B).
These ultrastructural studies strongly suggested that the pil locus governs pilus synthesis. To demonstrate that the pilLMNOPQRSUV cluster is sufficient for the biogenesis of a type IV pilus, a 10.8-kb NcoI fragment from cosmid pMM2.1, containing only the pil gene cluster and its putative promoter region, was subcloned into plasmid pACYC184 to yield pMM5. This plasmid was introduced into E. coli MC1061, and the bacterial cells were examined by TEM. The recombinant MC1061 strain, carrying only pACYC184, was used as a negative control. As previously observed with strain NM554(pMM2.1), bundles of filaments protruded from a polar end of strain MC1061(pMM5) whereas the control strain, MC1061(pACYC184), was nonpiliated (not shown).
The pil operon in Y. pseudotuberculosis is transcriptionally regulated by in vitro environmental factors.
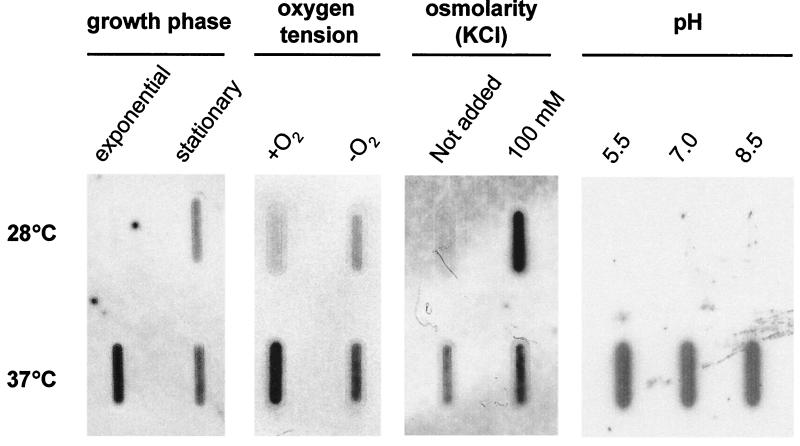
In addition to temperature shift, enteropathogenic Y. pseudotuberculosis encounters various environmental cues in the ileum lumen prior to invasion of intestinal mucosa: these include low pH, high osmolarity, and low oxygen tension (39). Hence, we investigated whether these signals influence Y. pseudotuberculosis pil operon expression. This was achieved by slot blot hybridization of pil transcripts from Yersinia grown in vitro under different environmental conditions (Fig. 4).
FIG. 4.
Transcription of the pil operon in Y. pseudotuberculosis grown under different environmental conditions. Total RNAs (15 μg) extracted from bacterial cultures in standard or modified LB broth were spotted onto nylon membranes and hybridized with a labeled 523-bp pilRS probe. As a positive control for constitutive gene expression, a ribosomal DNA 16S probe was used (not shown). No signal was detected when RNA samples were pretreated with RNase before hybridization. Experiments were repeated at least twice, and representative results are shown.
We initially assessed the phase of the Y. pseudotuberculosis growth cycle during which the pil gene cluster was expressed. Strain 32777 was cultivated in standard LB broth, and pil transcripts were detected only during the stationary phase at 28°C, whereas they were already synthesized as early as the exponential phase at 37°C. We then studied the influence of oxygen tension levels during exponential growth at 28 and 37°C in LB broth maintained aerobically or anaerobically. At 28°C, anaerobiosis induced expression of the pil operon. This was in contrast to pil transcription at 37°C. Overall, pil expression was both temperature and redox potential dependent.
Additionally, we tested whether, separately, osmotic and pH changes in growth media had effects on pil transcription. RNAs were extracted from exponentially growing cultures in LB broth enriched with large amounts of a salt (100 mM KCl) or a sugar (200 mM l-arabinose) to raise the osmolarity of the medium. As depicted in Fig. 4, 100 mM KCl increased pil expression at 28°C. Similar results were obtained with l-arabinose (data not shown). In contrast, pil transcription was not influenced by pH at either 28 or 37°C.
Deletion of the pil gene cluster in Y. pseudotuberculosis reduces bacterial pathogenicity in the mouse model.
The enteropathogenic species S. enterica, E. coli, and V. cholerae produce type IV pili, which contribute to their pathogenicity (1, 9, 48). To determine whether the pil gene cluster plays such a role in Y. pseudotuberculosis, an isogenic pil mutant was engineered from the wild-type strain. A DNA fragment encompassing almost all of the pil operon was deleted in the parental strain 32777 and replaced with a kanamycin resistance gene, aphA-Ia. This mutation was achieved by mating strain 32777 (Aps Kms) with E. coli strain SM10λpir harboring the suicide plasmid pMM4. Allelic exchange through homologous recombination, leading to mutant MIV, was carried out in two steps as previously described (4). The genotype of the pil mutant was further confirmed by Southern blot hybridization with appropriate DNA probes (not shown).
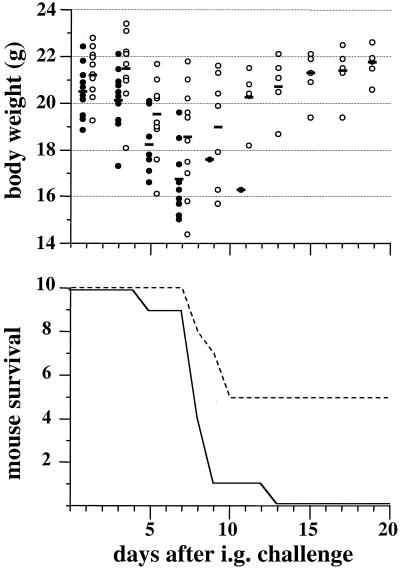
The virulence of the pil mutant was first assessed in an oral model of infection in the BALB/c mouse. The LD50 of this strain was slightly higher than that of the parental strain (108 versus 107.3). As depicted in Fig. 5, after i.g. inoculation of a lethal dose of Yersinia, the death of the animals was delayed when the infecting strain was pil deficient; this finding was well correlated with the body weights of mice throughout the infectious process. On the other hand, the pil mutant was found to be as virulent as the parental strain when administered intravenously (LD50, < 102).
FIG. 5.
Impact of the pil gene cluster on Y. pseudotuberculosis pathogenicity in the mouse model. BALB/c mice (groups of 10) were inoculated by the i.g. route with 108 pil+ or pil mutant bacteria. The body weights and survival times of the infected animals were recorded during a 20-day period after bacterial challenge. The experiments were repeated twice, and very similar results were found. Representative data are shown. The bars represent the mean body weights of infected animals. Symbols: • and solid line, wild-type strain 32777 (pil+); ○ and dashed line, isogenic mutant MIV (pil mutant).
The pil gene cluster is present in most pathogenic Y. pseudotuberculosis strains.
We investigated the distribution of the pil gene cluster in the Y. pseudotuberculosis species. We tested 91 Y. pseudotuberculosis strains originating from various countries by colony blot hybridization under stringent conditions with two PCR-generated pil probes from strain 32777 DNA: pilL (269 bp) and pilS (579 bp). Thirty-seven strains hybridized with each of the two probes: 14 of them belonged to serotype I (376/89, 347/89, 72/91, YPT6, YPT7, YPT9, YPT11, 2/91, 33036, 32849, 33053, 417/94, WS53/94, and WS34/94), 5 belonged to serotype II (MA, 2926, 2515, 2929, and WS25/91), 10 belonged to serotype III (304/89, 32945, 32977, 32975, 32992, YPT1, YPT5, 766/95, 1216/93, and 2887), 2 belonged to serotype IV (ST and NT), 4 belonged to serotype V (99/91, 199/90, 2821, and 2843), and 2 belonged to serotype VI (1553 and 1554).
We randomly chose 16 out of the 37 pil-positive strains (YPT6, YPT7, YPT9, 33036, WS53/94, MA, 2926, 2929, WS25/91, 304/89, YPT1, YPT5, 1216/93, 199/90, 1553, and 1554) and PCR amplified the entire pilS gene encoding a putative prepilin. The PCR products were sequenced, and the pilS sequence was found to be identical to the prototype sequence for all except four strains (2926, 2929, WS25/91, and 199/90) in which a nucleotide substitution led to replacement of Tyr137 by Ile137 in the pilS product. Taken together, these results show that at least the pilS gene is highly conserved among the species.
DISCUSSION
In this work, we cloned and sequenced an 11-kb chromosomal operon of 11 genes involved in type IV pilus biogenesis in Y. pseudotuberculosis. This novel locus, when introduced into E. coli, induced the emanation of appendages at one cellular pole that tended to form large bundles of pilus aggregates, an ultrastructural feature of type IV pili. The genetic locus that we discovered in Y. pseudotuberculosis strain 32777 was strongly homologous to the pil operon of S. enterica. In this enteropathogenic bacterial species, two closely related pil operons have been characterized: the first, called R64, includes 14 genes (pilIJKLMNOPQRSTUV) and was originally described in serovar Typhimurium (19); the second lacks the pilI, pilJ, and pilK genes and has been reported in serovar Typhi (48). No homologs of the Salmonella pilI, pilJ, pilK, and pilT genes were present in the Y. pseudotuberculosis pil operon, but as an E. coli strain trans-complemented with Y. pseudotuberculosis pilLMNOPQRSUV produced pili (Fig. 3), these genes do not seem to be required for pilus formation in this bacterial species. This is in agreement with the study of Yoshida and coworkers (47) showing that of the 14 pil genes (pilIJKLMNOPQRSTUV) carried by the Salmonella operon R64, pilI and pilJ were not required for R64 pilus biogenesis. In addition, as stated above, pilK is absent from the type IV pilus gene cluster of S. enterica serovar Typhi, even though pili are produced (48). On the other hand, the pilW gene present in the Y. pseudotuberculosis pil operon is missing in Salmonella; however, we demonstrated by a complementation assay in E. coli that this gene is not needed for pilus biogenesis. The homology of the pilW product with a putative transposase is consistent with the latter finding. Why pilW, whose G+C content is, surprisingly, the lowest of the gene cluster, is part of the pil operon remains to be elucidated. Sequencing of the flanking DNA environment of the pil operon, which is in progress, could shed light on this point.
Some Y. pseudotuberculosis Pil proteins (PilN, PilR, PilS, and PilV) are more similar to Pil homologs involved in R64 pilus biogenesis, whereas others (PilL, PilM, PilO, PilP, PilQ, and PilU) are more similar to the corresponding Pil proteins from serovar Typhi (Table 1). Bacterial localization of the Pil proteins involved in R64 pilus biogenesis have been specified, but the functions of most of these proteins are still poorly understood, with the exception of the products of pilS and pilV, which are prepilins, and that of pilU, a prepilin peptidase (19, 35). The putative length of the signal sequence (14 and 11 aa for prepilins S and V, respectively) and the nature of the first amino acid residue of the mature protein, which is a tryptophan rather than a phenylalanine for both Y. pseudotuberculosis PilS and PilV, identified these prepilins as members of the IVB subclass, to which pili from V. cholerae, E. coli pathovars EPEC and ETEC, and S. enterica belong. As for S. enterica serovars Typhi and Dublin, the amino acid at position +5 of the putative Y. pseudotuberculosis mature protein PilV is a serine. In type IV pilins, the +5 residue is almost always a glutamic acid (24). However, previous studies have shown that the replacement of glutamic acid by another amino acid did not affect prepilin processing by prepilin peptidase (31, 42). Unlike its homolog in S. enterica, the Y. pseudotuberculosis pilV gene does not contain a clustered inversion region (known as a shufflon) (22), which may generate several different products due to a rearrangement of the DNA in the region encoding the C terminus by a specific recombinase.
We also show in the present study that in vitro expression of the Y. pseudotuberculosis pil operon is transcriptionally regulated by temperature, cell density, osmolarity, and oxygen tension. Interestingly, pil gene transcription is upregulated under anaerobiosis or high osmolarity and at 37°C. These conditions are encountered by Y. pseudotuberculosis in the digestive tract of the host; hence, one can postulate a possible role for the fimbria-encoding gene cluster in bacterial enteropathogenicity. Indeed, inactivation of the pil gene cluster was associated with a reduction of Y. pseudotuberculosis virulence for the mouse in the oral, but not the systemic, model of infection. This is most likely associated with an impairment of bacterial colonization of the intestinal mucosa. Whether these pili can functionally substitute for other adhesins remains to be elucidated. Further studies of interactions of Y. pseudotuberculosis pil+ and pil mutant strains with reconstituted intestinal epithelial monolayers (38) should provide insight into this question and could indicate whether the pil gene cluster contributes to translocation of Yersinia across the intestinal barrier.
Two features suggest that Y. pseudotuberculosis may have acquired the pil operon by horizontal gene transfer. First, the pil locus is not uniformly present throughout the species, as only 38 of 92 (41%) nonepidemiologically related strains of diverse origins tested positive for pil by stringent hybridization with two specific probes. However, it should be noted that since both DNA probes used in the hybridization assay were extremities of the operon, we may have missed possible deletions in this operon in the different strains. Interestingly, both the strain 32953, which has been used for the Y. pseudotuberculosis genome project, and the extensively studied reference strain YPIII were pil negative under these detection conditions. Additionally, the pil locus was not found in the genome of strains CO92 and KIM5 of the closely genetically related species Y. pestis (http://www.sanger.ac.uk/Projects/Y_pestis and http://www.genome.wisc.edu/sequencing/pestis). Secondly, whereas the mean G+C content of the Y. pseudotuberculosis genome is 47%, that of the pil operon is 50.8%. This value is very close to the G+C contents of the genomes of the type IV pilus-producing species E. coli and Salmonella (51 to 52%). This suggests that Y. pseudotuberculosis, which occupies the same ecological niche as these two other enterobacteria, may have acquired the type IV pilus locus from one of these species by genetic exchange in the natural habitat. The two Salmonella pil operons have been found located either on a conjugative plasmid (in serovar Typhimurium) (19) or a large (118-kb) pathogenicity island (in serovar Typhi) (48). Other type IVB pilus gene clusters, in E. coli pathovars EPEC and ETEC and V. cholerae, were also found to be harbored on either a plasmid (11, 13) or a pathogenicity island (17). In Y. pseudotuberculosis 32777, in which the type IV pilus gene cluster has been discovered, no plasmid other than the virulence plasmid pYV was detected. Thus, given the scenarios in S. enterica, E. coli, and V. cholerae, it is tempting to speculate that the Y. pseudotuberculosis pil operon could constitute a novel “adaptation-pathogenicity” island, unknown until now, which, in addition to the already described high-pathogenicity island (2), contributes to the virulence of Y. pseudotuberculosis.
Acknowledgments
This work was partly supported by the Conseil Régional Nord-Pas de Calais, the Université René Descartes (Paris V), and the European Regional Development Fund. F. Collyn received a scholar fellowship from the Ministère de l'Enseignement Supérieur de la Recherche et de la Technologie.
We thank S. Aleksić, E. Carniel, D. Caugant, E. Falsen, N. Takeda, and R. Van Noyen for kindly supplying Y. pseudotuberculosis strains. We also thank C. Mullet for technical assistance and C. Carnoy for critical reading of the manuscript.
François Collyn and Marie-Annick Léty contributed equally to this work.
Editor: J. T. Barbieri
REFERENCES
- 1.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 2.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler, T. 1983. Plague and other Yersinia infection. Plenum Press, New York, N.Y.
- 4.Carnoy, C., C. Mullet, H. Müller-Alouf, E. Leteurtre, and M. Simonet. 2000. Superantigen YPMa exacerbates the virulence of Yersinia pseudotuberculosis in mice. Infect. Immun. 58:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dougherty, B. A., and H. O. Smith. 1999. Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology 145:401-409. [DOI] [PubMed] [Google Scholar]
- 7.Dubnau, D. 1997. Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins—a review. Gene 192:191-198. [DOI] [PubMed] [Google Scholar]
- 8.Ellington, A. 1990. Preparation of genomic DNA from bacteria, p. 241-245. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 9.Faast, R., M. A. Ogierman, U. H. Stroeher, and P. A. Manning. 1989. Nucleotide sequence of the structural gene, tcpA, for a major pilin subunit of Vibrio cholerae. Gene 85:227-231. [DOI] [PubMed] [Google Scholar]
- 10.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 11.Girón, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 12.Girón, J. A., M. M. Levine, and J. B. Kaper. 1994. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol. Microbiol. 12:71-82. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Duarte, O. G., A. Ruiz-Tagle, D. C. Gomez, G. I. Viboud, K. G. Jarvis, J. B. Kaper, and J. A. Girón. 1999. Identification of lngA, the structural gene of longus type IV pilus of enterotoxigenic Escherichia coli. Microbiology 145:1809-1816. [DOI] [PubMed] [Google Scholar]
- 14.Graupner, S., V. Frey, R. Hashemi, M. G. Lorenz, G. Brandes, and W. Wackernagel. 2000. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J. Bacteriol. 182:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermodson, M. A., K. C. Chen, and T. M. Buchanan. 1978. Neisseria pili proteins: amino-terminal amino acid sequences and identification of an unusual amino acid. Biochemistry 17:442-445. [DOI] [PubMed] [Google Scholar]
- 16.Hood, B. L., and R. Hirschberg. 1995. Purification and characterization of Eikenella corrodens type IV pilin. Infect. Immun. 63:3693-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karaolis, D. K., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 18.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183:4451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S. R., and T. Komano. 1997. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 179:3594-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirov, S. M., and K. Sanderson. 1996. Characterization of a type IV bundle-forming pilus (SFP) from a gastroenteritis-associated strain of Aeromonas veronii biovar sobria. Microb. Pathog. 21:23-34. [DOI] [PubMed] [Google Scholar]
- 21.Kirov, S. M., K. Sanderson, and T. C. Dickson. 1998. Characterisation of a type IV pilus produced by Aeromonas caviae. J. Med. Microbiol. 47:527-531. [DOI] [PubMed] [Google Scholar]
- 22.Komano, T., A. Kubo, and T. Nisioka. 1987. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 15:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaPointe, C. F., and R. K. Taylor. 2000. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 275:1502-1510. [DOI] [PubMed] [Google Scholar]
- 24.Manning, P. A., and T. F. Meyer. 1997. Type-4 pili: biogenesis, adhesins, protein export and DNA import. Proceedings of a workshop. Gene 192:1-198. [PubMed] [Google Scholar]
- 25.Marrs, C. F., G. Schoolnik, J. M. Koomey, J. Hardy, J. Rothbard, and S. Falkow. 1985. Cloning and sequencing of a Moraxella bovis pilin gene. J. Bacteriol. 163:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 27.Meyer, T. F., E. Billyard, R. Haas, S. Storzbach, and M. So. 1984. Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc. Natl. Acad. Sci. USA 81:6110-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunn, D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell. Biol. 9:402-408. [DOI] [PubMed] [Google Scholar]
- 29.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 30.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugsley, A. P. 1993. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol. Microbiol. 9:295-308. [DOI] [PubMed] [Google Scholar]
- 32.Raleigh, E. A., N. E. Murray, H. Revel, R. M. Blumenthal, D. Westaway, A. D. Reith, P. W. Rigby, J. Elhai, and D. Hanahan. 1988. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 16:1563-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed, L. J., and H. A. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 34.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 35.Sakai, D., and T. Komano. 2002. Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes. J. Bacteriol. 184:444-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sebbane, F., A. Devalckenaere, J. Foulon, E. Carniel, and M. Simonet. 2001. Silencing and reactivation of urease in Yersinia pestis is determined by one G residue at a specific position in the ureD gene. Infect. Immun. 69:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulte, R., S. Kerneis, S. Klinke, H. Bartels, S. Preger, J. P. Kraehenbuhl, E. Pringault, and I. B. Autenrieth. 2000. Translocation of Yersinia enterocolitica across reconstituted intestinal epithelial monolayers is triggered by Yersinia invasin binding to β1 integrins apically expressed on M-like cells. Cell. Microbiol. 2:173-185. [DOI] [PubMed] [Google Scholar]
- 39.Sleisenger, M. H. 1981. Pathophysiology of the gastrointestinal tract, p. 1526-1527. In L. H. Smith and S. O. Thier (ed.), Pathophysiology: the biological principles of disease. Saunders, Philadelphia, Pa.
- 40.Stone, B. J., and Y. A. Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stone, B. J., and Y. A. Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strom, M. S., and S. Lory. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266:1656-1664. [PubMed] [Google Scholar]
- 43.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 44.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 45.Weiss, R. L. 1971. The structure and occurrence of pili (fimbriae) on Pseudomonas aeruginosa. J. Gen. Microbiol. 67:135-143. [DOI] [PubMed] [Google Scholar]
- 46.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, T., S.-R. Kim, and T. Komano. 1999. Twelve pil genes are required for biogenesis of the R64 thin pilus. J. Bacteriol. 181:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y., J. M. Tennent, A. Ingham, G. Beddome, C. Prideaux, and W. P. Michalski. 2000. Identification of type 4 fimbriae in Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 189:15-18. [DOI] [PubMed] [Google Scholar]
- 50.Zucker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]