Abstract
We used high-resolution atomic force microscopy to image the compaction of linear and circular DNA by the yeast mitochondrial protein Abf2p, which plays a major role in packaging mitochondrial DNA. Atomic force microscopy images show that protein binding induces drastic bends in the DNA backbone for both linear and circular DNA. At a high concentration of Abf2p DNA collapses into a tight nucleoprotein complex. We quantified the compaction of linear DNA by measuring the end-to-end distance of the DNA molecule at increasing concentrations of Abf2p. We also derived a polymer statistical mechanics model that provides a quantitative description of compaction observed in our experiments. This model shows that sharp bends in the DNA backbone are often sufficient to cause DNA compaction. Comparison of our model with the experimental data showed excellent quantitative correlation and allowed us to determine binding characteristics for Abf2p. These studies indicate that Abf2p compacts DNA through a simple mechanism that involves bending of the DNA backbone. We discuss the implications of such a mechanism for mitochondrial DNA maintenance and organization.
INTRODUCTION
Mitochondria participate in many critical processes in the cell lifecycle. Aside from their primary role in ATP production, mitochondria also act as signaling centers through the regulation of calcium, iron, and metabolite levels in the cytosol. These organelles are also responsible for the main switch controlling apoptosis. Such critical responsibilities place stringent requirements on the integrity of the mitochondrial DNA (mtDNA). A variety of processes threatens mtDNA. The respiratory chain of mitochondrial metabolism produces large levels of oxygen radicals which can attack mtDNA. Oxidative damage to mtDNA often leads to several clinical disorders including Parkinson's, Hutchinson's, and Huntington's disease (Voet et al., 1999). Ironically, the very job that is required to keep the cell alive also yields dangerous byproducts. To operate under these harsh conditions mtDNA must be packaged in a way that protects it from damage, while not impairing the normal functions of mtDNA such as replication and transcription.
Mammals (Van Tuyle and McPherson, 1979; Satoh and Kuroiwa, 1991) and the budding yeast Saccharomyces cerevisiae (Wintersberger et al., 1975; Miyakawa et al., 1984, 1987) package mtDNA in compact globular structures similar to a bacterial nucleoid. These mt-nucleoid structures are distinctly different from the packaging of DNA into chromatin in the cell nucleus. Researchers have firmly established the role of histones in the formation of the nucleosome (Luger et al., 1997). However, very little data exists on the identity or function of the proteins that facilitate the formation of the mt-nucleoid.
Diffley and Stillman (1988) found that a particular 20-kDa protein was present in relatively high abundance among the various polypeptides isolated from mt-nucleoids. This protein, Abf2p (ARS binding factor 2), displays interesting DNA binding characteristics: it binds nonspecifically to general regions of DNA, but exhibits phased binding to replicating sequences such as ARS1 (Diffley and Stillman, 1988). Abf2p also induces negative supercoiling in DNA in the presence of topoisomerase. Although Abf2p is not required for mtDNA replication, changes in Abf2p levels alter mtDNA copy number (Zelenaya-Troitskaya et al., 1998), and null Abf2p mutants lose their wild-type (ρ+) mtDNA (Diffley and Stillman, 1991). Data also indicate that the level of Abf2p directly influences the number of recombination intermediates in vivo (MacAlpine et al., 1998). This information, coupled with the high abundance of Abf2p, led researchers to suggest that Abf2p is a primary mtDNA packaging protein. Abf2p is closely related to the HMG family; its sequence contains two HMG boxes linked by six amino acids (Diffley and Stillman, 1991). HMG proteins are, among other activities, involved in the structural organization of packaged DNA in higher ordered structures such as chromatin. However, no known DNA packaging mechanism uses HMG proteins as the fundamental packaging unit. Therefore, it is likely that the complete DNA packaging mechanism employed by mitochondria is different from other known DNA packaging processes. The establishment of such a mechanism should provide valuable information about the role of Abf2p in the overall mtDNA maintenance process.
Recent developments in molecular-scale imaging have enabled a number of unprecedented advances in biophysics. For example, high-resolution atomic force microscopy (AFM) can now visualize single biological molecules in native environments in real space (Hansma et al., 1997). Researchers have used AFM to image protein binding to DNA (Erie et al., 1994), virus particle surfaces, and cell surfaces, and determine the strength of protein-ligand interactions and elasticity of DNA molecules (Radmacher et al., 1992; Clausen-Schaumann et al., 2000). AFM also excels in visualizing the conformation of linear polymers such as DNA (Lyubchenko et al., 2001, 2002; Tiner et al., 2001; Bustamante and Rivetti, 1996; Rivetti et al., 1996). Early on, Balhorn and colleagues used AFM to investigate the mechanism of DNA compaction by protamine proteins from sperm and the packaging of DNA by nucleosomes (Allen et al., 1997, 1993). Others have used AFM to study the physical properties of DNA condensation by various ionic species which may in fact have biological significance in terms of the mechanism used by viruses to package DNA (Hoh and Fang, 1999; Fang and Hoh, 1999, 1998). Recent refinements in AFM imaging technology, such as new imaging modes and new sharper AFM probes, pushed the technique's limits even further (Dai et al., 1996; Hafner et al., 1999).
In this article we investigated binding of Abf2p to DNA using high-resolution AFM. We found that when Abf2p binds to DNA it induces pronounced structural distortions in DNA conformation. When we increased the protein coverage we observed a striking collapse of the DNA molecule into a dense globular complex. Our observations lead us to suggest that Abf2p compacts DNA by simply introducing a number of sharp bends into the DNA backbone. We also present a simple mathematical model that describes the compaction process quantitatively. Finally, we demonstrate that the Abf2p binding parameters, derived from application of the model to the AFM data, show excellent correlation with the results of an independent bulk protein binding assay.
MATERIALS AND METHODS
Abf2p isolation
The S. cerevisiae ABF2 gene (New England Biolabs, Beverly, MA) in a pMalc2X fusion vector (kindly provided by the Nunnari Lab, University of California at Davis) (Kao et al., 1993) was cloned into pET28b+ (Novagen, Madison, WI). Inverse PCR was used to fuse an initiator methionine and six histidine codons to the ABF2 reading frame encoding mature Abf2p, residues 21–177 with His tag, Mr = 19,484 Da. Protein was purified from BL21(DE3)(pET28b-His6ABF2) cultures (5–10 mg/L culture), essentially as described for Cre recombinase (Martin et al., 2002) except 500 mM NaCl was used in the initial lyses buffer, and the solution was dialyzed to low salt buffer before ion exchange chromatography. Concentrated and filtered samples, 20–70 mg/mL (Centricon 10, Millipore, Billerica, MA) in 20 mM Na-HEPES at pH 7.5, 1 mM Na-EDTA, 4 mM DTT, and 0.1 w/v Na-azide, were diluted with 10 mM Tris-Cl at pH 7.8, 4 mM DTT, and 1 mM Na-EDTA. Abf2p concentrations were determined using an extinction coefficient at 280 nm of 1.29 mg−1 mL, or 24,180 M−1. DNA binding activity was established with a gel-retardation assay (Cho et al., 2001) using either supercoiled pLitmus-38+ or linear λ-phage DNA (New England Biolabs) as a substrate.
pBR322 DNA preparation
Relaxed circular pBR322 DNA was purchased from TopoGen (Columbus, OH) and used as received. Linear DNA was prepared by digesting supercoiled pBR322 (Life Technologies, Rockville, MD) with the single cutter BamH1 (Invitrogen, Carlsbad, CA). The enzyme was then removed by washing the DNA in a Centricon 100 four times with 10 mM Tris and 1 mM EDTA at pH 8 followed by two buffer exchanges with 100 mM sodium bicarbonate at pH 8.
AFM imaging and analysis
Substrates were prepared for imaging by first applying 3 μl of 0.1% poly-L-lysine (PL) to freshly cleaved mica. After 1 min the mica was rinsed with copious amounts of water, and then dried with filtered N2 stream for 1 min. DNA/Abf2p complexes were prepared by mixing aliquots of DNA and protein stock solutions into buffer containing only 100 mM NaHCO3 at pH 9. We held DNA concentration constant at 1 μg/ml and varied the Abf2p concentrations. After we mixed the protein and DNA solutions we allowed them to equilibrate for 5 min; then 1 μl of sample solution was applied to the PL-coated mica substrate and allowed to adsorb for 1–2 min at room temperature. The substrate was then rinsed with water and finally dried with filtered N2 stream. All images were acquired in air using a Multimode Nanoscope IIIa Atomic Force Microscope (Digital Instruments, Santa Barbara, CA) operating in tapping mode. We used etched silicon FESP probes (NanoWorld, Santa Barbara, CA), spring constant ∼0.1 N/m, for all our images. We used a typical scan rate of 2 Hz for all our images, and collected ∼10–15 different images at each DNA/protein ratio with every image containing multiple DNA-protein complexes.
Measurements of DNA end-to-end distances and protein-induced bend angles were done using image analysis tools built into the Nanoscope IIIa software. We only measured the bend angles for the regions of DNA where we could clearly identify the protein bound to the DNA backbone. We assume the DNA bind to the PL-coated mica surface with little reorganization of the DNA strands in the plane parallel to the surface. Therefore we take the images to represent the two-dimensional projection of the molecule's native three-dimensional configuration in solution. The end-to-end distances of the DNA therefore represent a projection of the true distance and to compare them with a three-dimensional model we multiply our average measured distances 〈R〉m by a factor of π/2. The measured values that we obtained for DNA in absence of the bound protein match well the values predicted by the worm-like chain model, which indicates that the DNA conformation is trapped upon adsorption (Rivetti et al., 1996).
All model fits were performed by nonlinear χ2-minimization using the Levenberg-Marquardt algorithm built into the IGOR Pro 4.0 data analysis software (Wavemetrics, Lake Oswego, OR).
Circular dichroism
Linear DNA (pBR322) in buffer (100 mM NaHCO3 only, pH 9) at ∼50 μg/ml were titrated with aliquots of concentrated Abf2p in a 1-cm pathlength quartz cuvette. Circular dichroism (CD) spectra were taken using a J-715 spectropolarimeter (Jasco, Easton, MD) operated at room temperature. We first subtracted the baseline from the raw CD spectra and then smoothed them using a second-order, 11-point Savitzky-Golay algorithm in IGOR Pro 4.0 (Wavemetrics). For each protein concentration we subtracted protein CD signal at 275 nm from the corresponding DNA/protein CD signal. Fractional saturation of the CD data is fit by using a cooperative bimolecular binding model given by θ = (C/Kd)q/(1 + (C/Kd)q).
RESULTS AND DISCUSSION
AFM imaging of Abf2p-DNA complexes
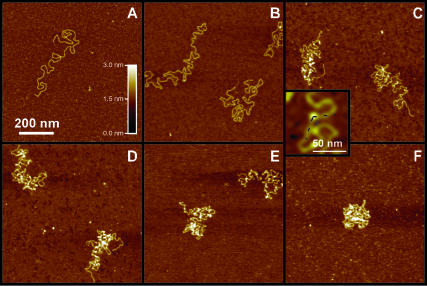
To investigate protein binding to DNA we imaged individual linearized pBR322 plasmid DNA molecules after we incubated them with Abf2p. After protein and DNA equilibrated in solution, we immobilized the DNA-protein complexes on atomically flat mica surfaces pretreated with PL. DNA concentration in solution was low enough to obtain single isolated molecules on the surface. AFM images of the DNA molecules before exposure to protein show smooth contours of the backbone, typically devoid of sharp bends or other distinguishing features (Fig. 1 A). In contrast, when we exposed DNA to the Abf2p we observed multiple sharp bends in the DNA backbone (Fig. 1, B–E). Images at low concentration of the Abf2p in most instances showed a protein molecule bound at the site of the bend (at least to within the limits of AFM resolution). Therefore we can conclude that binding of the protein causes the formation of these bends. This behavior is not entirely surprising, since other HMG family proteins often induce structural distortions in the DNA (Jones et al., 1994; Masse et al., 2002; Werner et al., 1995; Love et al., 1995). As we increased the concentration of the protein, the number of bends in the DNA backbone also increased. At higher concentrations of Abf2p we clearly started to observe the overall compaction of the molecule (Fig. 1 C). Finally, at a very high concentration of the protein, DNA collapsed into globular nucleoid-like structures (Fig. 1, E and F).
FIGURE 1.
AFM images of linearized pBR322 DNA molecules after exposure to increasing concentrations of Abf2p (ratio of Abf2p/bp in parentheses). (A) No Abf2p; (B) 1.5 μg/mL (1:20); (C) 3.5 μg/mL (1:8); (D) 7 μg/mL (1:4); (E) 15 μg/mL (1:2); and (F) 25 μg/mL (1:1). (Inset) Closeup image of a bend in the DNA backbone induced by the bound protein (bright dot).
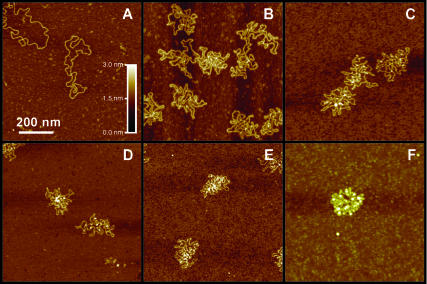
Diffley and Stillman (1991) reported that Abf2p induced negative supercoiling in DNA. It is unclear, however, if supercoiling plays a significant role in the compaction mechanism. To test this possibility we imaged Abf2p complexes with relaxed circular DNA. Use of relaxed circular pBR322 plasmid for these experiments allowed us to eliminate any possibility of sequence dependence influencing the outcome. Remarkably, AFM images of Abf2p complexed with circular DNA (Fig. 2) show almost identical behavior to what we observed for linear DNA. Protein binding induced large bends in the DNA backbone and the number of bends progressively increased with the increase in the protein concentration (Fig. 2, B–E). At high Abf2p concentration DNA again collapsed into globular structures (Fig. 2 F). This experiment strongly indicates that supercoiling does not play a significant role in the mechanism of Abf2p action. Rather, we believe that the negative supercoiling observed by Diffley and Stillman is simply a byproduct of the structural distortions that Abf2p introduces into the backbone of circular DNA.
FIGURE 2.
AFM images of relaxed circular pBR322 DNA molecules after exposure to increasing concentrations of Abf2p (ratio of Abf2p/bp in parentheses). (A) No Abf2p; (B) 1.5 μg/mL (1:20); (C) 3.5 μg/mL (1:8); (D) 7 μg/mL (1:4); (E) 15 μg/mL (1:2); and (F) 25 μg/mL (1:1).
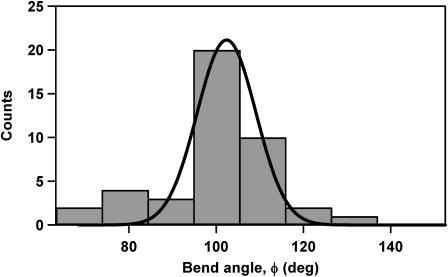
We can also use AFM images of the partially compacted DNA at low concentrations of Abf2p to estimate the angle of the DNA backbone bend induced by the protein. We have measured the bending angle for 43 different DNA molecules which had isolated protein units bound to DNA (Fig. 1, inset). Histogram of the measured angles shows a peak at 102° (Fig. 3). This value is clearly different from the mica lattice angle of 120°; therefore, we are confident that mica substrate does not cause this structural distortion. The bends caused by the Abf2p occur over very short distances, which also distinguishes them from the random smooth bends that sometimes appear when DNA adsorbs to the surface. Therefore, our images indicate that Abf2p bends DNA by ∼78° (if we adopt the angle measuring convention common to structural studies). Our measured value of the bend angle compares favorably with the literature values for the bend angles that other proteins from the HMG family induce in DNA. For example, SRY bends DNA by ∼70° (Tang and Nilsson, 1998), and SOX bends DNA by ∼83° (Scaffidi and Bianchi, 2001). However, a direct comparison between the literature values and our measured bend angle value is difficult since all the reported structures contain only one HMG box, whereas Abf2p contains two HMG boxes. At present, resolution constraints do not allow us to determine whether Abf2p binds to the DNA as a monomer or as a dimer, or whether the bend that we observe consists of two closely spaced sub-bends. Further investigations utilizing higher resolution probes, such as single-wall carbon nanotube AFM tips (Schnitzler et al., 2001) might provide more information about the binding mode for Abf2p.
FIGURE 3.
Histogram of 42 bend-angle values measured from images of individual DNA molecules exposed to Abf2p.
Our AFM images do not show any further type of aggregation or distortion besides the bend formation up until very high concentrations of protein, when it becomes difficult to discern individual protein molecules in the tightly compacted globular structure. We also have not found any clear evidence of cooperative binding. Mostly, the protein-DNA complexes showed random distribution of bends throughout the length of DNA, whereas cooperative binding would have forced a clustering of the bends in certain regions of DNA. Our images suggest that the major effect of the Abf2p binding to the DNA is the formation of sharp bends in the DNA backbone. Qualitatively, such bends would reduce the intrinsic stiffness of DNA and lead to the overall reduction of the DNA size in solution. However, it is unclear whether bending of the DNA backbone alone is sufficient to induce compaction. To answer this question quantitatively we need to consider how such bending changes the dynamics of a stiff polymer chain in solution. To address this question we developed a model that accounts for small, localized distortions in the DNA backbone while maintaining the nature of a DNA molecule elsewhere along its contour.
DNA-protein complex conformation: bent-worm-like chain model
We based our model on Kratky and Porod's worm-like chain (WLC) model which describes the statistical behavior of a random flight polymer chain that also has an intrinsic stiffness associated with it (Kratky and Porod, 1949). We modified this model to include a number P of fixed bends of angle φ. For simplicity we assumed that these bends are uniformly distributed along the DNA helix. We then derived the expression for the mean-squared end-to-end distance for such a polymer chain. We include a detailed mathematical derivation of this bent-worm-like chain (b-WLC) model in the Supplementary Materials, and here we just state the main premises and results of this model.
We start with a DNA chain of contour length L. The intrinsic stiffness of the DNA is governed by its persistence length A, which incorporates effects of temperature, charge, screening, and solute-solvent interactions. We assume that the bends that we introduce into the DNA do not affect its structure in the regions between the bends. In addition, we allow the dihedral angles to rotate freely in our model. We obtain the following exact analytical solution for the mean-squared end-to-end distance for an ensemble of linear polymers that incorporate P equidistant bends of angle φ,
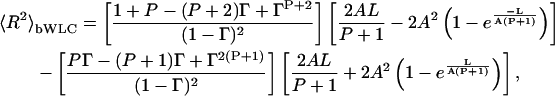 |
(1) |
where
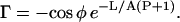 |
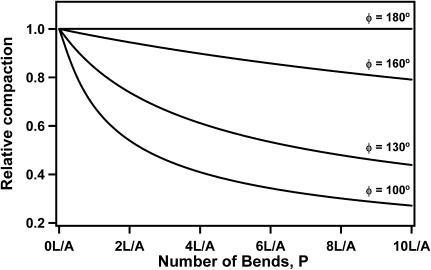
This result is consistent with the original Kratky-Porod model since it recovers the original WLC expression when no bends are present (P = 0). The most interesting aspect of this expression is that it predicts a significant drop in the average end-to-end distance as the number of bends increases (Fig. 4). This effect is most noticeable when the bend angle approaches 90° and it diminishes as the bend angles approach 180°. When the bend angle becomes equal to 180°, the bends vanish and we again recover the original WLC model behavior. Significantly, our model shows that the introduction of bends into the DNA backbone can alone cause significant compaction of the DNA. Therefore, it is consistent with the mechanism of Abf2p action that we propose.
FIGURE 4.
The b-WLC model predictions for DNA compaction as a function of the number of bends. We define compaction as decrease in the average end-to-end distance 〈R2〉1/2 relative to the average end-to-end distance for the free DNA.
As compaction proceeds, contributions to the DNA conformation from the steric repulsion between DNA segments will increase with added protein. We expect that in the last stages of compaction, when the segments are very close together, these contributions should become rather significant. Therefore, to make our model more realistic, we need to introduce a correction that would account for these excluded volume effects. It is reasonable to approximate the interaction between segments as a short-range repulsive force (Yamakawa, 1971) and apply perturbation theory to obtain the following first-order correction (Yamakawa, 1971; Grimley, 1953),
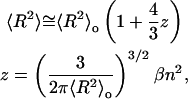 |
(2) |
where 〈R2〉o is the mean-squared end-to-end distance in the unperturbed state, n is the number of Kuhn statistical segments, and β is the binary cluster integral for a pair of segments, representing the effective volume excluded to one segment by another. Odijk and others estimated the excluded volume between two cylindrical Kuhn segments of length b and diameter D as πb2D/2 (Odijk, 1986; Onsager, 1949). As the protein binds to the double helix it increases the effective diameter of the cylinder from the 2-nm value characteristic of the free DNA. To calculate the excluded volume correction in this case we assumed that we can represent the areas of DNA covered with protein as helices of larger diameter and that the contribution of these larger helices is proportional to the amount of the bound protein (see Supplementary Material for details).
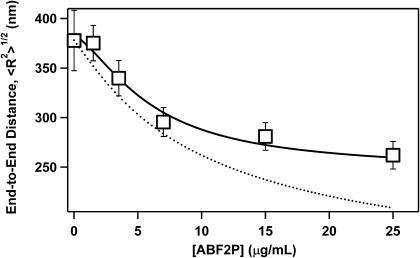
We tested our model by comparing average end-to-end distances obtained in the experiment with the model predictions. AFM images of individual compacted DNA molecules give us the ability to measure the end-to-end distance for a large number of molecules and collect necessary statistics. Fig. 5 shows that the measured end-to-end distance sharply decreases with the increase in the Abf2p concentration, as expected from images in Fig. 1. We can fit our data to the b-WLC model if we assume that 1), the number of bends, P, is proportional to the amount of bound protein and 2), Abf2p binding can be described with a simple binding constant Kd and a Hill constant q, such that
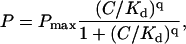 |
(3) |
where Pmax is the maximum number of bends that a protein can create in a given DNA length, and C is the total protein concentration. We used the 102° value for the bend angle that we measured in our experiments, and assumed that our DNA can contain the maximum of 145 bends, as determined by the pBR322 length (4361 basepairs) and Abf2p footprint (Diffley and Stillman, 1991) (30 basepairs). When we fit our experimental data to our model using these values we obtained an excellent fit to the experimental data (Fig. 5, solid line). For comparison, the b-WLC model without the excluded volume correction provided a much less satisfactory fit (Fig. 5, dotted line), especially at high protein concentration where steric repulsion makes the greatest contribution. In addition to the binding parameters summarized in Table 1, our model fit also yielded the value of the persistence length for free DNA of 48 nm, which matches the typical values for the standard WLC model (Bustamante et al., 1994; Baumann et al., 1997). These comparisons indicate that our model captures the essential physics of the compaction process induced by Abf2p.
FIGURE 5.
Plot of average end-to-end distance for linear pBR322 DNA molecules as a function of average Abf2p concentration. Each data point represents an average of at least 25 individual measurements. Lines represent the b-WLC model fits to the data without (dashed line) and with (solid line) excluded volume correction.
TABLE 1.
Abf2p binding parameters obtained from fitting our model to the end-to-end distances measured from AFM images and from CD measurements
| Binding parameter | AFM | CD |
|---|---|---|
| Dissociation constant, KD (μM) | 1.44 ± 0.43 | 1.38 ± 0.06 |
| Hill constant, q | 1.23 ± 0.24 | 1.48 ± 0.03 |
Abf2p binding assay
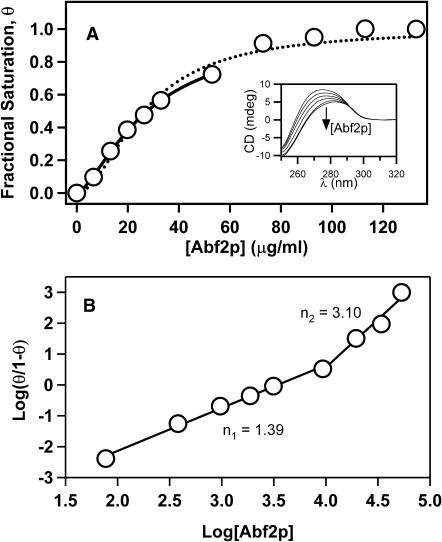
Finally, to compare the values of protein dissociation, KD, predicted by our model with the experimental KD value, we used CD spectroscopy to study binding of Abf2p to DNA. McAfee and co-workers used this technique to the study equilibrium binding of Sac7d and Sso7d to DNA (McAfee et al., 1996; Edmondson and Shriver, 2001). DNA CD in the region 250–310 nm is sensitive to conformational changes in the double helix (Baase and Johnson, 1979; Johnson et al., 1981). Conveniently, this band generally has the weakest CD signal for protein spectra. Therefore, this region represents a good choice for monitoring binding of a protein that distorts DNA, such as Abf2p. CD spectra obtained at increasing concentrations of Abf2p clearly showed an increase in DNA distortion that saturates at protein concentrations above 80 μg/mL (Fig. 6 A). The same data plotted in Hill coordinates (Fig. 6 B) indicate that Abf2p binding is weakly cooperative with Hill constant of 1.4. The Hill plot shows that at Abf2p concentrations above 60 μg/mL the cooperativity parameter sharply rises to 3.1; yet, we need to interpret these data with caution. As some literature examples show, rather than indicate the switch in protein binding mechanism this change might indicate a change in the structure of the protein-DNA complex that would have a stronger effect on the CD properties (Edmondson and Shriver, 2001). Significantly, the binding parameters that we obtained from the CD data in the initial regime (Fig. 6 A, solid line) show excellent agreement with the binding parameters that we obtained from the AFM data (Table 1). The values of the binding constant, KD, are virtually identical and the values of the Hill constant, q, are also very close. This comparison validates the b-WLC model and also confirms our hypothesis that Abf2p compacts DNA simply by placing sharp bends along the DNA double helix.
FIGURE 6.
(A) Plot of fraction of bound linearized pBR322 DNA molecules as a function of Abf2p concentration as determined from the CD spectra. Dashed line represents the fit to the whole data set with a cooperative bimolecular binding model. Solid line represents the fit to only the initial region of the Hill plot. Inset shows the CD spectra with an arrow indicating the direction of the spectral change as the protein concentration increases. (B) Hill plot of the data in A.
Our value of 1.44 ± 0.43 μM for the KD compares well with the literature value of 400 nM reported by Cho et al. (2001). Recently, Brewer and colleagues used optical trapping measurements to obtain an order-of-magnitude lower value (40 nM) for the binding constant of the Abf2p (Brewer et al., 2003). We believe that this lower value originated from the conditions inherent to the optical trapping measurement. In particular, in the flow cell environment, a trapped DNA molecule is constantly subjected to a stream of free protein, which may enhance protein binding and lower the KD value. Brewer and colleagues also used a slightly lower pH value, which could also influence the value of KD.
This mechanism can lead only to vaguely defined globular conformations of the DNA-protein complexes, which is quite similar to the appearance of the mt-nucleoid. It is also important to note that Abf2p action is strikingly different from other known compaction mechanisms, which typically involve much more ordered superstructures such as chromatin in the case of histone-induced packaging in the nucleus, or DNA toroids formed by protamines in sperm cells.
Further studies would be necessary to uncover the role of such a packaging arrangement in the regulation of DNA in mitochondria. However, we can speculate that such loose packing could simplify access of various regulatory proteins to DNA. Such an arrangement could allow the mitochondrion to avoid the need for a sophisticated DNA handling apparatus similar to the multistage machinery present in the eukaryotic cell nuclei.
CONCLUSIONS
We have used high-resolution AFM to observe binding of mitochondrial protein Abf2p to linear and circular DNA plasmids. Surprisingly, our images showed that the protein bends the DNA backbone sharply by ∼78°. AFM images clearly showed that Abf2p binding results in the compaction of DNA molecules for both linear and circular DNA. Moreover, at high protein concentration DNA molecules collapsed into compact globular structures reminiscent of a mitochondrial nucleoid. To analyze this compaction process we have developed a statistical model that describes the DNA-protein complex as a stiff polymer chain that has sharp bends incorporated throughout its length. Using this model, we have shown that incorporation of bends into the DNA backbone is alone sufficient to cause DNA compaction. End-to-end distances predicted by the model showed excellent agreement with the end-to-end distances that we measured from the AFM images. Moreover, binding parameters that we obtained from our model showed excellent agreement with the results of bulk studies. Significantly, the Abf2p compaction mechanism that we established appears to be distinctly different from common DNA packaging proteins.
Our findings have several important implications. First, we showed that high-resolution AFM imaging can provide quantitative characterization of protein-DNA interactions. Single molecule imaging not only can reveal the geometrical conformation of the protein-DNA complexes, but also can determine thermodynamic parameters for protein binding. We believe that AFM imaging will be an important part of the modern biophysics toolkit for studies of protein-DNA interactions. Second, we believe that our results will be important for establishing the mechanisms of mitochondrial DNA maintenance and regulation. The apparent loose packing of DNA by the Abf2p should provide important clues for the structure of the mitochondrial nucleoid and for possible access pathways for regulatory proteins. Further AFM studies using other mitochondrial proteins should provide a wealth of information about maintenance and regulation of mitochondrial DNA.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
Note added in proof: Bustamante and Rivetti have previously considered a worm-like chain model of DNA containing multiple bends; however, their general model assumed that all bends were coplanar (Rivetti et al., 1998).
We thank Professor C. Bustamante for pointing out some of the relevant work.
A.N. acknowledges Laboratory Directed Research and Development funding from the Science and Technology Office at Lawrence Livermore National Laboratory. R.W.F. is supported by the Lawrence Livermore National Laboratory Student Employee Graduate Research Fellowship. J.E.K. acknowledges support from Associated Western Universities and Lawrence Livermore National Laboratory. R.J.B. was supported by the National Science Foundation Center for Biophotonics Science and Technology (NSF agreement No. Phy-0120999). This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract #W 7405-Eng-48.
References
- Allen, M. J., E. M. Bradbury, and R. Balhorn. 1997. AFM analysis of DNA-protamine complexes bound to mica. Nucleic Acids Res. 25:2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M. J., X. F. Dong, T. E. O'Neill, P. Yau, S. C. Kowalczykowski, J. Gatewood, R. Balhorn, and E. M. Bradbury. 1993. Atomic force microscope measurements of nucleosome cores assembled along defined DNA sequences. Biochemistry. 32:8390–8396. [DOI] [PubMed] [Google Scholar]
- Baase, W. A., and W. C. Johnson. 1979. Circular dichroism and DNA secondary structure. Nucleic Acids Res. 6:797–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, C. G., S. B. Smith, V. A. Bloomfield, and C. Bustamante. 1997. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA. 94:6185–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, L., R. Friddle, A. Noy, E. Baldwin, S. Martin, M. Corzett, R. Balhorn, and R. Baskin. 2003. Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p. Biophys. J. 85. [DOI] [PMC free article] [PubMed]
- Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- Bustamante, C., and C. Rivetti. 1996. Visualizing protein-nucleic acid interactions on a large scale with the scanning force microscope. Annu. Rev. Biophys. Biomol. Struct. 25:395–429 (Review.). [DOI] [PubMed] [Google Scholar]
- Cho, J. H., Y. K. Lee, and C. B. Chae. 2001. The modulation of the biological activities of mitochondrial histone Abf2p by yeast PKA and its possible role in the regulation of mitochondrial DNA content during glucose repression. Biochim. Biophys. Acta Gene Struct. Expr. 1522:175–186. [DOI] [PubMed] [Google Scholar]
- Clausen-Schaumann, H., M. Seitz, R. Krautbauer, and H. E. Gaub. 2000. Force spectroscopy with single bio-molecules. Curr. Opin. Chem. Biol. 4:524–530. [DOI] [PubMed] [Google Scholar]
- Dai, H., J. Hafner, A. Rinzler, D. Colbert, and R. Smalley. 1996. Nanotubes as nanoprobes in scanning probe microscopy. Nature. 384:147–150. [Google Scholar]
- Diffley, J. F., and B. Stillman. 1988. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc. Natl. Acad. Sci. USA. 85:2120–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J. F., and B. Stillman. 1991. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl. Acad. Sci. USA. 88:7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson, S. P., and J. W. Shriver. 2001. DNA binding proteins Sac7d and Sso7d from Sulfolobus. Methods Enzymol. 334:129–145. [DOI] [PubMed] [Google Scholar]
- Erie, D., G. Yang, H. Schultz, and C. Bustamante. 1994. DNA bending by CRO protein in specific and nonspecific complexes: implications for protein site recognition and specificity. Science. 266:1562–1566. [DOI] [PubMed] [Google Scholar]
- Fang, Y., and J. H. Hoh. 1998. Early intermediates in spermidine-induced DNA condensation on the surface of mica. J. Am. Chem. Soc. 120:8903–8909. [Google Scholar]
- Fang, Y., and J. H. Hoh. 1999. Cationic silanes stabilize intermediates in DNA condensation. FEBS Lett. 459:173–176. [DOI] [PubMed] [Google Scholar]
- Grimley, T. B. 1953. The volume effect in polymer chains. J. Chem. Phys. 21:185–186. [Google Scholar]
- Hafner, J. H., C. L. Cheung, and C. M. Lieber. 1999. Growth of nanotubes for probe microscopy tips. Nature. 398:761–762. [Google Scholar]
- Hansma, H. G., K. J. Kim, D. E. Laney, R. A. Garcia, M. Argaman, M. J. Allen, and S. M. Parsons. 1997. Properties of biomolecules measured from atomic force microscope images: a review. J. Struct. Biol. 119:99–108. [DOI] [PubMed] [Google Scholar]
- Hoh, J. H., and Y. Fang. 1999. DNA condensation by cationic silanes. Mol. Biol. Cell. 10:179a. [DOI] [PubMed] [Google Scholar]
- Johnson, B. B., K. S. Dahl, I. J. Tinoco, V. I. Ivanov, and V. B. Zhurkin. 1981. Correlations between deoxyribonucleic acid structural parameters and calculated circular dichroism spectra. Biochemistry. 20:73–78. [DOI] [PubMed] [Google Scholar]
- Jones, D. N., M. A. Searles, G. L. Shaw, M. E. Churchill, S. S. Ner, J. Keeler, A. A. Travers, and D. Neuhaus. 1994. The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster. Structure. 2:609–627. [DOI] [PubMed] [Google Scholar]
- Kao, L. R., T. L. Megraw, and C. B. Chae. 1993. Essential role of the Hmg domain in the function of yeast mitochondrial histone Hm—functional complementation of Hm by the nuclear nonhistone protein Nhp6a. Proc. Natl. Acad. Sci. USA. 90:5598–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratky, O., and G. Porod. 1949. Röntgenuntersuchung gelöster Fadenmoleküle. Rec. Trav. Chim. Pays-Bas. 68:1106–1123. [Google Scholar]
- Love, J. J., X. Li, D. A. Case, K. Giese, R. Grosschedl, and P. E. Wright. 1995. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 376:791–795. [DOI] [PubMed] [Google Scholar]
- Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8-Ångstrom resolution. Nature. 389:251–260. [DOI] [PubMed] [Google Scholar]
- Lyubchenko, Y. L., A. A. Gall, and L. S. Shlyakhtenko. 2001. Atomic force microscopy of DNA and protein-DNA complexes using functionalized mica substrates. Meth. Mol. Biol. 148:569–578. [DOI] [PubMed] [Google Scholar]
- Lyubchenko, Y. L., L. S. Shlyakhtenko, M. Binus, C. Gaillard, and F. Strauss. 2002. Visualization of hemiknot DNA structure with an atomic force microscope. Nucleic Acids Res. 30:4902–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 1998. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl. Acad. Sci. USA. 95:6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. S., E. Pulido, V. C. Chu, T. S. Lechner, and E. P. Baldwin. 2002. The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J. Mol. Biol. 319:107–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse, J. E., B. Wong, Y. M. Yen, F. H. Allain, R. C. Johnson, and J. Feigon. 2002. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 323:263–284. [DOI] [PubMed] [Google Scholar]
- McAfee, J. G., S. P. Edmondson, I. Zegar, and J. W. Shriver. 1996. Equilibrium DNA binding of Sac7d protein from the hyperthermophile Sulfolobus acidocaldarius: fluorescence and circular dichroism studies. Biochemistry. 35:4034–4045. [DOI] [PubMed] [Google Scholar]
- Miyakawa, I., H. Aoi, N. Sando, and T. Kuroiwa. 1984. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci. 66:21–38. [DOI] [PubMed] [Google Scholar]
- Miyakawa, I., N. Sando, S. Kawano, S. Nakamura, and T. Kuroiwa. 1987. Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J. Cell Sci. 88:431–439. [DOI] [PubMed] [Google Scholar]
- Odijk, T. 1986. Theory of lyotropic polymer liquid crystals. Macromolecules. 19:2313–2329. [Google Scholar]
- Onsager, L. 1949. The effects of shape on the interaction of colloidal particles. Ann. N. Y. Acad. Sci. 51:627–659. [Google Scholar]
- Radmacher, M., R. W. Tillmann, M. Fritz, and H. E. Gaub. 1992. From molecules to cells: imaging soft samples with the atomic force microscope. Science. 257:1900–1905. [DOI] [PubMed] [Google Scholar]
- Rivetti, C., M. Guthold, and C. Bustamante. 1996. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 264:919–932. [DOI] [PubMed] [Google Scholar]
- Rivetti, C., C. Walker, and C. Bustamante. 1998. Polymer chain statistics and conformational analysis of DNA molecules with bends or sections of different flexibility. J. Mol. Biol. 280:41–59. [DOI] [PubMed] [Google Scholar]
- Satoh, M., and T. Kuroiwa. 1991. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 196:137–140. [DOI] [PubMed] [Google Scholar]
- Scaffidi, P., and M. Bianchi. 2001. Spatially precise DNA bending is an essential activity of the Sox2 transcription factor. J. Biol. Chem. 276:47296–47302. [DOI] [PubMed] [Google Scholar]
- Schnitzler, G. R., C. L. Cheung, J. H. Hafner, A. J. Saurin, R. E. Kingston, and C. M. Lieber. 2001. Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol. Cell. Biol. 21:8504–8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., and L. Nilsson. 1998. Interaction of human SRY protein with DNA: a molecular dynamics study. Proteins. 31:417–433. [DOI] [PubMed] [Google Scholar]
- Tiner, W. J. S., V. N. Potaman, R. R. Sinden, and Y. L. Lyubchenko. 2001. The structure of intramolecular triplex DNA: atomic force microscopy study. J. Mol. Biol. 314:353–357. [DOI] [PubMed] [Google Scholar]
- Van Tuyle, G. C., and M. L. McPherson. 1979. A compact form of rat liver mitochondrial DNA stabilized by bound proteins. J. Biol. Chem. 254:6044–6053. [PubMed] [Google Scholar]
- Voet, D., J. G. Voet, and C. W. Pratt. 1999. Fundamentals of Biochemistry. Wiley, New York.
- Werner, M. H., M. E. Bianchi, A. M. Gronenborn, and G. M. Clore. 1995. NMR spectroscopic analysis of the DNA conformation induced by the human testis determining factor SRY. Biochemistry. 34:11998–12004. [DOI] [PubMed] [Google Scholar]
- Wintersberger, U., M. Binder, and P. Fischer. 1975. Cytogenetic demonstration of mitotic chromosomes in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 142:13–17. [DOI] [PubMed] [Google Scholar]
- Yamakawa, H. 1971. Modern Theory of Polymer Solutions. Harper and Row, New York.
- Zelenaya-Troitskaya, O., S. M. Newman, K. Okamoto, P. S. Perlman, and R. A. Butow. 1998. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 148:1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.