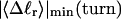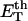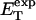Abstract
Mesoscopic models of unmelted and locally melted supercoiled DNAs in 20 mM ionic strength are simulated over a range of linking difference from Δℓ = 0 to −26 turns, or superhelix density from σ = 0 to −0.062. A domain containing m = 0, 28, or 56 melted basepairs (out of 4349 total) is modeled simply by a region of suitable length with substantially reduced torsion and bending elastic constants. Average structural properties are calculated from the saved configurations, and a reversible work protocol is used to calculate the supercoiling free energy, 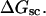 The cross-writhe between duplex and melted regions (defined herein) is found to be negligibly small. The total writhe, radius of gyration, and ordered elements of the diagonalized inertial tensor are found to be nearly universal functions of the residual linking difference (
The cross-writhe between duplex and melted regions (defined herein) is found to be negligibly small. The total writhe, radius of gyration, and ordered elements of the diagonalized inertial tensor are found to be nearly universal functions of the residual linking difference (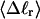 ) associated with the duplex region, independent of m. However, deformability of the tertiary structure, as manifested by the variance of those same properties, is not a universal function of
) associated with the duplex region, independent of m. However, deformability of the tertiary structure, as manifested by the variance of those same properties, is not a universal function of 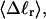 but depends upon m.
but depends upon m. 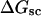 varies with
varies with 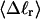 more strongly than
more strongly than 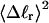 due to the low ionic strength. The twist energy parameter,
due to the low ionic strength. The twist energy parameter, 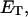 obtained from the simulated
obtained from the simulated 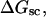
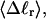 and net twisting strain of the melted region,
and net twisting strain of the melted region,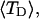 is found to be independent of m, hence also of the torsion and bending elastic constants of the melted region. However,
is found to be independent of m, hence also of the torsion and bending elastic constants of the melted region. However, 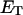 increases linearly with
increases linearly with 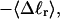 which leads to 1), a small overestimation of
which leads to 1), a small overestimation of 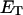 for any given
for any given 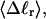 when
when 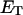 is determined from the observed
is determined from the observed 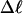 and
and 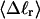 by the protocol of Bauer and Benham; and 2), a significant enhancement of the apparent slope,
by the protocol of Bauer and Benham; and 2), a significant enhancement of the apparent slope, 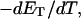 obtained via the protocol of Bauer and Benham, relative to the actual slope at fixed
obtained via the protocol of Bauer and Benham, relative to the actual slope at fixed 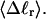 After taking these two effects into account, the theoretical and experimental
After taking these two effects into account, the theoretical and experimental 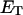 values and
values and 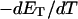 values agree rather well. For the larger
values agree rather well. For the larger 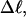 the melted regions are found preferentially in the linker domains between interwound arms, rather than in the apical regions at the ends of interwound arms.
the melted regions are found preferentially in the linker domains between interwound arms, rather than in the apical regions at the ends of interwound arms.
INTRODUCTION
Negative supercoiling generally raises the free energy of a duplex circular DNA relative to that of its melted intertwined single strands. In sufficiently low ionic strength (≲20 mM) at 37°C, supercoiled DNAs with sufficient superhelix density (σ) in the range 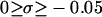 (native) will locally melt to an extent that depends upon the sequence (Benham, 1990, 1992; Bauer and Benham, 1993; Bauer et al., 1995). Even at higher, more physiological ionic strengths, where stable local melting is not observed up to native superhelix density (−0.05) (Kowalski et al., 1988), the free energy change required to locally melt the DNA is substantially reduced. This supercoiling-induced destabilization of duplex with respect to local melting has been suggested to facilitate certain biological processes, wherein melted regions are involved, such as the initiation of transcription, replication, and recombination (Benham, 1990; Bauer and Benham, 1993; Bauer et al., 1995; Sheridan et al., 1998).
(native) will locally melt to an extent that depends upon the sequence (Benham, 1990, 1992; Bauer and Benham, 1993; Bauer et al., 1995). Even at higher, more physiological ionic strengths, where stable local melting is not observed up to native superhelix density (−0.05) (Kowalski et al., 1988), the free energy change required to locally melt the DNA is substantially reduced. This supercoiling-induced destabilization of duplex with respect to local melting has been suggested to facilitate certain biological processes, wherein melted regions are involved, such as the initiation of transcription, replication, and recombination (Benham, 1990; Bauer and Benham, 1993; Bauer et al., 1995; Sheridan et al., 1998).
A quantitative understanding of supercoiling-induced local melting requires knowledge of 1), the decrease (ΔΔGsc)in deformational supercoiling free energy (ΔGsc) upon melting a sequence of m basepairs, which depends upon the unknown effective elastic constants for torsion and bending of the melted regions, and 2), the free energy increase upon melting m basepairs far below their normal melting temperature, TM. Until now, neither direct experimental measurements nor simulations of ΔGsc for a locally melted DNA have been reported. Likewise, direct experimental or theoretical assessments of ΔΔGsc for local melting of an initially unmelted supercoiled DNA are lacking. Nonetheless, the phenomenon of supercoiling-induced local melting has been extensively analyzed by Benham (1990, 1992)), who employed some of the known statistical thermodynamics of DNA melting together with several untested assumptions regarding both the structural properties of partially melted supercoiled DNAs and the thermodynamics of their supercoiling. After invoking an additional untested assumption pertaining to the gel mobilities of locally melted and unmelted supercoiled DNAs, Bauer and Benham (1993) and Bauer et al. (1995) investigated the magnitude and temperature dependence of the supercoiling free energy. We believe that it is now essential to address several questions pertaining to the structures and supercoiling thermodynamics of locally melted DNAs, and to test certain of the assumptions invoked by Benham and co-workers. This is done via Monte Carlo simulations of mesoscopic models of locally melted supercoiled DNAs, wherein the denatured (melted) regions are modeled simply as regions with much reduced torsion and bending elastic constants. The assumptions invoked by Benham and Bauer and Benham would be expected to be more applicable to our simple model molecules than to real DNAs. The results in this work significantly increase our knowledge regarding the structural properties and supercoiling thermodynamics of model circular DNAs with fixed numbers (28 or 56) of melted basepairs, and significantly deepen the level of analysis. Hopefully, our findings will lead to a further refinement of Benham's model, and ultimately lead to the extraction of increasingly reliable and accurate estimates of certain unknown quantities, including the effective elastic constants of the melted regions, by fitting various experimental data.
Topological and geometrical aspects of supercoiling
Every DNA topoisomer is characterized by its integral linking number 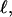 the number of turns of each single strand around the other. ℓ is a topological invariant that is unaltered by any change in tertiary or secondary structure of the DNA, including local melting. The extent of deformation of the DNA is characterized by the linking difference,
the number of turns of each single strand around the other. ℓ is a topological invariant that is unaltered by any change in tertiary or secondary structure of the DNA, including local melting. The extent of deformation of the DNA is characterized by the linking difference, 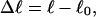 where
where 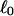 is the intrinsic twist of the unstrained DNA. For an unmelted supercoiled DNA comprising M basepairs (bp),
is the intrinsic twist of the unstrained DNA. For an unmelted supercoiled DNA comprising M basepairs (bp), 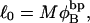 where
where 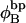 ((1/10.45) turns/bp) is the intrinsic succession angle between basepairs of the duplex DNA. For a locally melted DNA comprising M basepairs, of which m are melted,
((1/10.45) turns/bp) is the intrinsic succession angle between basepairs of the duplex DNA. For a locally melted DNA comprising M basepairs, of which m are melted, 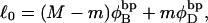 where
where 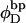 is the intrinsic succession angle between (open) basepairs of the melted, or denatured, DNA. Although
is the intrinsic succession angle between (open) basepairs of the melted, or denatured, DNA. Although 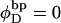 for a real DNA, it typically does not vanish for our simulated molecules, so is introduced at this point and carried along through the subsequent development. In the sequel, when a distinction is necessary, the properties of unmelted DNAs will be denoted by an overbar (e.g.,
for a real DNA, it typically does not vanish for our simulated molecules, so is introduced at this point and carried along through the subsequent development. In the sequel, when a distinction is necessary, the properties of unmelted DNAs will be denoted by an overbar (e.g., 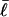 ) and those of locally melted DNAs will be denoted by a tilde (e.g.,
) and those of locally melted DNAs will be denoted by a tilde (e.g., 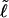 ). When considering unmelted and locally melted forms of the same molecule,
). When considering unmelted and locally melted forms of the same molecule, 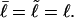 Then the change in linking difference upon local melting is
Then the change in linking difference upon local melting is
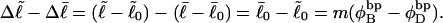 |
(1) |
For unmelted and locally melted molecules, the superhelix density is defined by 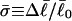 and
and 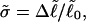 respectively.
respectively. 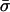 is generally negative for native supercoiled DNAs, since
is generally negative for native supercoiled DNAs, since 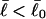 in that case.
in that case.
We assume that the linking number is partitioned between twist (t) and writhe (w) according to (White, 1969; Fuller, 1971):
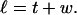 |
(2) |
Equation 2 was proved for ribbons of infinitesimal width with smooth edges. Its validity for ribbons of finite width might be restricted to the case of smooth inextensible edges. Moreover, its applicability to partially denatured DNAs is far from obvious. However, for the DNAs considered here, which exhibit a large linking difference and only a small locally denatured region, a large amount of linking difference is typically absorbed into the latter, which would be expected to wind that denatured DNA into a left-handed helical bundle, wherein the space curves of the single strands are reasonably smooth, and for which Eq. 2 is either valid or a good approximation.
The DNA is here regarded as a chain of N = M/ν subunits, each containing ν basepairs. These subunits are labeled consecutively by the index j, j = 1, … N. In each subunit (j) is fixed a coordinate frame (xj,yj,zj), the zj axis of which lies along the bond vector (bj) that extends from the origin of the jth frame to the origin of the succeeding (j + 1)th frame. The Euler rotation that carries a coordinate frame from coincidence with the jth frame to coincidence with the (j + 1)th frame is 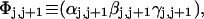 where the component rotations,
where the component rotations, 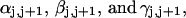 are taken sequentially around the body-fixed z, new body-fixed y′, and final body-fixed z″ axes (Edmonds, 1974). The net twist of the DNA is given by
are taken sequentially around the body-fixed z, new body-fixed y′, and final body-fixed z″ axes (Edmonds, 1974). The net twist of the DNA is given by
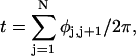 |
(3) |
where 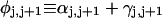 (radians) is the net twist of the Euler rotation from the jth to the (j + 1)th frame (Schurr, 1985). For a partially melted molecule, in which the N − n subunits, j = 1,2, … N − n, are B-helical duplex, but the n = m/ν subunits, j = N − n + 1, … ,N, are melted, the net twist can be partitioned among the two regions according to
(radians) is the net twist of the Euler rotation from the jth to the (j + 1)th frame (Schurr, 1985). For a partially melted molecule, in which the N − n subunits, j = 1,2, … N − n, are B-helical duplex, but the n = m/ν subunits, j = N − n + 1, … ,N, are melted, the net twist can be partitioned among the two regions according to
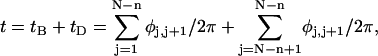 |
(4) |
where the first term on the far right-hand side (rhs) constitutes the twist (tB) of the B-helical duplex region and the second term constitutes that (tD) of the denatured (melted) region.
The writhe is commonly approximated by the discretized Gauss integral,
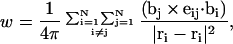 |
(5) |
where the 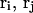 denote the positions of the origins of the ith and jth subunit frames, respectively, in the laboratory frame, and
denote the positions of the origins of the ith and jth subunit frames, respectively, in the laboratory frame, and 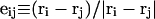 denotes a unit vector along ri − rj (Hao and Olson, 1989). We imagine that the single strands in a locally melted region are smoothly wound in such a way that their midline is well-defined, and can be taken as a line of discretized bond vectors for the purpose of a calculating w according to Eq. 5. It is useful to partition the writhe among the B-helical duplex and denatured regions according to
denotes a unit vector along ri − rj (Hao and Olson, 1989). We imagine that the single strands in a locally melted region are smoothly wound in such a way that their midline is well-defined, and can be taken as a line of discretized bond vectors for the purpose of a calculating w according to Eq. 5. It is useful to partition the writhe among the B-helical duplex and denatured regions according to
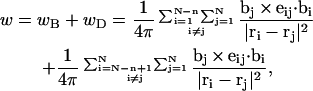 |
(6) |
where the first term on the far rhs constitutes the writhe (wB) associated with the B-helical duplex region and the second term constitutes the writhe (wD) associated with the denatured region. The writhe wB of the duplex region contains (N − n)2 self-terms in which both i and j lie within that same region (1, … N − n), plus (N − n)n cross-terms in which i lies within the duplex region but j lies within the denatured region (N − n + 1, … N). The writhe wD of the denatured region similarly contains n2 self-terms in which both i and j lie within the denatured region plus n(N − n) cross-terms in which i lies within the denatured region, but j lies within the duplex region. The number of cross-terms in wB and in wD is identical. Moreover, because 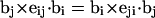 is invariant to interchange of i and j, every cross-term in wB has an identical counterpart in the cross-terms of wD. Thus, the entire cross-term contribution of the total writhe is divided evenly between the two regions, regardless of their relative sizes.
is invariant to interchange of i and j, every cross-term in wB has an identical counterpart in the cross-terms of wD. Thus, the entire cross-term contribution of the total writhe is divided evenly between the two regions, regardless of their relative sizes.
The linking number of locally melted DNA can be partitioned as 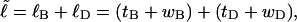 where
where 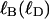 corresponds to the first (second) term on the far rhs. The intrinsic twist can also be partitioned as
corresponds to the first (second) term on the far rhs. The intrinsic twist can also be partitioned as 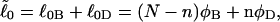 where
where 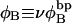 is the intrinsic succession angle (turns/subunit) between subunits of unmelted DNA and
is the intrinsic succession angle (turns/subunit) between subunits of unmelted DNA and 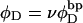 is that for melted DNAs, and
is that for melted DNAs, and 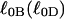 corresponds to the first (second) term on the far rhs. Finally, the linking difference,
corresponds to the first (second) term on the far rhs. Finally, the linking difference, 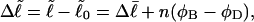 can be partitioned between the duplex and melted regions according to
can be partitioned between the duplex and melted regions according to
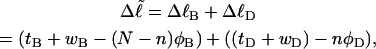 |
(7) |
where the first term on the rhs constitutes the linking difference 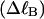 of the duplex region and the second term constitutes that
of the duplex region and the second term constitutes that 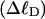 of the melted region. Although
of the melted region. Although 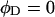 for a real DNA, it typically does not vanish for our model DNAs.
for a real DNA, it typically does not vanish for our model DNAs.
Questions pertaining to locally melted supercoiled DNAs
The following questions are relevant to the characterization and analysis of supercoiling-induced local melting:
How are the writhe, twisting strain, and linking difference of a locally melted topoisomer distributed between its duplex and melted regions, the latter of which exhibits much weaker torsional and bending rigidities than the former?
How does the presence of the elastically soft denatured region affect the structure (e.g., writhe and radius of gyration (
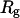 )), and fluctuations in structure of the molecule? In particular, how do such properties of a locally melted DNA compare with the corresponding properties of an unmelted topoisomer whose total
)), and fluctuations in structure of the molecule? In particular, how do such properties of a locally melted DNA compare with the corresponding properties of an unmelted topoisomer whose total 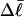 matches the
matches the 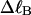 of the locally melted DNA?
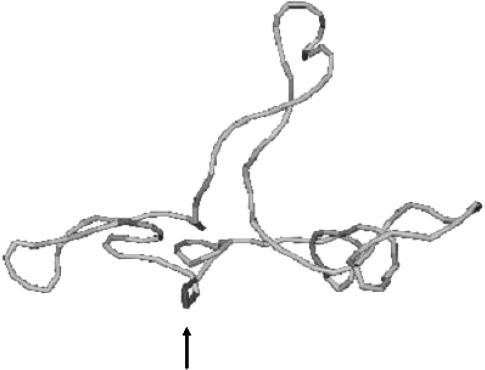
of the locally melted DNA?Are the more flexible melted regions found preferentially at certain locales or instead more or less uniformly distributed throughout all regions of the tertiary structure of the supercoiled DNA? Intrinsically curved sequences with normal duplex rigidity are found preferentially at the external ends, or apices, of interwound superhelical branches or domains (Bussiek et al., 2002; Pfannschmidt and Langowski, 1998; Chirico and Langowski, 1996). Supercoiled plasmids containing the SCA10 repeats, (ATTCT)n.(AGAAT)n, n = 5, … 29, were deposited on a mica surface treated with 3-aminopropyltriethoxysilane (APS) (Lyubchenko and Shlyakhtenko, 1997), rinsed, dried, and scanned by atomic force microscopy (AFM) in air (Potaman et al., 2003). Open regions were observed in these DNAs, and found not to be associated with apices. However, the extensive flattening, low prevailing water activity, and certain peculiarities in the data suggest that these are not equilibrium structures typical of normal locally melted supercoiled DNAs in solution. The open regions in the AFM images are specific to the SCA10 sequences, and are not observed for other DNAs with (A + T)-rich regions of comparable length, which also exhibit stable melting in two-dimensional gels. They also do not show the expected negative interwinding of the two single strands. Hence, the location(s) of locally melted regions in normal equilibrium supercoiled DNAs in solution remains an open question.
How does the (deformational) supercoiling free energy ΔGsc of a locally melted DNA vary with the linking difference of its unmelted parent topoisomer
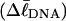 and with the number m of its melted basepairs, or equivalently the number n = m/ν of its melted subunits? From such information, the difference in supercoiling free energy, ΔΔGsc, between an unmelted parent topoisomer with linking difference
and with the number m of its melted basepairs, or equivalently the number n = m/ν of its melted subunits? From such information, the difference in supercoiling free energy, ΔΔGsc, between an unmelted parent topoisomer with linking difference 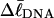 and its partially melted daughter molecule with m melted basepairs can be estimated. This latter quantity is important, because the reduction in supercoiling free energy upon partial melting provides the driving “force” for supercoiling-induced partial melting.
and its partially melted daughter molecule with m melted basepairs can be estimated. This latter quantity is important, because the reduction in supercoiling free energy upon partial melting provides the driving “force” for supercoiling-induced partial melting.Can the total supercoiling free energy, including the configurational entropy contribution, be partitioned between the duplex and melted regions and, if so, how does it vary with the average linking difference
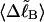 of the duplex region, and the average twisting strain of the melted region,
of the duplex region, and the average twisting strain of the melted region, 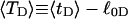 ?
?When the supercoiling free energy of the duplex region of a locally melted DNA is expressed in terms of the square of its associated linking difference,
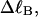 how does its effective torque constant, or equivalently its twist energy parameter (ET), compare with the ET for the corresponding unmelted DNA in regard to both magnitude and variation (if any) with linking difference?
how does its effective torque constant, or equivalently its twist energy parameter (ET), compare with the ET for the corresponding unmelted DNA in regard to both magnitude and variation (if any) with linking difference?
With the answers to these questions in hand, one could combine the estimated reduction in supercoiling free energy upon opening a region containing m basepairs with an estimate of the free energy increase upon melting m basepairs (below Tm) to estimate m for any given linking difference and temperature. By explicitly or implicitly assuming the answers to certain of the preceding questions, Benham (1990, 1992) formulated a protocol to do essentially that for sufficiently supercoiled DNAs in low ionic strength (≤20 mM) at temperatures 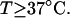 (There is currently no evidence for stable opening of a supercoiled DNA with
(There is currently no evidence for stable opening of a supercoiled DNA with 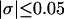 at
at 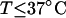 in ionic strengths ≥30 mM (Kowalski et al., 1988)). After invoking an additional assumption pertaining to the gel mobilities of unmelted and locally melted species, Bauer and Benham formulated and applied a protocol to analyze two-dimensional gels of an ensemble of topoisomers of supercoiled PBR322 DNA in ∼20 mM ionic strength at various temperatures,
in ionic strengths ≥30 mM (Kowalski et al., 1988)). After invoking an additional assumption pertaining to the gel mobilities of unmelted and locally melted species, Bauer and Benham formulated and applied a protocol to analyze two-dimensional gels of an ensemble of topoisomers of supercoiled PBR322 DNA in ∼20 mM ionic strength at various temperatures, 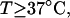 to estimate several quantities, including ET and the free energy, enthalpy, and entropy of supercoiling.
to estimate several quantities, including ET and the free energy, enthalpy, and entropy of supercoiling.
The primary objectives of this work are to address questions 1–6 above for a simple mesoscopic model of a locally melted DNA, to test or partially test certain assumptions of Bauer and Benham (1993), and to understand why such assumptions are, or are not, valid. It is first necessary to clarify those assumptions somewhat.
Assumptions in the model of Benham
Benham (1990, 1992) introduced a new quantity, called the residual linking difference (ΔLkr), for a locally denatured molecule. Bauer and Benham (1993) described ΔLkr as “that portion of the initial linking difference (i.e., of the unmelted parent) that is not accommodated either by local strand separation or by subsequent interstrand twisting in the denatured regions”. They further state that denaturation of a run of m basepairs leads to a change in the magnitude of ΔLkr equal to 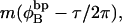 where τ is the “helicity” (i.e., twist) of the denatured regions in radians per basepair. Keeping our notation to distinguish denatured and nondenatured forms of the same molecule, we express the above statement as
where τ is the “helicity” (i.e., twist) of the denatured regions in radians per basepair. Keeping our notation to distinguish denatured and nondenatured forms of the same molecule, we express the above statement as
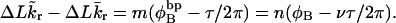 |
(8) |
This description of 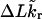 is incomplete, because Benham and Bauer and Benham did not consider the possibility that the denatured region might, owing to its rather low bending rigidity, absorb linking difference not only in the form of twist, but also in the form of writhe, perhaps by tight toroidal winding of the axis (i.e., midline) of the denatured region.
is incomplete, because Benham and Bauer and Benham did not consider the possibility that the denatured region might, owing to its rather low bending rigidity, absorb linking difference not only in the form of twist, but also in the form of writhe, perhaps by tight toroidal winding of the axis (i.e., midline) of the denatured region.
If the convex volume that just envelopes the minor denatured domain (D) is sufficiently small in all of its physical dimensions compared to that of the major unmelted domain (B), then in the vast majority of two-dimensional projections of the molecule taken from all orientations, the axis of the unmelted region will have no intersections with the axis of the unmelted region, so there will be no significant cross-term contributions to the total writhe. It will be explicitly demonstrated below that such a circumstance prevails for the relatively small denatured regions considered here. In this case, any significant writhe wD associated with the minor denatured domain D must stem primarily from its self-writhe 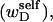 and would therefore be predominantly local. Unfortunately, such a local writhe,
and would therefore be predominantly local. Unfortunately, such a local writhe, 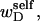 cannot be distinguished from local twist, tD, of that same region without some means of resolving the structure of the locally denatured region. Currently, there is no experimental method to ascertain unequivocally whether the linking difference associated with a small denatured region is present in the form of twist
cannot be distinguished from local twist, tD, of that same region without some means of resolving the structure of the locally denatured region. Currently, there is no experimental method to ascertain unequivocally whether the linking difference associated with a small denatured region is present in the form of twist 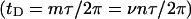 or self-writhe
or self-writhe 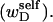 In any event, it will be shown rigorously below that Eq. 8 is incomplete whenever either
In any event, it will be shown rigorously below that Eq. 8 is incomplete whenever either 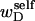 or the cross-writhe term
or the cross-writhe term 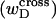 associated with the denatured region is nonvanishing.
associated with the denatured region is nonvanishing.
Benham (1990, 1992) and Bauer and Benham (1993) also assumed that the total supercoiling free energy, ΔGsc, of a single partially melted DNA consists of only two terms,
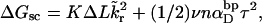 |
(9) |
where K is an effective torque constant for supercoiling of the duplex region and 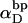 is the effective torsion elastic constant of each of the m = νn “torsion” springs between (open) basepairs of the denatured region. It is implicitly assumed in Eq. 9 that the intrinsic twist,
is the effective torsion elastic constant of each of the m = νn “torsion” springs between (open) basepairs of the denatured region. It is implicitly assumed in Eq. 9 that the intrinsic twist, 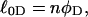 of the denatured regions vanishes, as it does for real DNAs. Our objective here is not to assess the validity of the assumed Hooke's law behavior of the melted region, but instead to assume such behavior to address other questions and test other assumptions. The quantity K in Eq. 9 is related to the twist energy parameter, ET, used previously in our laboratory by K = kT(ET/M), where k is Boltzmann's constant, T the absolute temperature, and M = νN the number of (duplex) basepairs (Wu et al., 1988; Clendenning and Schurr, 1994; Clendenning et al., 1994; Gebe et al., 1995, 1996; Schurr et al., 1995; Delrow et al., 1997a,b; Naimushin et al., 2001). For sufficiently large DNAs (M ≥ 2000 bp) ET is practically independent of M. (Bauer and Benham used the symbol q(T) instead of ET, and Cτ instead of αD). The partitioning of ΔGsc into separate contributions from the duplex and denatured regions, which is implied by Eq. 9, suggests that for a partially melted DNA, one should employ K = kT(ET/(M − m), where M is replaced by the number (M − m) of basepairs in the duplex region. However, both in the experiments of Bauer and Benham and in these simulations, m is only a tiny fraction of M (m/M ≲ 0.013), so the error in ΔGsc resulting from use of M in place of M − m in the expression for K is smaller than either the experimental or simulation errors. We test the assumption (Eq. 9) that was originally stated by Benham.
of the denatured regions vanishes, as it does for real DNAs. Our objective here is not to assess the validity of the assumed Hooke's law behavior of the melted region, but instead to assume such behavior to address other questions and test other assumptions. The quantity K in Eq. 9 is related to the twist energy parameter, ET, used previously in our laboratory by K = kT(ET/M), where k is Boltzmann's constant, T the absolute temperature, and M = νN the number of (duplex) basepairs (Wu et al., 1988; Clendenning and Schurr, 1994; Clendenning et al., 1994; Gebe et al., 1995, 1996; Schurr et al., 1995; Delrow et al., 1997a,b; Naimushin et al., 2001). For sufficiently large DNAs (M ≥ 2000 bp) ET is practically independent of M. (Bauer and Benham used the symbol q(T) instead of ET, and Cτ instead of αD). The partitioning of ΔGsc into separate contributions from the duplex and denatured regions, which is implied by Eq. 9, suggests that for a partially melted DNA, one should employ K = kT(ET/(M − m), where M is replaced by the number (M − m) of basepairs in the duplex region. However, both in the experiments of Bauer and Benham and in these simulations, m is only a tiny fraction of M (m/M ≲ 0.013), so the error in ΔGsc resulting from use of M in place of M − m in the expression for K is smaller than either the experimental or simulation errors. We test the assumption (Eq. 9) that was originally stated by Benham.
Another crucial assumption employed by Benham (1990, 1992) and Bauer and Benham (1993) is that the same value of K (or ET) applies to both the unmelted DNAs and the duplex regions of partially melted DNAs. Because K (or ET) sensitively reflects the bending rigidity and also the partitioning of  between twist and writhe in an unmelted supercoiled DNA, its use in Eq. 9 can be justified only if
between twist and writhe in an unmelted supercoiled DNA, its use in Eq. 9 can be justified only if  pertains exclusively to the unmelted region B, and contains no contribution from the torsionally and flexurally softer denatured region D.
pertains exclusively to the unmelted region B, and contains no contribution from the torsionally and flexurally softer denatured region D.
To better illuminate the assumptions of Benham (1990, 1992) and Bauer and Benham (1993) that are implied by Eqs. 8 and 9, we begin with 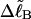 in Eq. 7, which is rigorously given by
in Eq. 7, which is rigorously given by
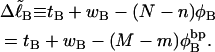 |
(10) |
We now define an alternative residual linking difference, 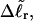 of the partially melted DNA as that part of the total linking difference that is associated with the unmelted duplex region. That is,
of the partially melted DNA as that part of the total linking difference that is associated with the unmelted duplex region. That is, 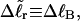 which gives exactly
which gives exactly
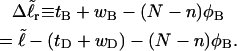 |
(11) |
For the unmelted parent molecule with the same linking number, 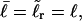 the residual linking difference is
the residual linking difference is
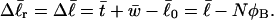 |
(12) |
After subtracting Eq. 12 from Eq. 11, and setting tD = νnτ/2π, we obtain
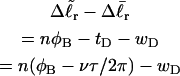 |
(13) |
for the change in 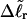 upon local melting of m = νn basepairs. Our Eq. 13 differs from the corresponding relation of Bauer and Benham (our Eq. 8) by the term, −wD. Our definition of the residual linking difference in Eq. 11 is complete and is the most appropriate choice for two main reasons. i), The value of our
upon local melting of m = νn basepairs. Our Eq. 13 differs from the corresponding relation of Bauer and Benham (our Eq. 8) by the term, −wD. Our definition of the residual linking difference in Eq. 11 is complete and is the most appropriate choice for two main reasons. i), The value of our 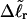 is unaffected by any interconversion between tD and
is unaffected by any interconversion between tD and 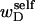 in the denatured region. This is important, because the exact form of the linking difference in that region is not known, and could well be self-writhe instead of twist. ii),
in the denatured region. This is important, because the exact form of the linking difference in that region is not known, and could well be self-writhe instead of twist. ii), 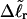 pertains only to the duplex region, and is therefore the appropriate quantity to employ in Eq. 9 for the supercoiling free energy, given that the same value of K (or ET) is assumed to apply for both unmelted DNAs and the duplex regions of locally melted DNAs. Consequently, we believe that Bauer and Benham either intended, or should have intended, to adopt our choice of the residual linking difference in Eq. 11. Thus, their adoption of Eq. 8 instead of Eq. 13 involves the implicit assumption that
pertains only to the duplex region, and is therefore the appropriate quantity to employ in Eq. 9 for the supercoiling free energy, given that the same value of K (or ET) is assumed to apply for both unmelted DNAs and the duplex regions of locally melted DNAs. Consequently, we believe that Bauer and Benham either intended, or should have intended, to adopt our choice of the residual linking difference in Eq. 11. Thus, their adoption of Eq. 8 instead of Eq. 13 involves the implicit assumption that 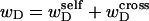 is negligibly small. Because
is negligibly small. Because 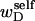 and the cross-writhe term
and the cross-writhe term 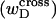 associated with the denatured region are expected to have the same sign, the assumption that wD is negligibly small is equivalent to assuming that
associated with the denatured region are expected to have the same sign, the assumption that wD is negligibly small is equivalent to assuming that 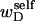 and
and 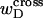 are both negligibly small.
are both negligibly small.
A possible alternative interpretation of the ansatz of Benham (1990, 1992) and Bauer and Benham (1993), contained in Eqs. 8 and 9, is that the total “helicity” of the denatured region (in turns) is actually the sum of the twist (tD) and the self-writhe 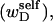 rather than simply the twist, so that
rather than simply the twist, so that 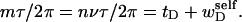 Then, if the ratio
Then, if the ratio 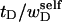 were to remain constant with increasing linking difference (as is the case for tB/wB in an unmelted DNA), then the free energy associated with the superhelicity in the denatured region could conceivably be quadratic in the sum,
were to remain constant with increasing linking difference (as is the case for tB/wB in an unmelted DNA), then the free energy associated with the superhelicity in the denatured region could conceivably be quadratic in the sum, 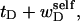 (as it is for an unmelted DNA). Because the effective torque constant αD is taken as an unknown adjustable parameter characteristic of denatured regions, it could conceivably apply as well to the square of
(as it is for an unmelted DNA). Because the effective torque constant αD is taken as an unknown adjustable parameter characteristic of denatured regions, it could conceivably apply as well to the square of 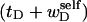 as to the square of tD. In this case, Eq. 13, which applies in all events, could be written as
as to the square of tD. In this case, Eq. 13, which applies in all events, could be written as
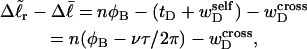 |
(14) |
where 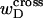 denotes the cross-writhe of the denatured region. Equation 8 is identical to Eq. 14 if, and only if,
denotes the cross-writhe of the denatured region. Equation 8 is identical to Eq. 14 if, and only if, 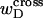 is negligibly small. In this interpretation, the implicit assumption of Benham is that
is negligibly small. In this interpretation, the implicit assumption of Benham is that 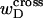 is negligibly small.
is negligibly small.
One might also ask whether Eqs. 8 and 9 would be reasonable assumptions, if the total “helicity” of the denatured region, nντ/2π turns, were taken to be its associated linking difference in turns, 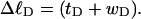 This would cause Eq. 8 to coincide with the first line of Eq. 13. However,
This would cause Eq. 8 to coincide with the first line of Eq. 13. However, 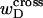 depends strongly on the space curve of the duplex B region, as well as that of the locally melted D region, and those regions have very different elastic constants. Consequently, it seems most unlikely that tD/wD would remain constant with increasing linking difference, or that the free energy of the denatured region would vary simply quadratically with
depends strongly on the space curve of the duplex B region, as well as that of the locally melted D region, and those regions have very different elastic constants. Consequently, it seems most unlikely that tD/wD would remain constant with increasing linking difference, or that the free energy of the denatured region would vary simply quadratically with 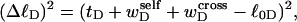 unless
unless 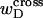 were negligibly small. Indeed, one cannot expect to decompose the supercoiling free energy of the entire molecule into separate local contributions of its unmelted and denatured domains, whenever cross-writhe terms make a significant contribution to either
were negligibly small. Indeed, one cannot expect to decompose the supercoiling free energy of the entire molecule into separate local contributions of its unmelted and denatured domains, whenever cross-writhe terms make a significant contribution to either 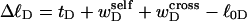 or
or 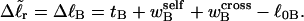 Hence, this interpretation would be sensible only if the identical cross-writhe terms,
Hence, this interpretation would be sensible only if the identical cross-writhe terms, 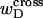 and
and 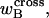 were negligibly small.
were negligibly small.
Thus, under any interpretation of the meaning of the “helicity” ascribed to the denatured region, an additional assumption is required, namely that 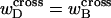 is negligibly small.
is negligibly small.
The assumptions of Benham's model that are tested in this work are summarized in our notation as follows:
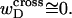
(15) 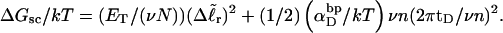
(16) ET is the same for unmelted and partially melted DNAs.
ET is a constant, independent of
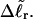
Our simulations of model DNAs containing m = 28 or 56 contiguous melted basepairs indicate that Assumption 1 above is an excellent approximation. However, the assumed quadratic variation of ΔGsc with 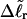 appears to be somewhat inaccurate, even for the unmelted DNA, at the prevailing low (20 mM) ionic strength. For the unmelted DNA, n = 0 and
appears to be somewhat inaccurate, even for the unmelted DNA, at the prevailing low (20 mM) ionic strength. For the unmelted DNA, n = 0 and 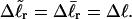 For the same value of
For the same value of 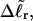 the same value of ET is obtained for all three DNAs (m = 0, 28, 56), so Assumption 3 appears to be surprisingly accurate. Assumption 4 is found to be somewhat inaccurate, again due to the nonquadratic variation of ΔGsc with
the same value of ET is obtained for all three DNAs (m = 0, 28, 56), so Assumption 3 appears to be surprisingly accurate. Assumption 4 is found to be somewhat inaccurate, again due to the nonquadratic variation of ΔGsc with 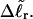
The final term in Eq. 16 reflects the assumption that the intrinsic twist of the melted region vanishes. Although that is indeed true for a real DNA, in our simulations the intrinsic twists of both duplex and melted regions are varied to alter the linking difference, as described below. In that case, the tD in Eq. 16 must be replaced by the net twisting strain of the melted region, 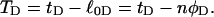 It is implicit in Benham's analysis that
It is implicit in Benham's analysis that 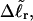 tD, and TD must all be regarded as ensemble average values.
tD, and TD must all be regarded as ensemble average values.
Additional assumptions in the protocol of Bauer and Benham
Bauer and Benham (1993) created a collection of topoisomers of pBR322 containing every linking difference from ∼+10 to −24 at an ionic strength ∼20 mM and investigated these at 40, 45, 50, 55 and 60°C. These topoisomers were analyzed by two-dimensional gels, where the prevailing temperatures for the first dimension were one or another of those just noted, which caused partial melting of the more negatively supercoiled topoisomers. The second dimension was run at 25°C in the presence of sufficient chloroquine to relieve the negative superhelical strain of every topoisomer, so that it is no longer locally melted, regardless of whether or not it was melted in the first dimension.
In addition to the assumptions implicit in their model, Bauer and Benham (1993) invoked an additional crucial assumption to interpret their two-dimensional gels, namely that any two topoisomers, one partially melted and the other unmelted, with the same gel mobility in the first dimension, have the same residual linking difference, 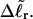 Under this gel mobility assumption, the residual linking difference
Under this gel mobility assumption, the residual linking difference 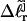 of the jth locally melted topoisomer can be equated to the interpolated
of the jth locally melted topoisomer can be equated to the interpolated 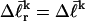 of the (hypothetical) unmelted topoisomer with the same mobility in the first dimension. The
of the (hypothetical) unmelted topoisomer with the same mobility in the first dimension. The 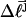 of the unmelted parent of the jth topoisomer, and the
of the unmelted parent of the jth topoisomer, and the 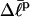 of all the other unmelted topoisomers, are obtained from their positions in the second dimension. Specifically,
of all the other unmelted topoisomers, are obtained from their positions in the second dimension. Specifically, 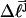 is reckoned by counting the sequence of (unmelted) topoisomers in the second dimension that extends from the topoisomer with the slowest mobility in the first dimension up to that with the mobility of the jth topoisomer in the first dimension. In the computational protocol of Benham and Bauer and Benham, the experimental value of
is reckoned by counting the sequence of (unmelted) topoisomers in the second dimension that extends from the topoisomer with the slowest mobility in the first dimension up to that with the mobility of the jth topoisomer in the first dimension. In the computational protocol of Benham and Bauer and Benham, the experimental value of 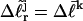 becomes the “target” for a statistical thermodynamic calculation conducted for the jth topoisomer with unmelted linking difference
becomes the “target” for a statistical thermodynamic calculation conducted for the jth topoisomer with unmelted linking difference 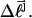 The relative probabilities for various partially melted states are calculated for a nearest-neighbor two-state (i.e., unmelted or melted) melting model that takes account of the supercoiling (deformational) free energy in a parameterized way, based upon Eq. 18 below. The calculation ideally treats all possible numbers,
The relative probabilities for various partially melted states are calculated for a nearest-neighbor two-state (i.e., unmelted or melted) melting model that takes account of the supercoiling (deformational) free energy in a parameterized way, based upon Eq. 18 below. The calculation ideally treats all possible numbers, 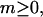 of open basepairs, which are arranged in all compatible numbers of melted domains of various sizes, but takes no direct account of the actual tertiary structures or fluctuations in tertiary structure that determine the thermodynamics of supercoiling and the reduction in supercoiling free energy upon melting. In each iteration, the three adjustable parameters, ET,
of open basepairs, which are arranged in all compatible numbers of melted domains of various sizes, but takes no direct account of the actual tertiary structures or fluctuations in tertiary structure that determine the thermodynamics of supercoiling and the reduction in supercoiling free energy upon melting. In each iteration, the three adjustable parameters, ET, 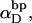 and an opening initiation free energy a, are fixed at appropriate trial values. For any given value of m,
and an opening initiation free energy a, are fixed at appropriate trial values. For any given value of m, 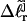 is eliminated in favor of τ and
is eliminated in favor of τ and 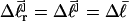 by using Eq. 13 with wD = 0. In the general case, where
by using Eq. 13 with wD = 0. In the general case, where 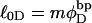 does not vanish, τ is related to the twisting strain of the denatured region,
does not vanish, τ is related to the twisting strain of the denatured region, 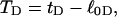 according to
according to 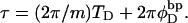 Using this relation in Eq. 13 with wD = 0 gives
Using this relation in Eq. 13 with wD = 0 gives 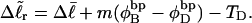 When this expression for
When this expression for 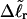 is substituted into Eq. 16, and tD in the final term of that equation is replaced by TD, then TD can also be eliminated in favor of
is substituted into Eq. 16, and tD in the final term of that equation is replaced by TD, then TD can also be eliminated in favor of 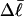 by minimizing ΔGsc with respect to TD, which gives (at the minimum)
by minimizing ΔGsc with respect to TD, which gives (at the minimum)
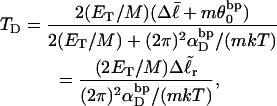 |
(17) |
where 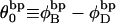 is the difference in the intrinsic succession angle between the duplex and denatured regions (
is the difference in the intrinsic succession angle between the duplex and denatured regions (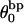 vanishes for our model filaments.)
vanishes for our model filaments.)
Now the supercoiling free energy in Eq. 16 can be written as
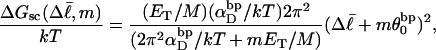 |
(18) |
which depends only on the fixed (known) 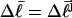 and the specified value of m. The total free energy of each state consists of
and the specified value of m. The total free energy of each state consists of 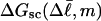 plus a free energy of melting term, which also depends upon m. The average value of
plus a free energy of melting term, which also depends upon m. The average value of 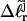 for the set of partially melted topoisomers with the same
for the set of partially melted topoisomers with the same 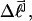 but different values of m, is computed by averaging the quantity
but different values of m, is computed by averaging the quantity
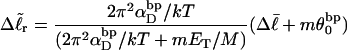 |
(19) |
from Eq. 17 over all of the various partially melted states, each of which is weighted according to the exponential function of its total free energy divided by −kT. In Benham's analysis, 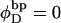 and
and 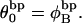 The experimental estimate of the average
The experimental estimate of the average 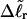 and its corresponding
and its corresponding 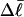 are fitted by Eq. 19, and the disposable parameters, ET,
are fitted by Eq. 19, and the disposable parameters, ET, 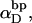 and a are adjusted until the computed average value of
and a are adjusted until the computed average value of 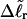 matches the experimental value. By simultaneously fitting the observed
matches the experimental value. By simultaneously fitting the observed 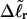 values for several (≥3) partially melted topoisomers at a given temperature, Bauer and Benham obtained a unique optimum fit for all three parameters at each temperature. Clearly, the gel mobility assumption that
values for several (≥3) partially melted topoisomers at a given temperature, Bauer and Benham obtained a unique optimum fit for all three parameters at each temperature. Clearly, the gel mobility assumption that 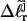 of the jth partially melted topoisomer is identical to the interpolated
of the jth partially melted topoisomer is identical to the interpolated 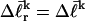 of the (hypothetical) unmelted topoisomer with the same gel mobility provides the crucial experimental estimate of
of the (hypothetical) unmelted topoisomer with the same gel mobility provides the crucial experimental estimate of 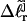 upon which the entire subsequent analysis is based.
upon which the entire subsequent analysis is based.
We wish to test this crucial gel mobility assumption insofar as possible. Unfortunately, a complete test is not possible, because we do not know how to calculate gel mobilities for individual topoisomers. Nevertheless, we can calculate equilibrium tertiary structural properties, such as the principal components of the inertial tensor, the radius of gyration Rg, and writhe wB, of the duplex region, upon which the gel mobility might depend, and with which it might be expected to vary in some smooth and continuous way. We then plot the average value of each structural property versus 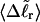 for each of our model DNAs, which contain m = 0, 28, or 56 melted basepairs, and compare the resulting “curves” to see whether they are superimposable. Coalescence of all three curves for each equilibrium tertiary structural property is regarded as a necessary, though not sufficient, condition for validity of the gel mobility assumption of Bauer and Benham. In fact, for several structural properties examined, all three curves for m = 0, 28, and 56 melted basepairs do coincide within the simulation error. Evidently, the global equilibrium tertiary structural properties of our model DNAs with very small denatured regions are determined almost entirely by the average residual linking difference,
for each of our model DNAs, which contain m = 0, 28, or 56 melted basepairs, and compare the resulting “curves” to see whether they are superimposable. Coalescence of all three curves for each equilibrium tertiary structural property is regarded as a necessary, though not sufficient, condition for validity of the gel mobility assumption of Bauer and Benham. In fact, for several structural properties examined, all three curves for m = 0, 28, and 56 melted basepairs do coincide within the simulation error. Evidently, the global equilibrium tertiary structural properties of our model DNAs with very small denatured regions are determined almost entirely by the average residual linking difference, 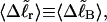 of the duplex region. The gel mobility assumption of Bauer and Benham clearly passes this test. However, it remains doubtful that these conclusions will extend to situations, wherein a much larger fraction of the total sequence is melted.
of the duplex region. The gel mobility assumption of Bauer and Benham clearly passes this test. However, it remains doubtful that these conclusions will extend to situations, wherein a much larger fraction of the total sequence is melted.
Electrophoretic migration of large supercoiled and linear DNAs in dilute agarose gels involves periodic extension of the DNA to highly elongated states far outside the normal equilibrium range, as it becomes temporarily trapped in extended hairpin configurations while sliding over physical constraints within the gel, followed by contraction to more normal dimensions, as it finally slides past those constraints (Smith et al., 1989; Song and Maestre, 1991). Hence, the gel mobility presumably reflects also the deformability of the DNA. In regard to small displacements from the average value of any particular structural property, the corresponding deformability is linearly related to the equilibrium variance of that same property (Kubo, 1957). Hence, we calculate the variance of each tertiary structural property and plot its standard deviation ((variance)1/2) versus 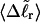 for each of our model DNAs with m = 0, 28, and 56 melted basepairs. Whenever the standard deviation is not negligibly small compared to the mean value, the coincidence of such curves for all three DNAs is regarded as a necessary, though probably still not sufficient, condition for validity of the gel mobility assumption of Bauer and Benham. In our simulations, the standard deviations in every tertiary structural property at the same value of
for each of our model DNAs with m = 0, 28, and 56 melted basepairs. Whenever the standard deviation is not negligibly small compared to the mean value, the coincidence of such curves for all three DNAs is regarded as a necessary, though probably still not sufficient, condition for validity of the gel mobility assumption of Bauer and Benham. In our simulations, the standard deviations in every tertiary structural property at the same value of 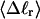 are found to be significantly larger for DNAs with 56 melted basepairs than for those with zero basepairs. In the case of Rg and principal moments of the inertial tensor, the standard deviations are negligibly small compared to the mean values, even for the partially melted DNA, and probably have an insignificant effect on the gel electrophoretic mobility. However, in the case of the writhe, the standard deviation is significant compared to its mean value, so the greater deformability of the writhe in the case of the partially melted DNA may well endow that DNA with a different gel electrophoretic mobility even at the same value of
are found to be significantly larger for DNAs with 56 melted basepairs than for those with zero basepairs. In the case of Rg and principal moments of the inertial tensor, the standard deviations are negligibly small compared to the mean values, even for the partially melted DNA, and probably have an insignificant effect on the gel electrophoretic mobility. However, in the case of the writhe, the standard deviation is significant compared to its mean value, so the greater deformability of the writhe in the case of the partially melted DNA may well endow that DNA with a different gel electrophoretic mobility even at the same value of 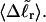 Thus, insofar as deformability of the writhe, or fluctuations therein, contributes significantly to the gel electrophoretic mobility, the gel mobility assumption of Bauer and Benham might be somewhat inaccurate.
Thus, insofar as deformability of the writhe, or fluctuations therein, contributes significantly to the gel electrophoretic mobility, the gel mobility assumption of Bauer and Benham might be somewhat inaccurate.
The analysis of experimental data by Bauer and Benham (1993) relies primarily on Eq. 18 for ΔGsc and Eq. 19 for 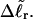 Both of these expressions were derived from Eq. 16 by minimizing ΔGsc with respect to TD (or τ) to eliminate TD in favor of
Both of these expressions were derived from Eq. 16 by minimizing ΔGsc with respect to TD (or τ) to eliminate TD in favor of 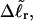 and ultimately to determine how the total linking difference
and ultimately to determine how the total linking difference 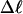 is apportioned between TD (or τ) and
is apportioned between TD (or τ) and 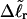 (i.e., between the melted and duplex regions). The specific protocol adopted by Benham to perform the minimization involves yet another assumption, namely that ET is a constant independent of
(i.e., between the melted and duplex regions). The specific protocol adopted by Benham to perform the minimization involves yet another assumption, namely that ET is a constant independent of 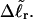 As described below, this assumption is found to be inaccurate at the prevailing relatively low ionic strength.
As described below, this assumption is found to be inaccurate at the prevailing relatively low ionic strength.
The plan of the article
In this work, we simulate supercoiled mesoscopic model DNAs with m = 0, 28, or 56 melted basepairs and selected linking differences over the range from 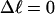 to −26 turns. The supercoiling free energies, ΔGsc, are reckoned as a function of
to −26 turns. The supercoiling free energies, ΔGsc, are reckoned as a function of 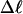 for each molecule with fixed m via a reversible work protocol. In addition, for each molecule at a given
for each molecule with fixed m via a reversible work protocol. In addition, for each molecule at a given 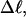 the ensemble average residual linking difference of its duplex region,
the ensemble average residual linking difference of its duplex region, 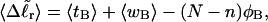 and net twisting strain of its melted region,
and net twisting strain of its melted region, 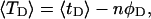 are calculated. Hence, we can directly assess the variation of ΔGsc with
are calculated. Hence, we can directly assess the variation of ΔGsc with 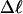 for each molecule, and for each value of
for each molecule, and for each value of 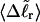 we can determine the effective ET value, since that is the only unknown quantity in Eq. 16, and examine its variation with
we can determine the effective ET value, since that is the only unknown quantity in Eq. 16, and examine its variation with 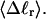 In addition, we can extract ET values from Eq. 19 by using the simulated
In addition, we can extract ET values from Eq. 19 by using the simulated 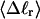 and
and 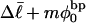 in a manner directly analogous to the way in which Bauer and Benham obtained ET. ET values can also be estimated from
in a manner directly analogous to the way in which Bauer and Benham obtained ET. ET values can also be estimated from 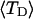 and the corresponding
and the corresponding 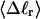 via Eq. 17 or from ΔGsc and the corresponding
via Eq. 17 or from ΔGsc and the corresponding 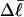 via Eq. 18. ET values obtained by these different methods are critically compared. The ET values determined from Eq. 16 are not necessarily identical to those extracted from Eq. 19, because the minimization protocol used to obtain the latter rests upon an additional assumption, as noted above, namely that ET does not vary with
via Eq. 18. ET values obtained by these different methods are critically compared. The ET values determined from Eq. 16 are not necessarily identical to those extracted from Eq. 19, because the minimization protocol used to obtain the latter rests upon an additional assumption, as noted above, namely that ET does not vary with 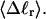 In fact, a nonquadratic variation of ΔGsc with
In fact, a nonquadratic variation of ΔGsc with 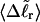 would imply a (possibly modest) variation of ET with
would imply a (possibly modest) variation of ET with 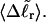 Because ET was assumed to be constant, when performing the minimization with respect to TD (or τ), Eqs. 17–19 will be somewhat inaccurate, whenever ET actually does vary with
Because ET was assumed to be constant, when performing the minimization with respect to TD (or τ), Eqs. 17–19 will be somewhat inaccurate, whenever ET actually does vary with 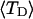 (or with
(or with 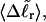 or
or 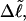 or
or 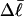 ). Fortunately, the errors in the extracted ET values that are associated with this minimization protocol are found to be relatively small (5–7%).
). Fortunately, the errors in the extracted ET values that are associated with this minimization protocol are found to be relatively small (5–7%).
THE MODEL
Our mesocopic model of a circular DNA comprises N = 155 rigid-rod subunits, each connected to its neighbors at either end by Hookean torsion and bending springs. The subunit length, b = 95.4 Å, corresponds to ν = 28.06 bp, and the overall chain length corresponds to 4349 bp, which is similar in size to the pBR322 plasmid DNA (4363 bp) investigated by Bauer and Benham (1993). The total potential energy is given by
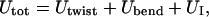 |
(20) |
where Utwist is torsion potential energy, Ubend is the bending potential energy, and UI is the repulsive potential energy of interaction between nonnearest-neighbor subunits. Expressions for Utwist, Ubend, and UI, and the selections of input parameters therein, are presented and discussed in the Appendix.
In this work, we simulate model DNAs containing small torsionally and flexurally weak regions. This is done by designating n = 0, 1, or 2 consecutive springs as weak. We take the (155,1) spring for n =1, and the (154,155) and (155,1) springs for n = 2 as weak springs, which are assigned reduced torque constants for torsion and bending. For melted regions comprising only one or two rigid-rod subunits, 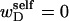 (see Appendix), so
(see Appendix), so 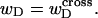 With a subunit length of 95.4 Å, the n = 0, 1, and 2 weak springs correspond to denatured regions with m = 0, 28.06, and 56.12 bp. For convenience, these m values are rounded to the nearest integer in the text, though not in the data analysis. The torsion elastic constant for a duplex subunit corresponds to a torsional rigidity, C = 2.0 × 10−19 dyne cm2, and its bending elastic constant is chosen to yield a persistence length, P = 500 Å, after taking account of the intersubunit repulsions. The torsion elastic constant of a melted subunit was determined by extrapolating the corresponding results of Bauer and Benham to 37°C, and is 54.6-fold smaller than that for the duplex region. The bending elastic constant of a melted subunit was taken to be 10-fold smaller than for a duplex subunit, and yields a 2.9-fold smaller persistence length after taking account of the intersubunit repulsions. The intersubunit potential consists of a screened Coulomb interaction plus a hard-cylinder interaction with a 24 Å diameter. The locations and magnitudes of the effective charges appropriate for the prevailing ionic strength are described in the Appendix. The simulation temperature is 298 K.
With a subunit length of 95.4 Å, the n = 0, 1, and 2 weak springs correspond to denatured regions with m = 0, 28.06, and 56.12 bp. For convenience, these m values are rounded to the nearest integer in the text, though not in the data analysis. The torsion elastic constant for a duplex subunit corresponds to a torsional rigidity, C = 2.0 × 10−19 dyne cm2, and its bending elastic constant is chosen to yield a persistence length, P = 500 Å, after taking account of the intersubunit repulsions. The torsion elastic constant of a melted subunit was determined by extrapolating the corresponding results of Bauer and Benham to 37°C, and is 54.6-fold smaller than that for the duplex region. The bending elastic constant of a melted subunit was taken to be 10-fold smaller than for a duplex subunit, and yields a 2.9-fold smaller persistence length after taking account of the intersubunit repulsions. The intersubunit potential consists of a screened Coulomb interaction plus a hard-cylinder interaction with a 24 Å diameter. The locations and magnitudes of the effective charges appropriate for the prevailing ionic strength are described in the Appendix. The simulation temperature is 298 K.
SIMULATION PROTOCOLS
Our simulation protocols, relevant computational procedures, and statistical methods are described in the Appendix. Average values of the various quantities are plotted versus 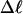 or
or 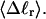 Typically, either straight lines or quadratic polynomials are fitted to the data to guide the eye.
Typically, either straight lines or quadratic polynomials are fitted to the data to guide the eye.
RESULTS AND DISCUSSION
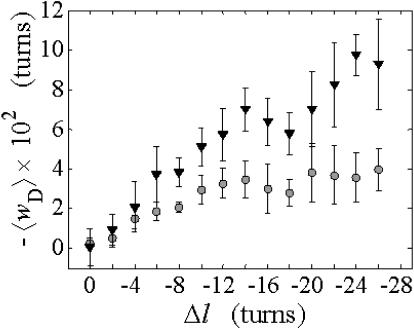
The calculated 〈wD〉 are negligibly small for n = 1, 2
The ensemble average value 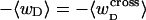 is plotted versus
is plotted versus 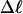 for n = 1 and 2 in Fig. 1. Although
for n = 1 and 2 in Fig. 1. Although 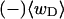 increases overall between
increases overall between 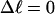 and
and 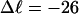 turns, it never exceeds 0.05 turns for n = 1 or 0.10 turns for n = 2, even though the total writhe
turns, it never exceeds 0.05 turns for n = 1 or 0.10 turns for n = 2, even though the total writhe 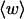 rises from −0 to −13 turns over the same range of
rises from −0 to −13 turns over the same range of 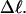 The average fraction,
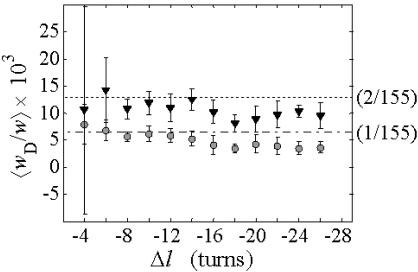
The average fraction, 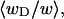 of the total writhe contained in wD is plotted versus
of the total writhe contained in wD is plotted versus 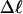 in Fig. 2. Clearly,
in Fig. 2. Clearly, 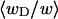 is typically less than or equal to the fraction of bond vectors, 1/155 or 2/155, that are contained in the weak region. Consequently,
is typically less than or equal to the fraction of bond vectors, 1/155 or 2/155, that are contained in the weak region. Consequently, 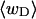 can be regarded as negligibly small compared to
can be regarded as negligibly small compared to 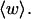 Thus, the implicit assumption of Benham that
Thus, the implicit assumption of Benham that 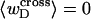 is a rather good approximation, and our Eq. 13 becomes practically identical to the relation assumed by Benham and Bauer and Benham (our Eq. 8). Because the fraction of cross-writhe in the melted region evidently increases with size of that region, this approximation very likely fails for much larger melted regions.
is a rather good approximation, and our Eq. 13 becomes practically identical to the relation assumed by Benham and Bauer and Benham (our Eq. 8). Because the fraction of cross-writhe in the melted region evidently increases with size of that region, this approximation very likely fails for much larger melted regions.
FIGURE 1.
Negative writhe of the melted region, 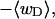 versus linking difference of the entire molecule,
versus linking difference of the entire molecule, 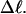 Simulated molecules have either n = 1 (shaded circles) or n = 2 (▾) contiguous melted subunits out of 155 total subunits. In both cases,
Simulated molecules have either n = 1 (shaded circles) or n = 2 (▾) contiguous melted subunits out of 155 total subunits. In both cases, 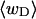 is purely cross-writhe between the melted and duplex regions.
is purely cross-writhe between the melted and duplex regions.
FIGURE 2.
Ratio 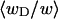 of the writhe of the melted region, wD, to the total writhe of the entire molecule,
of the writhe of the melted region, wD, to the total writhe of the entire molecule, 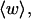 versus linking difference of the entire molecule,
versus linking difference of the entire molecule, 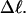 Simulated molecules have either n = 1 (shaded circles) or n = 2 (▾) contiguous melted subunits out of 155 total subunits. The dashed lined has the value 1/155, and the dotted line has the value 2/155.
Simulated molecules have either n = 1 (shaded circles) or n = 2 (▾) contiguous melted subunits out of 155 total subunits. The dashed lined has the value 1/155, and the dotted line has the value 2/155.
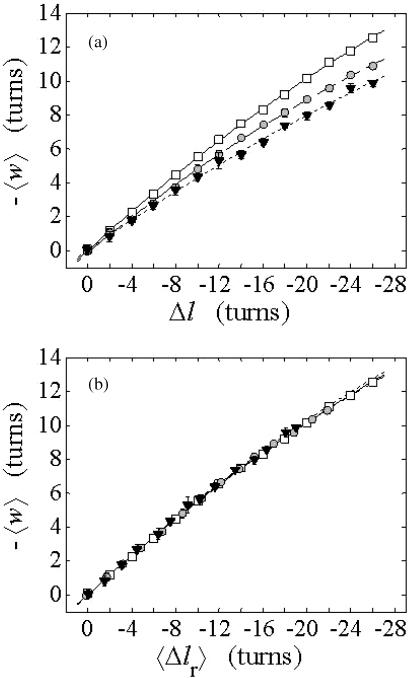
The mean tertiary structural properties are nearly universal functions of 〈Δℓr〉
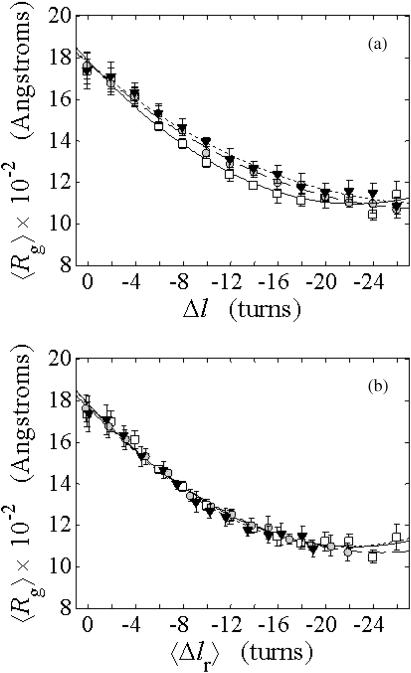
The total mean writhe 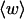 for n = 0, 1, and 2 is plotted versus
for n = 0, 1, and 2 is plotted versus 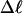 in Fig. 3 a and versus
in Fig. 3 a and versus 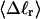 in Fig. 3 b. The three separate curves in Fig. 3 a have coalesced to a single curve in Fig. 3 b, which indicates that
in Fig. 3 b. The three separate curves in Fig. 3 a have coalesced to a single curve in Fig. 3 b, which indicates that 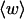 is a nearly universal function of
is a nearly universal function of 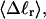 independent of n. The evident curvature shows that this universal relation is not strictly a proportionality.
independent of n. The evident curvature shows that this universal relation is not strictly a proportionality.
FIGURE 3.
(a) Negative total writhe, 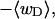 versus linking difference of the entire molecule,
versus linking difference of the entire molecule, 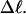 (b)
(b) 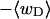 versus residual linking difference associated with the duplex region,
versus residual linking difference associated with the duplex region, 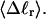 Simulated molecules have n = 0 (□), n = 1 (shaded circles), or n = 2 (▾) melted subunits out of 155 total subunits.
Simulated molecules have n = 0 (□), n = 1 (shaded circles), or n = 2 (▾) melted subunits out of 155 total subunits. 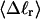 is calculated according to the first line of Eq. 11.
is calculated according to the first line of Eq. 11.
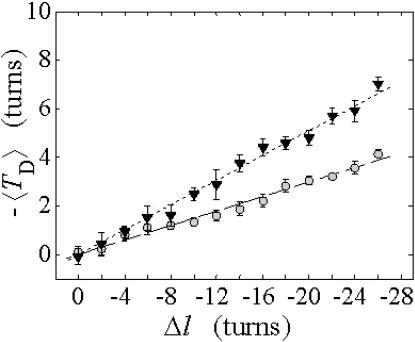
The net twisting strain in the melted region, 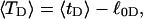 is plotted versus
is plotted versus 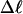 for n = 1 and 2 in Fig. 4. These two data sets do not coalesce even when plotted versus
for n = 1 and 2 in Fig. 4. These two data sets do not coalesce even when plotted versus 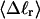 (not shown), so
(not shown), so 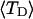 is not a universal function of
is not a universal function of 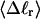 independent of n. Of course,
independent of n. Of course, 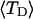 is also not a tertiary structural property. Within simulation error,
is also not a tertiary structural property. Within simulation error, 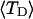 is proportional to both
is proportional to both 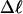 and
and 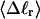 (not shown) in agreement with Eq. 17, when
(not shown) in agreement with Eq. 17, when 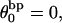 as it does here. This simultaneous proportionality of
as it does here. This simultaneous proportionality of 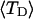 to both
to both 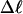 and
and 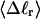 implies that
implies that 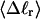 is also proportional to
is also proportional to 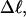 in agreement with Eq. 19 (when
in agreement with Eq. 19 (when 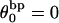 ). Because
). Because 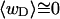 for n = 1 and 2, Eq. 7 gives
for n = 1 and 2, Eq. 7 gives 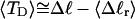 for our model DNAs.
for our model DNAs.
FIGURE 4.
Negative twisting strain per subunit of the melted region, 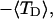 versus linking difference,
versus linking difference, 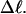 Simulated molecules have either n = 1 (shaded circles) or n = 2 (▾) melted subunits out of 155 total. The dashed and dotted lines are the respective best-fit proportionality relations.
Simulated molecules have either n = 1 (shaded circles) or n = 2 (▾) melted subunits out of 155 total. The dashed and dotted lines are the respective best-fit proportionality relations.
The fraction of the total linking difference that resides in the melted region is approximately 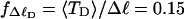 for m = 1 and 0.26 for m = 2. These values apply over the full range of
for m = 1 and 0.26 for m = 2. These values apply over the full range of 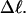 When an unmelted parent DNA with σ = −0.05 and
When an unmelted parent DNA with σ = −0.05 and 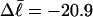 turns melts 56 bp, then its linking difference drops to
turns melts 56 bp, then its linking difference drops to 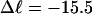 turns, of which −4.0 are contained in the melted region. The −4.0 turns are −0.75 times the intrinsic twist of that same region, when it is present as duplex. Thus, very substantial negative interwinding of the single strands of the melted region is expected, whenever the molecule is at least moderately supercoiled, provided Benham's torsion elastic constant for the melted region applies, as assumed here. The fraction of the total twisting strain in the denatured region is
turns, of which −4.0 are contained in the melted region. The −4.0 turns are −0.75 times the intrinsic twist of that same region, when it is present as duplex. Thus, very substantial negative interwinding of the single strands of the melted region is expected, whenever the molecule is at least moderately supercoiled, provided Benham's torsion elastic constant for the melted region applies, as assumed here. The fraction of the total twisting strain in the denatured region is 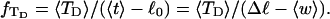 Because w is not strictly proportional to
Because w is not strictly proportional to 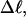
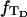 is not independent of
is not independent of 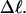 However, for
However, for 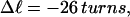
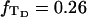 for n = 1 and 0.42 for n = 2.
for n = 1 and 0.42 for n = 2.
The mean radius of gyration, 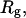 for n = 0, 1, and 2, is plotted versus
for n = 0, 1, and 2, is plotted versus 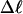 in Fig. 5 a and versus
in Fig. 5 a and versus 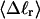 in Fig. 5 b. Again, the three curves in Fig. 5 a have largely coalesced in Fig. 5 b. The ensemble average values of
in Fig. 5 b. Again, the three curves in Fig. 5 a have largely coalesced in Fig. 5 b. The ensemble average values of 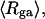
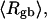 and
and 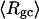 for n = 0, 1, and 2 also do not coalesce when plotted versus
for n = 0, 1, and 2 also do not coalesce when plotted versus 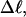 but are significantly more coalesced, when plotted versus
but are significantly more coalesced, when plotted versus 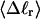 (not shown) (Sucato, 2001). This coalescence is most pronounced for the largest element,
(not shown) (Sucato, 2001). This coalescence is most pronounced for the largest element, 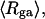 which is most sensitive to n at fixed
which is most sensitive to n at fixed 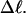 The values of the smaller elements,
The values of the smaller elements, 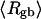 and
and 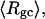 are considerably less sensitive to n at fixed
are considerably less sensitive to n at fixed 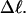
FIGURE 5.
(a) Radius of gyration, Rg, versus linking difference of the entire molecule, 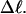 (b) Rg versus residual linking difference associated with the duplex region,
(b) Rg versus residual linking difference associated with the duplex region, 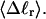 Simulated molecules have n = 0 (□), 1 (shaded circles), or 2 (▾) melted subunits out of 155 total subunits.
Simulated molecules have n = 0 (□), 1 (shaded circles), or 2 (▾) melted subunits out of 155 total subunits. 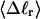 is calculated according to the first line of Eq. 11.
is calculated according to the first line of Eq. 11.
These results indicate that 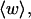
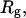 and
and 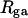 depend primarily upon the residual linking difference of the duplex region,
depend primarily upon the residual linking difference of the duplex region, 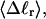 independent of the size of the melted region. This conclusion most likely does not extend to DNAs with very much larger melted regions, for reasons noted above.
independent of the size of the melted region. This conclusion most likely does not extend to DNAs with very much larger melted regions, for reasons noted above.
The fluctuations in structural properties are not universal functions of 〈Δℓr〉
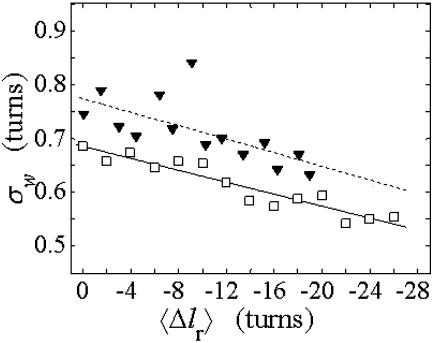
The standard deviation of the total writhe, σw, is plotted versus 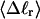 in Fig. 6 for n = 0 and 2. σw clearly depends upon n even at the same
in Fig. 6 for n = 0 and 2. σw clearly depends upon n even at the same 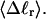 Specifically, at the same
Specifically, at the same 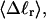 σw increases with the size of the melted region. In addition, σw declines with increasing
σw increases with the size of the melted region. In addition, σw declines with increasing 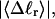 which presumably reflects the diminished deformability of the more tightly writhed structures prevailing at larger
which presumably reflects the diminished deformability of the more tightly writhed structures prevailing at larger 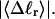 For the molecule with n = 2 melted subunits, σw exceeds 10% of
For the molecule with n = 2 melted subunits, σw exceeds 10% of 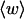 at all
at all 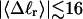 turns. These fluctuations in the writhe imply an enhanced deformability of the writhe that increases with size of the melted region at constant
turns. These fluctuations in the writhe imply an enhanced deformability of the writhe that increases with size of the melted region at constant 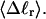 To the extent that such writhe deformability affects the gel mobility, the latter will not be a universal function of
To the extent that such writhe deformability affects the gel mobility, the latter will not be a universal function of 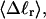 independent of n. This finding leaves open the possibility that the gel mobility assumption of Bauer and Benham is not entirely correct, despite the fact that the average structural properties of the unperturbed DNAs are universal functions of
independent of n. This finding leaves open the possibility that the gel mobility assumption of Bauer and Benham is not entirely correct, despite the fact that the average structural properties of the unperturbed DNAs are universal functions of 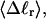 independent of n.
independent of n.
FIGURE 6.
Standard deviation of the writhe, σw, versus linking difference associated with the duplex region, 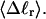 Simulated molecules have n = 0 or 2 melted subunits out of 155 total subunits.
Simulated molecules have n = 0 or 2 melted subunits out of 155 total subunits.
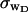 for the denatured region increases with increasing
for the denatured region increases with increasing 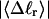 (data not shown). This trend is in the opposite direction to that for σw. Because
(data not shown). This trend is in the opposite direction to that for σw. Because 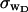 pertains only to the cross-writhe,
pertains only to the cross-writhe, 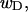 which is the same as the cross-writhe in wB, we conclude that fluctuations in cross-writhe increase with increasing
which is the same as the cross-writhe in wB, we conclude that fluctuations in cross-writhe increase with increasing 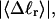 whereas fluctuations in self-writhe of the B-helix region decrease with increasing
whereas fluctuations in self-writhe of the B-helix region decrease with increasing 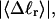 as expected. The increase in cross-writhe fluctuations with increasing
as expected. The increase in cross-writhe fluctuations with increasing 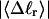 may simply be a consequence of compaction of the dimensions of the B-helical region, which may allow greater fluctuations in cross-writhe for the same range of variations in local structure. In any case,
may simply be a consequence of compaction of the dimensions of the B-helical region, which may allow greater fluctuations in cross-writhe for the same range of variations in local structure. In any case, 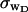 is much smaller than σw, and is insignificant compared to
is much smaller than σw, and is insignificant compared to 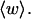
It was similarly found that 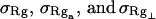 vary significantly with n at constant
vary significantly with n at constant 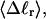 but these standard deviations are negligibly small compared to the corresponding average values (data not shown). Hence, their effect on the gel mobility is probably negligible.
but these standard deviations are negligibly small compared to the corresponding average values (data not shown). Hence, their effect on the gel mobility is probably negligible.
Results pertaining to the supercoiling free energy
When a real unmelted supercoiled DNA with linking difference 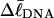 undergoes melting of m basepairs, its linking difference becomes
undergoes melting of m basepairs, its linking difference becomes 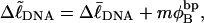 and the supercoiling free energy declines due to both the decline in magnitude of the (negative) linking difference and the presence of elastically weak regions. However, for our model molecules,
and the supercoiling free energy declines due to both the decline in magnitude of the (negative) linking difference and the presence of elastically weak regions. However, for our model molecules, 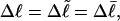 because in our simulations we have for convenience taken
because in our simulations we have for convenience taken 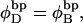 as noted above. Nevertheless, for each of our simulated molecules, we can reckon the linking difference
as noted above. Nevertheless, for each of our simulated molecules, we can reckon the linking difference 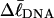 of that real unmelted DNA, whose linking difference after melting m basepairs,
of that real unmelted DNA, whose linking difference after melting m basepairs, 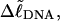 is precisely equal to the linking difference of our simulated molecule,
is precisely equal to the linking difference of our simulated molecule, 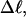 by setting
by setting 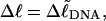 which gives
which gives 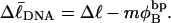
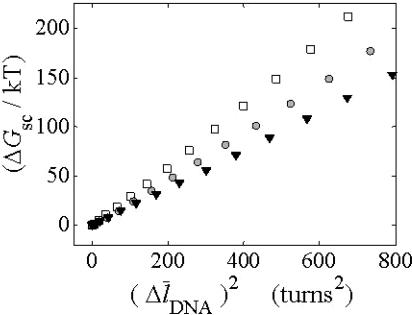
Computed values of ΔGsc/kT are plotted versus 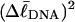 in Fig. 7. The drop from the top curve (n = 0) to the middle (n = 1) or lower (n = 2) curve at fixed
in Fig. 7. The drop from the top curve (n = 0) to the middle (n = 1) or lower (n = 2) curve at fixed 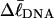 gives the decrease in supercoiling free energy,
gives the decrease in supercoiling free energy, 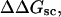 upon opening m = 28 or 56 basepairs, respectively. The evident curvature indicates that ΔGsc is not a quadratic function of
upon opening m = 28 or 56 basepairs, respectively. The evident curvature indicates that ΔGsc is not a quadratic function of 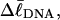 even for n = 0 (m = 0), where
even for n = 0 (m = 0), where 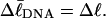 This curvature far exceeds that (negligibly small curvature) predicted and observed for 0.1 M ionic strength (Gebe et al., 1995), and is attributed to the greater magnitude and range of the electrostatic repulsions prevailing at this lower ionic strength.
This curvature far exceeds that (negligibly small curvature) predicted and observed for 0.1 M ionic strength (Gebe et al., 1995), and is attributed to the greater magnitude and range of the electrostatic repulsions prevailing at this lower ionic strength. 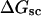 rises more rapidly than the second power of
rises more rapidly than the second power of 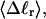 because at 20 mM ionic strength the intersubunit electrostatic repulsions not only contribute significantly to the supercoiling free energy, but also contribute a larger fraction of the total at the larger
because at 20 mM ionic strength the intersubunit electrostatic repulsions not only contribute significantly to the supercoiling free energy, but also contribute a larger fraction of the total at the larger 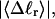 where the DNA is more compact. When plotted versus
where the DNA is more compact. When plotted versus 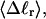 the ΔGsc/kT values do not coalesce to a single curve (not shown). Hence, ΔGsc/kT is not a universal function of
the ΔGsc/kT values do not coalesce to a single curve (not shown). Hence, ΔGsc/kT is not a universal function of 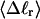 independent of n, nor was it expected to be so.
independent of n, nor was it expected to be so.
FIGURE 7.
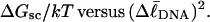
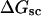 is the supercoiling free energy of a simulated molecule with linking difference,
is the supercoiling free energy of a simulated molecule with linking difference, 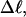 and
and 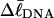 is the linking difference of the unmelted real DNA that would have the linking difference of the simulated molecule after melting m basepairs
is the linking difference of the unmelted real DNA that would have the linking difference of the simulated molecule after melting m basepairs 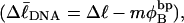 and
and 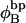 is the intrinsic twist per basepair of the duplex region. Simulated molecules contain n = 0 (□), n = 1 (shaded circles), or n = 2 (▾) melted subunits out of 155 total melted subunits.
is the intrinsic twist per basepair of the duplex region. Simulated molecules contain n = 0 (□), n = 1 (shaded circles), or n = 2 (▾) melted subunits out of 155 total melted subunits.
Values of ET that are determined directly from the simulated ΔGsc, 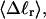 and
and 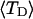 via Eq. 16 (with
via Eq. 16 (with 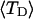 in place of
in place of 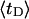 ) are denoted by
) are denoted by 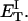 In Fig. 8,
In Fig. 8, 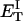 is plotted versus
is plotted versus 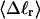 for m = 0, 28, and 56. Although the
for m = 0, 28, and 56. Although the 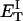 values are very noisy at small
values are very noisy at small 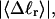 they nearly coalesce at large
they nearly coalesce at large 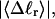 where the simulated
where the simulated 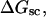
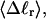 and
and 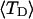 are better converged. This coalescence confirms that ET is independent of n, as assumed by Benham (1990, 1992). However, the upward slope of
are better converged. This coalescence confirms that ET is independent of n, as assumed by Benham (1990, 1992). However, the upward slope of 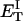 with
with 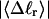 implies a nonquadratic variation of ΔGsc/kT with
implies a nonquadratic variation of ΔGsc/kT with 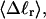 which was not anticipated by Benham (1990, 1992) or Bauer and Benham (1993).
which was not anticipated by Benham (1990, 1992) or Bauer and Benham (1993).
FIGURE 8.
Twist energy parameter, 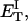 versus residual linking difference of the duplex region,
versus residual linking difference of the duplex region, 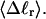
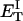 is determined directly from the simulated
is determined directly from the simulated 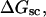
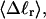 and
and 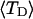 via Eq. 16 with
via Eq. 16 with 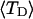 in place of
in place of 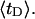 Simulated molecules have n = 0 (□), n = 1 (shaded circles), or n = 2 (▾) melted subunits out of 155 total subunits.
Simulated molecules have n = 0 (□), n = 1 (shaded circles), or n = 2 (▾) melted subunits out of 155 total subunits.
ET values can also be determined from Eq. 19 by using the values of 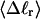 and
and 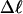 from the simulations in each case. This corresponds most closely to the protocol of Bauer and Benham (1993). These values are denoted by
from the simulations in each case. This corresponds most closely to the protocol of Bauer and Benham (1993). These values are denoted by 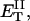 and are coplotted with the
and are coplotted with the 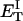 values versus
values versus 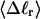 in Fig. 9. These
in Fig. 9. These 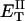 values are much noisier than the
values are much noisier than the 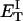 values. This is due to the fact that modest relative errors in
values. This is due to the fact that modest relative errors in 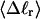 produce five- to sixfold larger relative errors in ET via Eq. 19 and in
produce five- to sixfold larger relative errors in ET via Eq. 19 and in 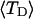 via Eq. 17. For the larger
via Eq. 17. For the larger 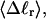 where the relative errors in
where the relative errors in 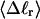 are smallest, the
are smallest, the 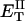 values systematically exceed the corresponding
values systematically exceed the corresponding 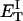 values. This systematic overestimation of
values. This systematic overestimation of 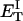 by
by 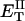 at the larger
at the larger 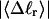 arises because the variation of ET with
arises because the variation of ET with 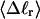 (or
(or 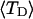 ) was neglected, when taking the derivative of ΔGsc/kT with respect to
) was neglected, when taking the derivative of ΔGsc/kT with respect to 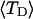 in the minimization step. When the simulated slope,
in the minimization step. When the simulated slope, 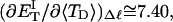 is actually taken into account in the minimization step for n = 1, the corrected
is actually taken into account in the minimization step for n = 1, the corrected 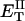 values lie 5–7% lower than those obtained directly from Eq. 19, and are in reasonable, if noisy, accord with the
values lie 5–7% lower than those obtained directly from Eq. 19, and are in reasonable, if noisy, accord with the 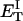 values in Figs. 8 and 9. These observations indicate that the ET values obtained by Bauer and Benham from Eq. 19 are likely to be systematically several percent too large, and to be quite sensitive to errors in the experimental estimates of
values in Figs. 8 and 9. These observations indicate that the ET values obtained by Bauer and Benham from Eq. 19 are likely to be systematically several percent too large, and to be quite sensitive to errors in the experimental estimates of 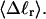
FIGURE 9.
Twist energy parameter, ET, versus residual linking difference of the duplex region. The solid or shaded symbols apply to 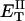 values that are determined from the
values that are determined from the 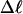 and
and 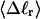 of the simulated molecules according to Eq. 19. These
of the simulated molecules according to Eq. 19. These 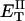 values apply for n = 1 (shaded circles) or n = 2 (▾) melted subunits. The open symbols represent the same
values apply for n = 1 (shaded circles) or n = 2 (▾) melted subunits. The open symbols represent the same 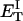 values that appear in Fig. 8 for n = 0 (□), n = 1 (○), or n = 2 (▿) melted subunits out of 155 total subunits.
values that appear in Fig. 8 for n = 0 (□), n = 1 (○), or n = 2 (▿) melted subunits out of 155 total subunits.
ET values obtained from the simulated 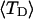 and
and 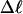 via Eq. 17 are similar to, and behave like, those from Eq. 19. This is expected, because Eqs. 17 and 19 are simply two different realizations of the same equation. In contrast, ET values reckoned from Eq. 18 (with the simulated ΔGsc/kT and
via Eq. 17 are similar to, and behave like, those from Eq. 19. This is expected, because Eqs. 17 and 19 are simply two different realizations of the same equation. In contrast, ET values reckoned from Eq. 18 (with the simulated ΔGsc/kT and 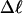 ) lie much closer to the corresponding
) lie much closer to the corresponding 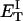 values. It is unfortunate that the particular expression (Eq. 19) required to analyze the
values. It is unfortunate that the particular expression (Eq. 19) required to analyze the 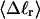 versus
versus 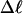 data of Bauer and Benham manifests to a much greater extent the effects of the unexpected slope,
data of Bauer and Benham manifests to a much greater extent the effects of the unexpected slope, 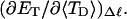
A systematic error in the apparent slope, −dET/dT, obtained by the protocol of Bauer and Benham
With increasing temperature, the spans of the experimental 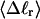 versus
versus  data that are fitted by Bauer and Benham (1993) are centered about progressively smaller
data that are fitted by Bauer and Benham (1993) are centered about progressively smaller  values. This is because topoisomers with lower
values. This is because topoisomers with lower 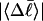 undergo melting at higher temperature. If
undergo melting at higher temperature. If 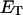 decreases with decreasing
decreases with decreasing 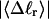 at fixed temperature, as predicted by the present simulations, then the decrease in typical
at fixed temperature, as predicted by the present simulations, then the decrease in typical 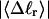 values that are sampled with increasing T should act to enhance the slope,
values that are sampled with increasing T should act to enhance the slope, 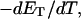 above the intrinsic value that would apply, if
above the intrinsic value that would apply, if 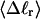 were held constant. The magnitude of this enhancement can be estimated in the following way. First, we note that the simulated
were held constant. The magnitude of this enhancement can be estimated in the following way. First, we note that the simulated 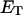 values in Fig. 8 for
values in Fig. 8 for 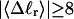 can be satisfactorily approximated by the empirical linear relation,
can be satisfactorily approximated by the empirical linear relation, 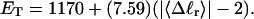 The
The 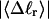 values at each temperature in Fig. 4 of Bauer and Benham can be characterized by the minimum value,
values at each temperature in Fig. 4 of Bauer and Benham can be characterized by the minimum value, 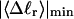 in each case. These
in each case. These 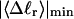 values range from 13.25 turns at 313 K to 4.5 turns at 333 K. We now assume that the simulated slope,
values range from 13.25 turns at 313 K to 4.5 turns at 333 K. We now assume that the simulated slope, 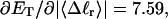 is independent of T. The simulated
is independent of T. The simulated 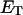 value at 298 K for
value at 298 K for 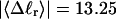 is
is 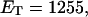 and that for
and that for 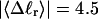 is
is 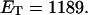 The shift in
The shift in 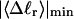 from 13.25 to 4.5, as T increases from 313 to 333 K, then contributes to the apparent slope,
from 13.25 to 4.5, as T increases from 313 to 333 K, then contributes to the apparent slope, 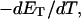 a term −(1189 − 1255)/20 = 3.3 K−1. This value should be subtracted from the negative slope of the ET values reported by Bauer and Benham, which are given in Table 1. A least-squares fit of a straight line
a term −(1189 − 1255)/20 = 3.3 K−1. This value should be subtracted from the negative slope of the ET values reported by Bauer and Benham, which are given in Table 1. A least-squares fit of a straight line 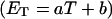 to those data yields the experimental slope, −a = −dET/dT = 9.36 K−1. After subtracting the preceding correction, 3.3 K−1, we obtain
to those data yields the experimental slope, −a = −dET/dT = 9.36 K−1. After subtracting the preceding correction, 3.3 K−1, we obtain 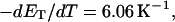 which should apply to any fixed
which should apply to any fixed 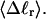
TABLE 1.
Comparison of theoretical and experimental ET values
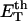 is calculated for the appropriate
is calculated for the appropriate 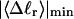 according to Eq. 21.
according to Eq. 21.
Reported as q by Bauer and Benham.
Statistical errors in ET and −dET/dT
Bauer and Benham (1993) reported no standard deviations for ET. Determinations of ET by the topoisomer distribution method require typically 10 replicate measurements of ET to reduce the standard deviation of the mean to the 3% level, or ±30, when ET is ∼1000. This 3% uncertainty is comparable to the spacing between reported ET values at the different temperatures in the study of Bauer and Benham. In addition, problems with reproducibility of ET measurements and slow temporal shifts in behavior of pBR322 have been noted previously (Song et al., 1990; Naimushin et al., 1994). Although the protocol of Bauer and Benham is a very different approach to ET measurements that is possibly less prone to statistical and reproducibility errors, in the absence of any evidence to support such a conjecture, that must be regarded as unlikely. Indeed, in a subsequent application of the same protocol to pSM1 DNA, the scatter in the plot of ET versus 1/T is ∼±30 around the best-fit curve (Bauer et al., 1995). If we assume a standard deviation of ±30 in the ET measurements, then a routine error propagation analysis yields a standard deviation, 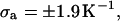 for the best-fit slope, a = −9.36 K−1, of the experimental data. The above corrected experimental slope for fixed
for the best-fit slope, a = −9.36 K−1, of the experimental data. The above corrected experimental slope for fixed 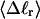 can now be restated as
can now be restated as 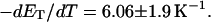 The 31% relative uncertainty in the corrected slope is considerably greater than that (∼8%) reported for the uncorrected slope by Bauer and Benham, who evidently assumed much smaller relative errors,
The 31% relative uncertainty in the corrected slope is considerably greater than that (∼8%) reported for the uncorrected slope by Bauer and Benham, who evidently assumed much smaller relative errors, 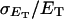 <1%, in their measurements. We suspect that the fourfold larger relative errors in both ET and
<1%, in their measurements. We suspect that the fourfold larger relative errors in both ET and 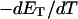 found for pSM1 are more realistic.
found for pSM1 are more realistic.
The predicted variation of ET with 〈Δℓr〉 and T
The present simulations for n = 0 were performed using a torsion elastic constant, 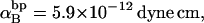 between basepairs, a persistence length, P = 500 Å, and repulsive interactions for 20 mM ionic strength at 298 K. The line defined by the linear parts of the coalesced ET versus
between basepairs, a persistence length, P = 500 Å, and repulsive interactions for 20 mM ionic strength at 298 K. The line defined by the linear parts of the coalesced ET versus 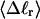 curves in Fig. 8 is empirically given by
curves in Fig. 8 is empirically given by 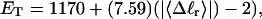 as noted above, and takes the value 1170 at
as noted above, and takes the value 1170 at 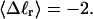
Previously, ET values at various temperatures were simulated for a model unmelted DNA in a 55 mM ionic strength buffer containing 5 mM Mg2+ by using the measured temperature dependent torsion elastic constants of an 1876 bp pBR322 fragment under those same conditions (Delrow et al., 1997b). In addition, the persistence length was taken as 500 Å, and the intersubunit repulsions were appropriate for the 55 mM ionic strength buffer containing 5 mM Mg2+. Simulations were performed only for 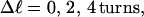 so any variation of
so any variation of  with
with 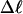 was not discernible. Although the presence of the Mg2+ significantly raised the torsion elastic constant to
was not discernible. Although the presence of the Mg2+ significantly raised the torsion elastic constant to 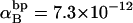 dyne cm at 293 K, it also significantly decreased the repulsive interactions in comparison to those in the present simulation. The best-fit line of the simulated ET versus T, namely
dyne cm at 293 K, it also significantly decreased the repulsive interactions in comparison to those in the present simulation. The best-fit line of the simulated ET versus T, namely 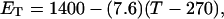 takes the value ET = 1187 at 298 K, which is close to the corresponding value, ET = 1170, obtained from the empirical line of ET versus
takes the value ET = 1187 at 298 K, which is close to the corresponding value, ET = 1170, obtained from the empirical line of ET versus 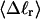 for
for 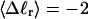 at 298 K in our simulations. Evidently, for small
at 298 K in our simulations. Evidently, for small 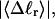 the differences in torsion elastic constant and intersubunit repulsions between the two simulations have almost exactly canceling effects on ET. The slope of the torsion elastic constant,
the differences in torsion elastic constant and intersubunit repulsions between the two simulations have almost exactly canceling effects on ET. The slope of the torsion elastic constant, 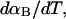 was previously found to be insensitive to either removal of the 5.5 mM Mg2+ or an increase in ionic strength. We now assume that the temperature dependence of ET at small fixed
was previously found to be insensitive to either removal of the 5.5 mM Mg2+ or an increase in ionic strength. We now assume that the temperature dependence of ET at small fixed 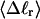 under conditions of these simulations would be practically the same as found in the previous simulations. Then the combined variation of the simulated ET with both T and
under conditions of these simulations would be practically the same as found in the previous simulations. Then the combined variation of the simulated ET with both T and 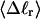 in 20 mM ionic strength should be closely approximated by the relation,
in 20 mM ionic strength should be closely approximated by the relation,
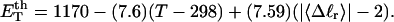 |
(21) |
The theoretical slope, 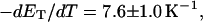 agrees with the above corrected experimental slope,
agrees with the above corrected experimental slope, 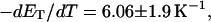 within their combined uncertainties.
within their combined uncertainties.
Equation (21) enables theoretical predictions of the absolute magnitude of ET for arbitrary T and 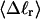 for pBR322 in 20 mM ionic strength.
for pBR322 in 20 mM ionic strength.
A test of the theory versus experiment
We assume that the minimum value, 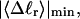 of the fitted data at each temperature in Fig. 4 of Bauer and Benham carries the most weight in their fitting procedure, and is maximally responsible for determining the best-fit value of ET. If we further suppose that their protocol yields reliable values of both
of the fitted data at each temperature in Fig. 4 of Bauer and Benham carries the most weight in their fitting procedure, and is maximally responsible for determining the best-fit value of ET. If we further suppose that their protocol yields reliable values of both 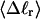 and ET, then the experimental ET values reported by Bauer and Benham should match the theoretical values computed from Eq. 21 by using the various
and ET, then the experimental ET values reported by Bauer and Benham should match the theoretical values computed from Eq. 21 by using the various 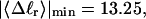 11.0, 8.8, 6.8, and 4.5 turns at T = 313, 318, 323, 328, and 333 K, respectively. This comparison is given in Table 1. The experimental values exceed the theoretical values by ∼2–5%. As noted above, a ∼5% overestimation of the experimental ET values is expected due to the use of Eq. 19, when ET increases with
11.0, 8.8, 6.8, and 4.5 turns at T = 313, 318, 323, 328, and 333 K, respectively. This comparison is given in Table 1. The experimental values exceed the theoretical values by ∼2–5%. As noted above, a ∼5% overestimation of the experimental ET values is expected due to the use of Eq. 19, when ET increases with 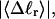 as predicted in 20 mM ionic strength. After scaling the experimental values by 0.95 to compensate for their overestimation, the agreement is remarkably good, with discrepancies of 3.4, 1.6, 0.1, −0.5, and −0.1% between the theoretical and the downward scaled experimental values (not shown) at, respectively, 313, 318, 323, 328, and 333 K. Such good agreement argues for the approximate validity of both Eq. 21 and the 0.95 scaling of the experimental ET values.
as predicted in 20 mM ionic strength. After scaling the experimental values by 0.95 to compensate for their overestimation, the agreement is remarkably good, with discrepancies of 3.4, 1.6, 0.1, −0.5, and −0.1% between the theoretical and the downward scaled experimental values (not shown) at, respectively, 313, 318, 323, 328, and 333 K. Such good agreement argues for the approximate validity of both Eq. 21 and the 0.95 scaling of the experimental ET values.
To what extent does Eq. 21 depend upon parameters of the melted region, such as its torsion and bending elastic constants? The coalescence of the ET versus 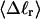 curves in Fig. 8 establishes that ET is independent of n (or m). Because ET for n = 0 is necessarily independent of the torsion and bending elastic constants of the melted region, it must be presumed that ET depends only on parameters of the duplex region, even in locally melted DNAs. Thus, the slope,
curves in Fig. 8 establishes that ET is independent of n (or m). Because ET for n = 0 is necessarily independent of the torsion and bending elastic constants of the melted region, it must be presumed that ET depends only on parameters of the duplex region, even in locally melted DNAs. Thus, the slope, 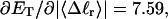 in Eq. 21 should be practically independent of the values of the torsion and bending elastic constants of the melted region. Equation 21 may prove useful in the prediction of ET values at various temperatures for the duplex regions of both unmelted and locally melted supercoiled DNAs with lengths comparable to 4349 bp in 20 mM ionic strength.
in Eq. 21 should be practically independent of the values of the torsion and bending elastic constants of the melted region. Equation 21 may prove useful in the prediction of ET values at various temperatures for the duplex regions of both unmelted and locally melted supercoiled DNAs with lengths comparable to 4349 bp in 20 mM ionic strength.
A possible enhancement of the slope, −dαD/dT, in the protocol of Bauer and Benham
It seems likely that the elastic constant for interwinding single strands, corresponding to 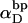 in our model, increases with increased interwinding, or linking difference,
in our model, increases with increased interwinding, or linking difference, 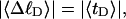 in the melted region. This phenomenon should be similar to the increase in ET with
in the melted region. This phenomenon should be similar to the increase in ET with 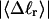 of the duplex region, and should occur for a similar reason, namely an increase in electrostatic free energy relative to the interwinding free energy for a hard filament upon increasing
of the duplex region, and should occur for a similar reason, namely an increase in electrostatic free energy relative to the interwinding free energy for a hard filament upon increasing 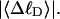 If this conjectured increase of
If this conjectured increase of 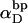 with increasing
with increasing 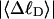 were true, that would provide an additional contribution to
were true, that would provide an additional contribution to 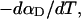 because the typical
because the typical 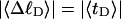 (or
(or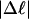 ) of the topoisomer with a given fixed number of melted basepairs decreases with increasing T. Such an artifactual enhancement of
) of the topoisomer with a given fixed number of melted basepairs decreases with increasing T. Such an artifactual enhancement of 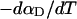 may be partly responsible for the extrapolation of the experimental
may be partly responsible for the extrapolation of the experimental 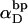 to unphysical negative values for temperatures above 79°C, as discussed in the Appendix.
to unphysical negative values for temperatures above 79°C, as discussed in the Appendix.
Locations of the melted regions
More than 100 configurations were examined visually with the aid of a modeling program. The writhe was found predominantly in interwound domains, as expected, and the superhelix axis was commonly branched especially at the larger 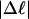 values. To determine where the melted regions occurred, each molecule was subdivided into the following four rough categories:
values. To determine where the melted regions occurred, each molecule was subdivided into the following four rough categories:
An arm domain, which is the interior of a well-defined interwound superhelix.
An apex domain, which is the external end loop of an interwound superhelix.
A linker domain at a “node”, where two or more superhelix branches, or arms, meet.
Other domains not easily characterized.
Detailed inspections were carried out for randomly chosen configurations from three regular simulations:
n = 0,
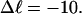
n = 2,
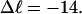
n = 2,
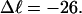
Subsets a and b have very similar 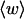 and
and 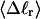 values despite the difference in n, whereas subsets b and c have the same n, but very different
values despite the difference in n, whereas subsets b and c have the same n, but very different 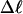 and
and 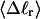 values. Twenty configurations were inspected in each case to determine the locations of subunits 154 and 155, which are those possessing the weak bending and torsion springs. The numbers of occurrences in each kind of domain are shown in Table 2. For n = 0, there are no elastically weak regions, and the results simply provide a noisy sampling of the fraction of the molecule within each kind of domain. For the n = 2 molecules with intermediate linking difference (
values. Twenty configurations were inspected in each case to determine the locations of subunits 154 and 155, which are those possessing the weak bending and torsion springs. The numbers of occurrences in each kind of domain are shown in Table 2. For n = 0, there are no elastically weak regions, and the results simply provide a noisy sampling of the fraction of the molecule within each kind of domain. For the n = 2 molecules with intermediate linking difference (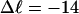 ), the melted regions occupy about equally the apex and linker domains, both of which are apparently preferred to arms. However, at larger linking difference (
), the melted regions occupy about equally the apex and linker domains, both of which are apparently preferred to arms. However, at larger linking difference (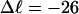 ), where more superhelix branching is evident, the melted regions strongly prefer linker and to a lesser extent arm domains to apex domains. A typical location for a melted domain is shown in Fig. 10. Substantial bends are typically associated with the melted regions in the linker domains. This behavior contrasts sharply with that found for directional permanent bends (Bussiek et al., 2002; Pfannschmidt and Langowski, 1998; Chirico and Langowski, 1996), which strongly prefer apex domains. The strong preference of a melted region for linker domains in the typically branched molecules at large
), where more superhelix branching is evident, the melted regions strongly prefer linker and to a lesser extent arm domains to apex domains. A typical location for a melted domain is shown in Fig. 10. Substantial bends are typically associated with the melted regions in the linker domains. This behavior contrasts sharply with that found for directional permanent bends (Bussiek et al., 2002; Pfannschmidt and Langowski, 1998; Chirico and Langowski, 1996), which strongly prefer apex domains. The strong preference of a melted region for linker domains in the typically branched molecules at large 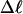 is likely due in part to the entropy associated with the ability of the molecule to bend in any direction at that point, which allows for many relative orientations of the two superhelix branches that are connected by that linker domain.
is likely due in part to the entropy associated with the ability of the molecule to bend in any direction at that point, which allows for many relative orientations of the two superhelix branches that are connected by that linker domain.
TABLE 2.
Numbers of occurrences of subunits 154 and 155 in different regions for various simulated molecules
| Simulated molecules
|
|||||
|---|---|---|---|---|---|
| n = 0 | n = 2 | n = 2 | n = 1* | n = 1* | |
| Domain | 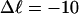 |
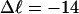 |
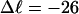 |
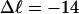 |
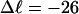 |
| Arm | 8 | 0 | 5 | 2 | 3 |
| Apex | 2 | 5 | 2 | 5 | 1 |
| Linker | 2 | 6 | 11 | 6 | 10 |
| Other | 8 | 9 | 2 | 7 | 6 |
The (154,155) spring has the weak bending elastic constant of a melted subunit, but also has the torsion elastic constant of a normal duplex subunit.
FIGURE 10.
Typical structure of a simulated model DNA with n = 2 melted subunits and 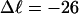 turns. The location of the melted region is indicated by the arrow, and lies in the linker region between interwound arms.
turns. The location of the melted region is indicated by the arrow, and lies in the linker region between interwound arms.
To determine whether the softer bending potential or instead the softer torsion potential was primarily responsible for the preference of the melted regions for linker regions at large 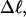 two additional simulations were performed for models containing a single subunit with a weak bending spring typical of a melted region, but also a normal duplex torsion spring. The results for this model are presented in the fourth and fifth columns of Table 2. These results are very similar to those in columns three and four, respectively. We conclude that the softer torsion elastic constant contributes little or nothing to the preference of melted regions for linker domains.
two additional simulations were performed for models containing a single subunit with a weak bending spring typical of a melted region, but also a normal duplex torsion spring. The results for this model are presented in the fourth and fifth columns of Table 2. These results are very similar to those in columns three and four, respectively. We conclude that the softer torsion elastic constant contributes little or nothing to the preference of melted regions for linker domains.
The contrasting preferences of flexurally weak melted regions for linker domains on one hand and of intrinsically curved duplex sequences for apex regions on the other may stem largely from the strong directionality of the latter bends, which would prevent such a region from realizing the full entropy of bending in different directions, if it were to occupy a linker domain. This interpretation is supported by the fact that the strong preference of flexurally weak regions for linker domains over apex regions occurs only at the larger linking differences, where superhelix branching is both more common and more extensive.
Acknowledgments
This work was supported in part by grants MCB-9982735 from the National Science Foundation and R01 GM61685 from the National Institutes of Health.
APPENDIX: POTENTIALS, INPUT PARAMETERS, SIMULATION PROTOCOLS, AND STATISTICAL METHODS
Potential functions
The torsion potential is given by
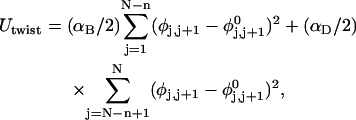 |
(A1) |
where N − n and n are the numbers of subunits in the duplex and melted regions, respectively, αB is the torsion elastic constant of those springs for which 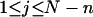 lies in the duplex region,
lies in the duplex region, 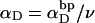 is the torsion elastic constant of those springs for which
is the torsion elastic constant of those springs for which 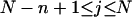 lies in the melted region,
lies in the melted region, 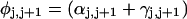 is the sum of the first and last angles of the Euler rotation,
is the sum of the first and last angles of the Euler rotation, 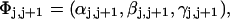 which orients the (j + 1)th subunit in the frame of the jth subunit, and
which orients the (j + 1)th subunit in the frame of the jth subunit, and 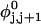 (radians) is the intrinsic twist from the jth to (j + 1)th subunit. In this study,
(radians) is the intrinsic twist from the jth to (j + 1)th subunit. In this study, 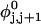 is not identified with the intrinsic subunit succession angle,
is not identified with the intrinsic subunit succession angle, 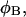 of a real DNA, but instead is a parameter that is employed to continuously vary the linking difference from 0 to −26 turns, whereas the linking number remains fixed at zero turns.
of a real DNA, but instead is a parameter that is employed to continuously vary the linking difference from 0 to −26 turns, whereas the linking number remains fixed at zero turns.
The simulation temperature is taken to be 298 K. For duplex regions, we take 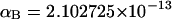 dyne cm, which corresponds to a torsional rigidity,
dyne cm, which corresponds to a torsional rigidity, 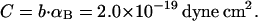 Numerous measurements of the torsional rigidities of pBR322 and other plasmids of comparable size by time-resolved fluorescence polarization anisotropy yielded values in the range (1.9–2.2) × 10−19 dyne cm2 in the ionic strength range 10–100 mM at ambient temperature (Schurr et al., 1992; Heath et al., 1996; Naimushin et al., 2000, 2001). When such measured values of C were employed along with a persistence length, P = 500 A, and appropriate intersubunit repulsions in simulations of supercoiled p30δ DNA (4752 bp) in 0.1 M ionic strength, the predicted values of the twist energy parameter (ET), structure factor (S(q)) over a range of scattering vectors from q = 0 to 2.4 × 105 cm−1, and translational diffusion coefficient (DT) agreed well with the corresponding measured values over a wide range of superhelix density from zero to native (Gebe et al., 1995, 1996; Clendenning et al., 1994). Likewise, simulations of surface-flattened pSA509 DNA (3760 bp) in 161 mM ionic strength, using C = 2.0 × 10−19 dyne cm2, yielded structures similar to those seen in AFM (Fujimoto and Schurr, 2002; Lyubchenko and Shlyakhtenko, 1997).
Numerous measurements of the torsional rigidities of pBR322 and other plasmids of comparable size by time-resolved fluorescence polarization anisotropy yielded values in the range (1.9–2.2) × 10−19 dyne cm2 in the ionic strength range 10–100 mM at ambient temperature (Schurr et al., 1992; Heath et al., 1996; Naimushin et al., 2000, 2001). When such measured values of C were employed along with a persistence length, P = 500 A, and appropriate intersubunit repulsions in simulations of supercoiled p30δ DNA (4752 bp) in 0.1 M ionic strength, the predicted values of the twist energy parameter (ET), structure factor (S(q)) over a range of scattering vectors from q = 0 to 2.4 × 105 cm−1, and translational diffusion coefficient (DT) agreed well with the corresponding measured values over a wide range of superhelix density from zero to native (Gebe et al., 1995, 1996; Clendenning et al., 1994). Likewise, simulations of surface-flattened pSA509 DNA (3760 bp) in 161 mM ionic strength, using C = 2.0 × 10−19 dyne cm2, yielded structures similar to those seen in AFM (Fujimoto and Schurr, 2002; Lyubchenko and Shlyakhtenko, 1997).
The torsion elastic constant between subunits (containing 28.06 bp/subunit) for locally melted regions is here taken to be 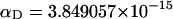 dyne cm, which is 54.5-fold smaller than αB for the duplex regions at 298 K. This choice of
dyne cm, which is 54.5-fold smaller than αB for the duplex regions at 298 K. This choice of 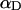 corresponds to
corresponds to 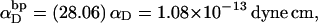 which is obtained by extrapolating the results of Bauer and Benham to 37°C. A longer extrapolation to 25°C, some 15° below their lowest measurement, cannot be justified, because unphysical negative values of
which is obtained by extrapolating the results of Bauer and Benham to 37°C. A longer extrapolation to 25°C, some 15° below their lowest measurement, cannot be justified, because unphysical negative values of 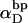 are predicted by their best-fit straight line
are predicted by their best-fit straight line 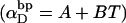 for T ≥ 79°C, and presumably arise from an overestimated negative slope (−B), as noted in the main text.
for T ≥ 79°C, and presumably arise from an overestimated negative slope (−B), as noted in the main text.
The electrostatic part of the nonnearest-neighbor interaction energy is reckoned by placing three spheres, each containing Z electronic charges, on the axis of each subunit rod. These charged spheres are spaced (95.4)/3 = 31.8 Å apart and are centered at positions rj, rj + (1/3)bj, and rj + (2/3)bj, where rj is the position of the coordinate frame of the jth subunit at the end of the (j − 1)th rod, and bj is the bond vector from the jth frame to the (j + 1)th frame. The electrostatic interaction energy is taken over all pairs of charged spheres that do not lie on the same or adjacent subunit rods. The subunit interaction energy also includes a hard-cylinder repulsion energy, UHC, between nonnearest neighbor units. The total interaction energy between two charged spheres, i and j, on nonadjacent subunits is taken as (Fujimoto and Schurr, 2002; Delrow et al., 1997a)
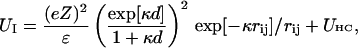 |
(A2) |
where e is the electronic charge, ɛ is the dielectric constant, rij is the distance between charge centers, d is the charged sphere radius, and κ(cm−1) is the Debye-Hückel screening parameter, given in cgs units by 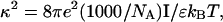 where NA is Avogadro's number,
where NA is Avogadro's number, 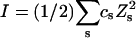 is the ionic strength, cs is the concentration (mole/L), and Zs is the valence of the sth kind of ion. The dielectric constant is taken to be ɛ = 78.54 at 25°C. The radius of the charged spheres is taken as d = 12 Å. UHC is infinite, when the axes of the two subunit rods approach more closely than 24 Å (corresponding to a hard-cylinder diameter, 2d = 24 Å), and zero otherwise. Screened Coulomb interactions are ignored, when
is the ionic strength, cs is the concentration (mole/L), and Zs is the valence of the sth kind of ion. The dielectric constant is taken to be ɛ = 78.54 at 25°C. The radius of the charged spheres is taken as d = 12 Å. UHC is infinite, when the axes of the two subunit rods approach more closely than 24 Å (corresponding to a hard-cylinder diameter, 2d = 24 Å), and zero otherwise. Screened Coulomb interactions are ignored, when 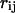 exceeds a cut-off distance, which is chosen so that the first term on the right-hand side of Eq. A2 equals (7.3 × 10−4)kT at the cut-off distance. UHC is implemented by a protocol that performs the test and rejects any Monte Carlo move leading to a configuration that violates the minimum distance of closest approach (Gebe et al., 1995).
exceeds a cut-off distance, which is chosen so that the first term on the right-hand side of Eq. A2 equals (7.3 × 10−4)kT at the cut-off distance. UHC is implemented by a protocol that performs the test and rejects any Monte Carlo move leading to a configuration that violates the minimum distance of closest approach (Gebe et al., 1995).
The screening parameter κ is calculated from the prevailing ionic strength in the 90 mM Tris-borate solution of Bauer and Benham (1993). The reckoning of this ionic strength is complicated, because the measured pH of the 90 mM Tris-borate solution (8.4 at 25°C) differed from that expected (8.65) from the pKa's of Tris·H+ (8.069) and boric acid (9.236) alone. This discrepancy is presumably attributable to the formation of complexes between the neutral Tris and borate anion (Taylor et al., 1996). Three different approximate estimates of the ionic strength clustered around 20 mM, which is the value adopted here. Finally, the effective valence of the charged spheres, Z = −9.19, was chosen so that the electrostatic potential surrounding the middle of a linear array of 2001 charged spheres with 31.8 Å spacing closely matched the solution of the nonlinear Poisson-Boltzmann equation for a uniformly charged cylinder with 12 Å radius and the linear charge density of DNA at all distances beyond the hard-cylinder diameter, d = 24 Å, as described by Delrow et al. (1997a).
The bending potential energy is given by
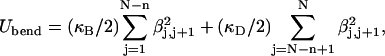 |
(A3) |
where κB and κD are the torque constants for bending of the duplex and denatured regions, respectively. βj,j+1 is the second rotation in the composite Euler rotation,  The value of κB was chosen to yield a persistence length, P = 500 Å, in the following manner. Simulations of both 10- and 20-subunit (b = 95.4 Å) chains were performed (10 million attempted moves per simulation) with several different values of
The value of κB was chosen to yield a persistence length, P = 500 Å, in the following manner. Simulations of both 10- and 20-subunit (b = 95.4 Å) chains were performed (10 million attempted moves per simulation) with several different values of  but using always the same values of the parameters in Utwist and UI that were discussed above. For each simulation, the persistence length was calculated according to
but using always the same values of the parameters in Utwist and UI that were discussed above. For each simulation, the persistence length was calculated according to  for each model, where the average,
for each model, where the average, 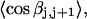 was taken over all subunits, j = 1, … ,N − 1, in all configurations. The value κB = 1.806 × 10−13 dyne cm was found to yield P = 500 Å (within the simulation errors) for both 10- and 20-subunit chains, and was adopted for the simulations reported here.
was taken over all subunits, j = 1, … ,N − 1, in all configurations. The value κB = 1.806 × 10−13 dyne cm was found to yield P = 500 Å (within the simulation errors) for both 10- and 20-subunit chains, and was adopted for the simulations reported here.
To our knowledge, the effects of flexurally soft regions on the tertiary structure and energetics of supercoiled DNAs have not previously been simulated. In the absence of any precedent, we choose 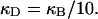 However, due to the large size and valence of the charged spheres, the electrostatic interactions make an overly large contribution to the persistence length (Delrow et al., 1997a). Thus, despite the fact that
However, due to the large size and valence of the charged spheres, the electrostatic interactions make an overly large contribution to the persistence length (Delrow et al., 1997a). Thus, despite the fact that 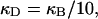 the persistence length associated with κD is ∼175 Å, which is only ∼2.9-fold smaller than that associated with
the persistence length associated with κD is ∼175 Å, which is only ∼2.9-fold smaller than that associated with 
Calculation of tB, wB, tD, TD, wD, and Δℓ̃r
For each selected configuration, the net twist, t, is calculated according to Eq. 3), and tB and tD are reckoned from the first and second terms, respectively, on the far right-hand side of Eq. 4. Finally, TD = tD − nφD.
Due to the long subunit length in this work, the writhe computed according to Eq. 5 is insufficiently accurate, and leads to fluctuations in the computed linking difference, 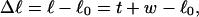 of as much as 4% around the starting value. To improve the accuracy of the writhe calculations, each subunit rod is subdivided into 10 coaxial subrods of equal length, b/10. The total writhe is now calculated according to
of as much as 4% around the starting value. To improve the accuracy of the writhe calculations, each subunit rod is subdivided into 10 coaxial subrods of equal length, b/10. The total writhe is now calculated according to
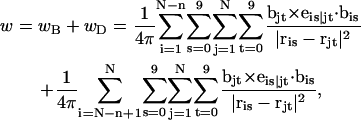 |
(A4) |
where 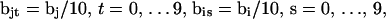
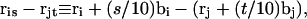 and
and 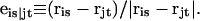 wB and wD correspond to the first and second terms, respectively, in Eq. A4. Use of this higher-resolution writhe calculation reduced the fluctuations in computed linking difference around the starting value to less than one part in 103. The writhe is calculated for only one configuration in every 10,000 moves.
wB and wD correspond to the first and second terms, respectively, in Eq. A4. Use of this higher-resolution writhe calculation reduced the fluctuations in computed linking difference around the starting value to less than one part in 103. The writhe is calculated for only one configuration in every 10,000 moves.
Nonvanishing self-writhe requires a minimum of three bond vectors, sufficient to create a nonplanar bend. In these models, wherein the denatured region contains only n = 1 or 2 rigid-rod subunits, 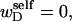 so
so 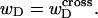
The total linking difference of any simulated DNA, whether partially melted or not, is reckoned according to 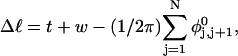 where
where 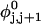 is the intrinsic twist of the torsion spring between the jth and(j + 1)th subunits, and is defined explicitly below.
is the intrinsic twist of the torsion spring between the jth and(j + 1)th subunits, and is defined explicitly below.
The residual linking difference of the duplex region in a partially melted molecule is calculated by 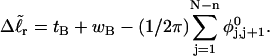
Models with total linking differences of {0, −2, −4, −6, … , −26} turns are simulated by using the corresponding uniform values of 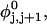 namely
namely 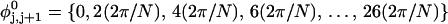 for all j = 1, … , N, together with a starting configuration that is a planar polygon with initial twist angles, φj,j+1 = 0, so w = 0 and t = 0. In this convention,
for all j = 1, … , N, together with a starting configuration that is a planar polygon with initial twist angles, φj,j+1 = 0, so w = 0 and t = 0. In this convention, 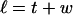 remains at the value 0 in all of our simulations, but the linking difference is decreased stepwise, as
remains at the value 0 in all of our simulations, but the linking difference is decreased stepwise, as 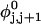 is raised from each step to the next. In other words, the actual DNA comprising a duplex region with intrinsic subunit succession angle,
is raised from each step to the next. In other words, the actual DNA comprising a duplex region with intrinsic subunit succession angle, 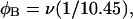 a melted region with intrinsic succession angle, φD = 0, and a nonvanishing linking number,
a melted region with intrinsic succession angle, φD = 0, and a nonvanishing linking number, 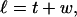 is replaced by a model that exhibits a linking number,
is replaced by a model that exhibits a linking number, 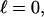 and a uniform intrinsic twist,
and a uniform intrinsic twist, 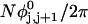 (turns), that is chosen to yield the desired linking difference. Although the intrinsic twist is chosen to be uniform over the entire molecule, the torsion and bending elastic constants differ greatly between duplex and melted regions. Such a procedure is valid because the state of torsional strain of the molecule depends only upon the difference between the actual twist
(turns), that is chosen to yield the desired linking difference. Although the intrinsic twist is chosen to be uniform over the entire molecule, the torsion and bending elastic constants differ greatly between duplex and melted regions. Such a procedure is valid because the state of torsional strain of the molecule depends only upon the difference between the actual twist 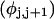 and intrinsic twist
and intrinsic twist 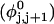 of each spring, not upon either value separately. In principle, the
of each spring, not upon either value separately. In principle, the 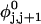 for a given subunit could be assigned any value, provided that
for a given subunit could be assigned any value, provided that 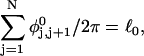 the total intrinsic twist of our model DNA. Here,
the total intrinsic twist of our model DNA. Here, 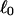 is used as an adjustable parameter to vary the linking difference, so for each of our DNAs with n = 0, 1, or 2 weak springs (i.e., “melted” subunits), the initial value is taken to be
is used as an adjustable parameter to vary the linking difference, so for each of our DNAs with n = 0, 1, or 2 weak springs (i.e., “melted” subunits), the initial value is taken to be 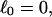 and
and 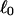 is increased stepwise to 2, 4, … , 26 turns to generate total linking differences,
is increased stepwise to 2, 4, … , 26 turns to generate total linking differences, 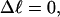 −2, −4, … , −26 turns, respectively. The unmelted parent of a locally melted model molecule is not simulated. However, its linking difference
−2, −4, … , −26 turns, respectively. The unmelted parent of a locally melted model molecule is not simulated. However, its linking difference 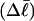 is reckoned from the
is reckoned from the 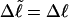 of its locally melted model daughter DNA via Eq. 1,
of its locally melted model daughter DNA via Eq. 1, 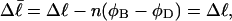 since
since 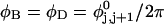 in these simulations. For our simulated molecules, then, it is always true that
in these simulations. For our simulated molecules, then, it is always true that 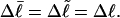
Calculation of the inertial tensor elements and the radius of gyration
The squared radius of gyration of a single configuration is calculated according to 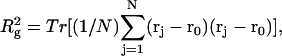 where
where 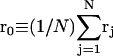 is the center of “mass”. The quantity in square brackets is the dyadic inertial tensor, which can be expressed as a 3 × 3 symmetric matrix. This symmetric matrix is diagonalized to yield its three positive eigenvalues, which are designated in order from largest to smallest as
is the center of “mass”. The quantity in square brackets is the dyadic inertial tensor, which can be expressed as a 3 × 3 symmetric matrix. This symmetric matrix is diagonalized to yield its three positive eigenvalues, which are designated in order from largest to smallest as 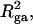
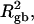 and
and 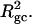 Each of these three quantities and also
Each of these three quantities and also 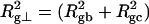 is averaged over the saved molecular configurations. Invariance of the trace to similarity transformation gives
is averaged over the saved molecular configurations. Invariance of the trace to similarity transformation gives 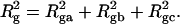
Simulation protocols
The Metropolis Monte Carlo protocol is similar to that described previously by Schurr et al. (1995). In particular, the full potential energy function, rather than just the reduced potential energy function, is evaluated on every step and used in the Metropolis acceptance criterion. Moreover, the 3N Euler angles that orient each subunit in the laboratory frame are kept to define each configuration. The topological constraint in Eq. 2 is never explicitly enforced, except for the initial configuration, but instead is automatically obeyed to acceptable numerical accuracy (better than 1 part in 103). Our protocol employs the sampling, ring closure, hard-cylinder exclusion, knot detection, and reversible work algorithms of Gebe et al. (1995), Gebe and Schurr (1996), and Schurr et al. (1995), and the electrostatic interaction algorithm of Delrow et al. (1997a). When supplied with appropriate input parameters, simulations using these algorithms satisfactorily reproduce experimental values of the supercoiling free energy, translational diffusion coefficient, and structure factor of the p30δ DNA over a wide range of superhelix density in 0.1 M ionic strength, and also yield typical observed structures of a surface-flattened native supercoiled plasmid in 0.161 M ionic strength, as noted above. However, these same protocols and potentials are known not to work so well at lower ionic strength, specifically at 0.01 M (Gebe et al., 1996; Fujimoto and Schurr, 2002). There is considerable evidence that certain supercoiled DNAs in 0.01 M ionic strength undergo one or more long-range allosteric transitions in secondary structure with increasing superhelix density (Shibata et al., 1984; Wu et al., 1988, 1991; Song et al., 1990; Schurr et al., 1992, 1997; Heath et al., 1996; Delrow et al., 1997b; Naimushin et al., 2000). This phenomenon may be at least partially responsible for the observed shortcomings of these protocols and potentials for 0.01 M ionic strength. Such complications are ignored here to assess the properties of the very simplest idealized model of a supercoiled DNA.
The starting configuration for the simulation of each model supercoiled DNA with n = 0, 1, or 2 weak springs was taken to be a planar polygon with zero intrinsic twist, 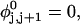 j = 1, … , N, and zero actual twist,
j = 1, … , N, and zero actual twist, 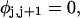 j = 1, … , N. Thus, the initial values of the writhe, w; net twist, t; linking number,
j = 1, … , N. Thus, the initial values of the writhe, w; net twist, t; linking number, 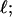 intrinisic twist,
intrinisic twist, 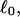 and linking difference all vanish. The system was then “equilibrated” for 12 million attempted moves before saving any configurations. The final configuration at the end of this “equilibration” period was used as the initial configuration for eight additional runs of 4 million attempted moves, totaling 32 million attempted moves. During these latter data collection runs, a configuration was saved once every 10,000 moves (3200 saved configurations for each linking difference). From these saved configurations, the average values and variances are calculated for t, tB, tD, TD, w, wB, wD,
and linking difference all vanish. The system was then “equilibrated” for 12 million attempted moves before saving any configurations. The final configuration at the end of this “equilibration” period was used as the initial configuration for eight additional runs of 4 million attempted moves, totaling 32 million attempted moves. During these latter data collection runs, a configuration was saved once every 10,000 moves (3200 saved configurations for each linking difference). From these saved configurations, the average values and variances are calculated for t, tB, tD, TD, w, wB, wD, 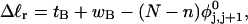
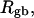
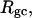
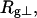 and
and 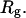 The total linking difference
The total linking difference 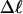 is initially selected in the simulation. The values of
is initially selected in the simulation. The values of 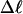 computed according to Eq. A5 fluctuate only very slightly (by less than 1 part in 103) from the initial value, due to numerical errors primarily in the writhe calculation. The ensemble average values
computed according to Eq. A5 fluctuate only very slightly (by less than 1 part in 103) from the initial value, due to numerical errors primarily in the writhe calculation. The ensemble average values 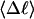 lie even closer to the initially selected values. For that reason, we take
lie even closer to the initially selected values. For that reason, we take 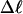 as the initially selected value, rather than the practically identical average value,
as the initially selected value, rather than the practically identical average value, 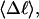 in the discussion of results.
in the discussion of results.
The squared standard deviation (variance) of each property, x, is calculated in the usual way, 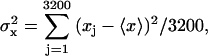 where the average value,
where the average value, 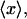 is reckoned from all 3200 sample values.
is reckoned from all 3200 sample values.
The squared standard deviation of the mean of each property is estimated according to 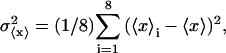 where
where 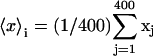 is the average over individual values of the ith data collection run, of which there are eight (for each selected value of
is the average over individual values of the ith data collection run, of which there are eight (for each selected value of 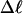 and n). This relation provides an accurate estimate, when the system relaxes well within the 4 million moves of each data collection run, which is the case here.
and n). This relation provides an accurate estimate, when the system relaxes well within the 4 million moves of each data collection run, which is the case here.
After completion of the data collection runs, the intrinsic succession angle, 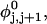 is incremented by 4π/N for all j = 1, … , N, which decreases the linking difference by two turns. The starting configuration for this new linking difference is taken to be the final configuration at the end of the “equilibration” period for the previous linking difference. The system is then “equilibrated” at the new linking difference for 12 million attempted moves. Again, the final configuration at the end of the “equilibration” period was taken as the starting configuration for each of eight additional runs of 4 million moves, and the entire procedure outlined above is repeated.
is incremented by 4π/N for all j = 1, … , N, which decreases the linking difference by two turns. The starting configuration for this new linking difference is taken to be the final configuration at the end of the “equilibration” period for the previous linking difference. The system is then “equilibrated” at the new linking difference for 12 million attempted moves. Again, the final configuration at the end of the “equilibration” period was taken as the starting configuration for each of eight additional runs of 4 million moves, and the entire procedure outlined above is repeated.
References
- Bauer, W. R., and C. J. Benham. 1993. The free energy, enthalpy, and entropy of native and of partially melted supercoiled DNA. J. Mol. Biol. 234:1184–1196. [DOI] [PubMed] [Google Scholar]
- Bauer, W. R., H. Ohtsubo, E. Ohtsubo, and C. J. Benham. 1995. Energetics of coupled twist and writhe changes in closed circular pSM1 DNA. J. Mol. Biol. 253:438–452. [DOI] [PubMed] [Google Scholar]
- Benham, C. J. 1990. Theoretical analysis of heteropolymeric transitions in superhelical DNA molecules of specified sequence. J. Chem. Phys. 92:6294–6305. [Google Scholar]
- Benham, C. J. 1992. Energetics of the strand separation transition in superhelical DNA. J. Mol. Biol. 225:835–847. [DOI] [PubMed] [Google Scholar]
- Bussiek, M., K. Klenin, and J. Langowski. 2002. Kinetics of site-site interactions in supercoiled DNA with bent sequences. J. Mol. Biol. 322:707–718. [DOI] [PubMed] [Google Scholar]
- Chirico, G., and J. Langowski. 1996. Brownian dynamics simulations of supercoiled DNAs with bent sequences. Biophys. J. 71:955–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendenning, J. B., U.-S. Kim, L. Song, B. S. Fujimoto, D. W. Stewart, and J. M. Schurr. 1994. Effect of ethidium binding and superhelix density on the supercoiling free energy and torsion and bending constants of p30δ DNA. Biophys. Chem. 52:219–226. [DOI] [PubMed] [Google Scholar]
- Clendenning, J. B., and J. M. Schurr. 1994. Circularization of small DNAs in the presence of ethidium. A theoretical analysis. Biopolymers. 34:849–868. [DOI] [PubMed] [Google Scholar]
- Delrow, J. J., J. A. Gebe, and J. M. Schurr. 1997a. Comparison of hard-cylinder and screened Coulomb interactions in the modeling of supercoiled DNAs. Biopolymers. 42:455–470. [DOI] [PubMed] [Google Scholar]
- Delrow, J. J., P. J. Heath, and J. M. Schurr. 1997b. On the origin of the temperature dependence of the supercoiling free energy. Biophys. J. 73:2688–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds, A. R. 1974. Angular Momentum in Quantum Mechanics. Princeton University Press, Princeton, NJ.
- Fujimoto, B. S., and J. M. Schurr. 2002. Monte Carlo simulations of DNAs confined to a plane. Biophys. J. 82:944–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, F. B. 1971. The writhing number of a space curve. Proc. Natl. Acad. Sci. USA. 68:815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebe, J. A., S. A. Allison, J. B. Clendenning, and J. M. Schurr. 1995. Monte Carlo simulations of supercoiling free energies of unknotted and trefoil knotted DNAs. Biophys. J. 68:619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebe, J. A., and J. M. Schurr. 1996. Thermodynamics of the first transition in writhe of a small circular DNA by Monte Carlo simulation. Biopolymers. 38:493–508. [DOI] [PubMed] [Google Scholar]
- Gebe, J. A., J. J. Delrow, P. J. Heath, D. W. Stewart, and J. M. Schurr. 1996. Effects of Na+ and Mg2+ ions on the structures of supercoiled DNAs. Comparison of simulations with experiments. J. Mol. Biol. 262:105–128. [DOI] [PubMed] [Google Scholar]
- Hao, M., and W. K. Olson. 1989. Global equilibrium configurations of supercoiled DNA. Macromolecules. 22:3292–3303. [Google Scholar]
- Heath, P. J., J. B. Clendenning, B. S. Fujimoto, and J. M. Schurr. 1996. Effect of bending strain on the torsion elastic constant of DNA. J. Mol. Biol. 260:718–730. [DOI] [PubMed] [Google Scholar]
- Kowalski, D., D. Natale, and M. Eddy. 1988. Stable DNA unwinding, not breathing, accounts for single-strand-specific hypersensitivity of specific A + T-rich sequences. Proc. Natl. Acad. Sci. USA. 85:9464–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, R. 1957. Statistical mechanical theory of irreversible processes. I. General theory and simple applications to magnetic and conduction problems. J. Physical Soc. Japan. 12:570–586. [Google Scholar]
- Lyubchenko, Y., and L. S. Shlyakhtenko. 1997. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl. Acad. Sci. USA. 94:496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimushin, A. N., J. B. Clendenning, U.-S. Kim, L. Song, B. S. Fujimoto, D. W. Stewart, and J. M. Schurr. 1994. Effect of ethidium binding and superhelix density on the supercoiling free energy and torsion constant of pBR322 DNA. Biophys. Chem. 52:219–226. [DOI] [PubMed] [Google Scholar]
- Naimushin, A. N., B. S. Fujimoto, and J. M. Schurr. 2000. Dynamic bending rigidity of a 200 bp DNA in 4 mM ionic strength. Biophys. J. 78:1498–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimushin, A. N., N. Quach, B. S. Fujimoto, and J. M. Schurr. 2001. Effect of polyethylene glycol on the supercoiling free energy of DNA. Biopolymers. 58:204–217. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, C., and J. Langowski. 1998. Superhelix organization by DNA curvature as measured by site specific labeling. J. Mol. Biol. 275:601–611. [DOI] [PubMed] [Google Scholar]
- Potaman, V. N., J. J. Bissler, V. I. Hashem, E. A. Oussatcheva, L. Lu, L. S. Shlyakhtenko, Y. L. Lyubchenko, T. Matsuura, T. Ashizawa, M. Leffak, C. J. Benham, and R. R. Sinden. 2003. Unpaired structures in SCA10 (ATTCT)n (AGAAT)n repeats. J. Mol. Biol. 326:1095–1111. [DOI] [PubMed] [Google Scholar]
- Schurr, J. M. 1985. Effect of anisotropic bending rigidity and finite twisting rigidity on statistical properties of DNA model filaments. Biopolymers. 24:1233–1246. [DOI] [PubMed] [Google Scholar]
- Schurr, J. M., H. P. Babcock, and J. A. Gebe. 1995. Effect of anisotropy of the bending rigidity on the supercoiling free energy of small circular DNAs. Biopolymers. 36:633–641. [DOI] [PubMed] [Google Scholar]
- Schurr, J. M., J. J. Delrow, B. S. Fujimoto, and A. S. Benight. 1997. The question of long-range allosteric transitions in DNA. Biopolymers. 44:283–308. [DOI] [PubMed] [Google Scholar]
- Schurr, J. M., B. S. Fujimoto, P.-G. Wu, and L. Song. 1992. Fluorescence studies of nucleic acids: Dynamics, rigidities and structures. In Topics in Fluorescence Spectroscopy, Vol. 3: Biochemical Applications. J. R. Lakowicz, editor. Plenum Press, New York. 137–229.
- Sheridan, S. D., C. J. Benham, and G. W. Hatfield. 1998. Activation of gene expression by a novel DNA structural transmission mechanism that requires supercoiling induced DNA destabilization in an upstream activating sequence. J. Biol. Chem. 273:21298–21308. [DOI] [PubMed] [Google Scholar]
- Shibata, J. H., J. Wilcoxon, J. M. Schurr, and V. Knauf. 1984. Structures and dynamics of a supercoiled DNA. Biochemistry. 23:1188–1194. [DOI] [PubMed] [Google Scholar]
- Smith, S. B., P. K. Aldridge, and J. B. Callis. 1989. Observation of individual DNA molecules undergoing gel electrophoresis. Science. 243:203–206. [DOI] [PubMed] [Google Scholar]
- Song, L., B. S. Fujimoto, P.-G. Wu, J. C. Thomas, J. H. Shibata, and J. M. Schurr. 1990. Evidence for allosteric transitions in secondary structure induced by superhelical stress. J. Mol. Biol. 214:307–326. [DOI] [PubMed] [Google Scholar]
- Song, L., and M. F. Maestre. 1991. Unhooking dynamics of U-shaped DNA molecules undergoing gel electrophoresis. J. Biomol. Struct. Dyn. 8:643–655. [DOI] [PubMed] [Google Scholar]
- Sucato, C. 2001. Monte Carlo simulations of locally denatured, closed double-stranded DNA at low salt concentration. MS thesis. University of Washington, Seattle, WA.
- Taylor, M. C., J. A. Grigg, and I. H. Laban. 1996. Triolborates and the aminoalcohol derivatives of boric acid: Their formation and hydrolysis. Polyhedron. 15:3261–3270. [Google Scholar]
- White, J. H. 1969. Self-linking and the Gauss integral in higher dimensions. Am. J. Math. 91:693–728. [Google Scholar]
- Wu, P.-G., B. S. Fujimoto, L. Song, and J. M. Schurr. 1991. Effect of ethidium on the torsion constants of linear and supercoiled DNAs. Biophys. Chem. 41:217–236. [DOI] [PubMed] [Google Scholar]
- Wu, P.-G., and J. M. Schurr. 1989. Effects of chloroquine on the torsional dynamics and rigidities of linear and supercoiled DNAs at low ionic strength. Biopolymers. 28:1695–1703. [DOI] [PubMed] [Google Scholar]
- Wu, P.-G., L. Song, J. B. Clendenning, B. S. Fujimoto, A. S. Benight, and J. M. Schurr. 1988. Interaction of chloroquine with linear and supercoiled DNAs. Effect on the torsional dynamics, rigidity, and twist energy parameter. Biochemistry. 27:8128–8144. [DOI] [PubMed] [Google Scholar]