Abstract
Tendons are composed of collagen and other molecules in a highly organized hierarchical assembly, leading to extraordinary mechanical properties. To probe the cross-links on the lower level of organization, we used a cantilever to pull substructures out of the assembly. Advanced force probe technology, using small cantilevers (length <20 μm), improved the force resolution into the sub-10 pN range. In the force versus extension curves, we found an exponential increase in force and two different periodic rupture events, one with strong bonds (jumps in force of several hundred pN) with a periodicity of 78 nm and one with weak bonds (jumps in force of <7 pN) with a periodicity of 22 nm. We demonstrate a good correlation between the measured mechanical behavior of collagen fibers and their appearance in the micrographs taken with the atomic force microscope.
INTRODUCTION
Tendons are among the most important stress-carrying structures in mammals. They have a key role in musculoskeletal biomechanics. The knowledge of the mechanical properties of tendons is important for the development of new materials based on biological polymers and also for an understanding of collagen-related diseases. Furthermore, the properties of collagen and associated polymers are very important to understand the structure and functional mechanisms of biocomposites such as bone and cartilage on the microscopic level.
Tendons have a hierarchical structure consisting of smaller entities called fascicles. These fascicles consist of fibrils, which again consist of sub- and/or microfibrils. The fibrils consist mainly of two components, collagen and proteoglycan (Scott, 1991). Type I collagen, the most abundant form of collagen (∼90%), is present in many tissues, such as bone, tendon, and skin. The type I collagen molecule is a triple-stranded coil consisting of two α1-chains and one α2-chain. Three of the left-handed coiled collagen molecules form a right-handed coiled triple helix called tropocollagen (Ramachandra and Karthan, 1955). For the structural organization of tropocollagen, there are different models discussed in literature. Most of the research on the structure and the mechanical properties of collagen has been done on type I collagen, which has the most well-organized structure of all collagen types. Type I collagen fibrils in their native form typically display a banding pattern with a periodicity of 67 nm, called D period, when visualized with transmission electron microscopy (Schmitt et al., 1942; Chapman and Hulmes, 1984) or AFM (Baselt et al., 1993; Revenko et al., 1994). The structural organization of the 67-nm banding is still a source of discussion. Petruska and Hodge developed a widely accepted model of the organization of collagen molecules into fibrils that was based on the results they obtained from transmission electron microscopy micrographs of negatively stained samples. The 67-nm periodicity was explained by a repetition of overlap and gap regions (Petruska and Hodge, 1964). In addition to the 67-nm banding, smaller and larger periodicities were also observed (Venturoni et al., 2003).
Collagen is described as an exceptional design for elastic-energy storage and as a modest design for strength and toughness (Gosline et al., 2002). Therefore, the knowledge of the function of collagen fibrils is of general interest for material researchers. The stress versus strain curve of tendons (Fig. 1) can be divided into five distinct regions (for review see Fratzl et al., 1998):
Toe region: A small strain leads to the removal of macroscopic crimps in the collagen fibrils, which is visible in the light microscope (Diamant et al., 1972).
Heel region: There is a straightening of kinks in the collagen structure at strains beyond 3%. This occurs first at the fibrillar and then at the molecular level by reducing the disorder in the lateral molecular packing.
Linear region: Higher strains lead to a stretching of collagen triple helices or of the cross-links between helices. This stretching increases the length of the gap region with respect to the length of the overlap region. Moreover, it was proposed that molecular gliding within the fibrils also plays a role because not all of the elongation could be explained by stretching of the fibrils.
Plateau region: This region is pronounced in cross-link deficient collagen and indicates creep behavior caused by slippage within fibrils (Puxkandl et al., 2002).
Rupture: Ultimately, a higher strain leads to a disruption of the structure of the fibril.
FIGURE 1.
Stress versus strain curve of a rat tail tendon. The stress-strain curve is divided into five regions: (A) Toe region, (B) heel region, (C) linear region, (D) plateau region, and (E) the rupture of the tendon.
Collagen fibrils are capable of reversible deformation. This property makes collagen an elastic protein (Gosline et al., 2002) with a resilience of ∼90% (Shadwick, 1990). Besides collagen, other molecules, like proteoglycans (Cribb and Scott, 1995), also play a role in the mechanical properties of tendon fibrils. It was proposed that these other molecules are important for the gliding of fibrils in the matrix (Puxkandl et al., 2002). All common models are based on the assumption of laterally homogeneous close packing of a collagen fibril, however, we recently showed that a collagen fibril is an inhomogeneous structure composed of a relatively hard shell and a less dense core (Gutsmann et al., 2003). Thus, the mechanical properties of the whole fibril depend on the properties of the shell and the core.
Besides the macroscopic stretching of whole tendons, Sun et al. (2002) used the optical tweezer technique to determine mechanical properties of single collagen monomers. The persistence length of collagen I monomers was determined to be 14.5 nm and the contour length was 309 nm (Sun et al., 2002). However, there is a big gap in determining mechanical properties between the stretching of single molecules and the stretching of whole tendons. The atomic force microscope has proven to be a powerful tool for the characterization of biological macromolecules through high-resolution topography imaging (Bustamante et al., 1997; Hansma et al., 1997) and single molecule force spectroscopy (Clausen-Schaumann et al., 2000; Zlatanova et al., 2000; Carrion-Vazquez et al., 2000). Force spectroscopy experiments have been performed on single molecules like titin (Oberhauser et al., 2001), DNA (Strunz et al., 1999), and polysaccharides (Marszalek et al., 2002). Becker et al. recently published data on pulling spider capture-silk molecules out of original fibers (Becker et al., 2003). We previously used an atomic force microscope to perform force spectroscopy experiments on collagen molecules that were not structured in fibrils (Thompson et al., 2001).
Here we present force spectra (pulling curves) of molecules pulled out of native fibrils from rat tail tendons. To get a high-force resolution we used a small-cantilever prototype AFM. The force curves show various characteristics such as two distinct periodicities in the rupture events, which we correlated with topography images of collagen fibrils.
MATERIAL AND METHODS
Tails from rats, sacrificed for other experiments, were frozen at −20°C typically for weeks, before our experiments. Rat tail tendons were removed from thawed rat tails and stored in a buffer containing 150 mM NaCl and 2 mM Tris adjusted to a pH of 7.4 for several hours before sample preparation.
Fibrils from rat tail tendons were prepared wet on a glass disc, and dried with a stream of filtered air. They were then imaged with an AFM (Veeco/Digital Instruments, Santa Barbara, CA) in contact mode using commercial cantilevers (CSC12, MikroMasch, Portland, OR).
The dried tendons were cut into pieces (∼7 mm long) and the two ends were glued to the glass disc with epoxy (2-Ton Clear Epoxy, Devcon, Danvers, MA). The fibrils were rehydrated in a buffer containing 150 mM NaCl, 2 mM Tris adjusted to a pH of 7.4. The pulling experiments were conducted using different types of conventional and small cantilevers (length <20 μm; width <8 μm) and different types of AFM: a prototype small-cantilever AFM, a Veeco/Digital Instruments multimode, and a molecular force puller MFP1 (Asylum Research, Santa Barbara, CA). We used different controls for the z-position, such as strain gauges and the sensored z axis of the MFP1. All data shown in this article was conducted using the prototype small-cantilever AFM. The cantilevers that were typically used had spring constants of ∼25 mN/m, and resonance frequencies of ∼40 kHz, which were determined by the measurement of the thermal fluctuations of the cantilevers. On the cantilevers, a 0.5-μm-long tip was deposited by electron beam deposition. The pulling curves shown were taken with loading rates of 7 or 14 μm/s.
For the histogram in Fig. 3 C we used data from 25 randomly chosen pulling curves. The distances between consecutive significant rupture events (rupture events with a change in force of >5 pN) were collected. No distances below 16 nm were included because the evaluation of some of these short distances was not precise enough due to noise, bending of the cantilever, and other influences. No events were collected in the very complex initial phase (200–300 nm), where several uncontrollable factors make an interpretation of the data unreliable.
FIGURE 3.
Force spectroscopy of rat tail tendon fibrils. Different shapes of curves can be found in the force versus extension curves. (A) A reversible increase to high force values shows hysteresis; (B) steep increase in force that may be due to stretching individual molecules. (C) The histogram of the distances between rupture events shows two peaks, one around 23 (arrow a) and one around 77 nm (arrow b). The inset shows the histogram of the same data, but with twice the bin size to demonstrate the significance of the two peaks. Most of these two distinct distances were observed in repeating patterns.
RESULTS
Before performing the pulling experiments, we imaged the dry samples and found two different banding periodicities of the collagen fibrils: the 65-nm D-spacing (of dried collagen fibrils) that is typical for collagen fibrils and a small 23-nm spacing (Fig. 2), which occurred in <3% of the collagen fibrils. For the pulling experiments the collagen fibrils were rehydrated.
FIGURE 2.
AFM images of rat tail tendon collagen. (A) A typical collagen fiber showing the 67-nm D-spacing; (B) fiber showing a 23-nm banding pattern, which is observable in <3% of the fibrils. These images show the AFM deflection signals.
Most of the force (F) versus extension (z) curves showed very complex structures containing different features. Fig. 3 A shows a force-extension curve recorded after pressing the cantilever tip into the collagen fibril for >10 s. In many cases, the force increased up to several nN and showed a small hysteresis effect (see Fig. 3 A). Due to the relatively high forces, we propose that these force-extension curves resulted from pulling on structural elements that were higher in the hierarchy of the collagen fibril than a microfibril.
In few force-extension curves, we observed relatively steep spikes (Fig. 3 B). The force increased over a distance of 10 nm by a force of ∼400 pN. Because of the complex initial phase of each of these spikes, it is not possible to perform a reliable worm-like chain (WLC) or other fit.
Most curves showed a superposition of different features making it impossible to describe all of them in detail. However, a histogram of the distances between rupture events (Fig. 3 C) shows two distinct distances, one around 23 and one around 77 nm. In most cases the two distinct distances appear in repeating patterns that are described below. It is important to notice that the observed ruptures are irreversible; once a bond breaks it will not reform again. A histogram of the respective peak forces did not show specific levels of forces (data not shown).
In Fig. 4 the shape of the force-extension curve resembles an exponential increase. In numerous previous force spectroscopy studies, the WLC model was used to characterize mechanical unfolding of proteins (Oberhauser et al., 1998; Rief et al., 1999; Fisher et al., 1999; Oroudjev et al., 2002). The WLC describes the relationship between the protein extension and the entropic force generated as a result of such extension. However, fitting our data with this model led to values of persistence lengths of <10 pm, which is below the length of a single atom, thus making this model not suitable. This is also true for the (extended) Langevin function (Rief et al., 1997). The force curve F(z) is well described by an exponential function:
 |
(1) |
with F0 being the initial force starting the stretching on one individual part of the structure, FP being the rupture force, zP being the peak position, and D being a characteristic length. We introduced F0 because when pulling on assemblies of molecules, it is sometimes possible that parallel substructures of the whole assembly will only begin to stretch at forces starting higher than zero. For the pulling curve in Fig. 4 B we determined the characteristic length D to be 80 ± 3 nm. The jumps down to zero force were not reversible, thus, they were ruptures and not a reversible unfolding of proteins like titin. However, we do not know whether this rupture originates from the breaking of bonds between the tip and the molecule or from the breaking of bonds between two or more molecules. It is unlikely that covalent bonds in the molecules break.
FIGURE 4.
Exponential increase in force versus extension curves. Some of the observed force versus extension curves can be fitted by an exponential function with a characteristic length D of ∼80 nm. (A) Overview of one pulling experiment on lin/log scale with the same exponential function fitted to different events showing that most of the events have the same characteristics. (B) Detail of the same experiment on lin/lin scale with an exponential fit.
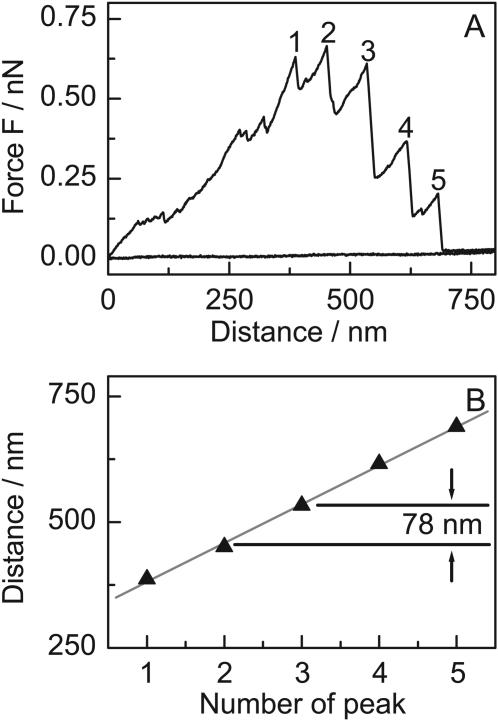
Interestingly, we found two different patterns of rupture events showing a distinct periodicity that clearly show up in the histogram (Fig. 3 C). The first pattern shows a periodicity of 78 nm and is easily visible in the histogram, an example is shown in Fig. 5. After an increase in force up to >500 pN, repeating rupture events occurred. They resulted in a stepped decrease in the maximum force reached before the breaking of each bond. The average distance between the rupture events was 78 ± 3 nm (Fig. 5 B). In several hundred pulling curves, we never observed more then five of these repeating events in one pulling curve.
FIGURE 5.
Stepwise rupture of bonds in the collagen fiber. In some pulling curves, a regular pattern with an average distance between the rupture events of 78 ± 3 nm can be observed. This pattern is statistically significant and is clearly visible in the distance histogram (Fig. 3 C). The jumps in force are not reversible and they always lead to a stepwise decrease of the force versus distance curve.
The second periodicity is barely detectable because the jumps in force were <7 pN. These measurements became possible with the use of low spring-constant, high-resonance frequency, small cantilevers (Viani et al., 1999). These rupture events occurred in a quasilinear region of the force-extension curve (Fig. 6 A). The average distance between these rupture events was 22 ± 2 nm (Fig. 6 B) and the average increase in force between two consecutive events was 53 ± 4 pN. The overall linear increase does not allow the direct conclusion that it resulted from a linear stretching of the molecules. The shape of the curves between two rupture events appears to be more linear than exponential. However, the distance between the ruptures is too short to perform a precise analysis, even with the high-force resolution achieved by using a small cantilever.
FIGURE 6.
Small rupture events in the linear-like region. In most of the experiments, the linear-like increase in force is due to very small jumps in force. These jumps are ∼7 pN every 22 nm corresponding to every 53 pN. This 22-nm periodicity is also statistically significant and appears as a spike in the distance histogram (Fig. 3 C).
DISCUSSION
Collagen fibrils are important for the function of musculoskeletal biomechanics in various species. The mechanical properties of whole tendons, as well as the chemical composition and the topography of collagen fibrils, were studied in detail. However, very little is known about the mechanical properties of the fibrils on the molecular level. Knowledge of this will lead to a better understanding of the binding between tropocollagen molecules, how they assemble to fibrils, and with that an understanding of the topography as well as the macroscopic mechanical behavior. Later, we will discuss our results along with the topography of the fibrils and the macroscopic stress versus strain curves of tendon.
Exponential increase in force
The extension of collagen fibrils inside the tendon is considerably less than the total extension of the tendon. Puxkandl et al. proposed that the deformation must occur outside the collagen fibrils, presumably in the proteoglycan-rich matrix (Puxkandl et al., 2002). Thus, the tendon is considered as a composite material with collagen fibrils embedded in a proteoglycan-rich matrix.
There are relatively few theoretical models describing the rupture of multiple parallel bonds under dynamic loading (Seifert, 2000) and most of them simplify the model by using Hookean springs with zero rest length. Puxkandl et al. introduced a Voight-Kelvin mechanical model consisting of parallel elastic (spring) and viscous (dashpot) portions for the collagen fibrils, as well as for the proteoglycan matrix (Puxkandl et al., 2002). This model leads to an exponential stress versus strain function. This is in agreement with the exponential function we observed in the pulling curves. The most likely explanation on the molecular level is a stretching of whole assemblies of collagen and other components of the fibrils, e.g., proteoglycan. The stretching of individual molecules, molecular cross-links and matrix shearing all probably contribute to the pulling forces. However, without a suitable model, it is difficult to make an unambiguous connection between the characteristic length D and the average distance between the repeating rupture pattern of ∼80 nm. We suggest that the described mechanical behavior on the molecular level corresponds mainly to the heel and/or linear region on the macroscopic level (Fig. 1). We exclude the other regions because the toe region corresponds to the removal of macroscopic crimps in the collagen fibrils and the plateau region corresponds to the slippage within fibrils, neither of which lead to an exponential increase in force.
Repeating rupture pattern of 78 nm
The periodicity of 78 ± 3 nm between rupture events in the force spectra (Fig. 5) shows that there are bonds between molecules in the fibrils with a distance of ∼78 nm. We believe that this pattern results from pulling an assembly of molecules out of fibrils, followed by a sequential rupture of individual subunits. Most likely the molecules are tropocollagen, however, it is possible that other molecules, like proteoglycans, participate. Therefore, the decrease of the peak forces with each successive rupture event can be explained by the fact that when a bond ruptures, the force on the remaining bonds increases. Thus, the overall force that needs to be applied to a bundle of molecules to break the weakest bond in the bundle decreases with the number of force-carrying subunits in the bundle. For the interpretation of our data, the precise knowledge of the involved molecules is not so important because we focused on the structure of entire fibrils.
The measured periodicity of 78 nm is ∼16% longer as compared to the 67-nm D-banding obtained from the topography of wet collagen fibrils. The 67 nm is the projection of the wavy fibril structure onto the x-y plane and, with a height corrugation of at least 3–7 nm. The surface distance is therefore longer than 67 nm by at least 6 nm (Venturoni et al., 2003; Lin and Goh, 2002). Furthermore, it was proposed that the collagen molecules are tilted by ∼16° to the fibril's axis leading to a length of 70 nm. These two effects lead to a length of at least 76 nm. Katsura and Ono proposed that the precoiled length of D in the α-chain is ∼81 nm and the length of D in the tropocollagen triple helix is ∼75 nm (Katsura and Ono, 1998). This is in agreement with our data suggesting a periodicity of bonds between the molecules every 78 nm. We propose that there is a direct correlation between the topography of the collagen fibrils and the force spectra.
In vivo, the enzyme lysyl oxidase converts certain lysine and hydroxylysine amino groups into aldehydes. Reactions that occur between the aldehydic groups and other amino acids of adjoining molecules lead to cross-links between the molecules and the self-assembly into fibers (Tanzer, 1976). However, the cross-link locations are still under discussion. Based on our data, we propose that one distinct distance between two binding positions of stretched tropocollagen is ∼78 nm. It was proposed that in the linear region of the macroscopic pulling experiments (Fig. 1) a higher strain results from the stretching of collagen triple helices or of the cross-links between helices. Thus, in the molecular pulling curve, the increase in force before a rupture event corresponds to this macroscopic linear region.
Based on our results, we can only speculate if the observation that tendons rupture at a strain of ∼15% and the observed periodicity in rupture events of 78 nm being 16% longer than the 67-nm D-banding is only a coincidence. And with that the question if these molecular ruptures occur in the linear region, plateau region, or when the tendon ruptures, remains open (Fig. 1).
The force necessary to pull molecules out of a fibril is similar to the force necessary to pull a single proteomer out of a hexagonally packed intermediate layer, which is 312 ± 43 pN (Müller et al., 1999).
Repeating rupture pattern of 22 nm
The small jumps in force of <7 pN in the linear-like increase region of the force-extension curves, are remarkable because of two reasons: first, they modulate the increase in force, and second, the jumps in force are at the low end of the forces determined. The distance between the rupture events is ∼22 ± 2 nm (Fig. 6), which correlates to the banding pattern of ∼23 nm we found in a few images (Fig. 2 B). As discussed earlier by Venturoni et al. (2003), we propose that there is a major binding distance leading to the 67-nm D-banding and, in <3% of the fibrils, a minor distance of weaker bonds leading to the 23-nm banding. Perhaps, this small banding pattern and the periodicity in the rupture events arise from the same weak interactions between molecules of the fibrils.
Because of the very small jump forces of <7 pN, we propose that these rupture events occur due to a gliding within the fibrils, as proposed for the linear region of macroscopic pulling experiments (Fig. 1). Collagen, like lustrin in abalone shell, spider dragline silk, or titin in muscles, appears to derive much of its combination of strength and toughness from its modular sacrificial bonds (Fisher et al., 1999; Oberhauser et al., 2001; Oroudjev et al., 2002; Smith et al., 1999; Thompson et al., 2001). Electrostatic interactions between the molecules in fibrils, and perhaps other forces, could play a role in the cross-linking between the molecules. However, a mechanism of “hidden length” due to three-dimensional structure, as in titin, is very unlikely.
Single molecule stretching
The steepness of the spikes we observed (Fig. 3 B) indicates that they originate from the stretching of very stiff molecules or small units of the collagen fibrils. It is most likely that they occurred while stretching tropocollagen molecules. The force values for stretching the molecules are high as compared to other structural proteins, such as tenascin (Oberhauser et al., 1998), titin (Fisher et al., 1999), and pS(4 + 1) spider silk protein, which is 176 ± 73 pN (Oroudjev et al., 2002). The rupture force for the helix bundles of spectrin are only between 25 and 35 pN (Rief et al., 1999), which is much lower than the rupture force we determined.
SUMMARY
In summary, the extraordinary mechanical properties of collagen fibers can be investigated by performing force spectroscopy experiments. The most important result of our investigation is the correlation between periodic patterns in the topography of rat tail tendon collagen fibrils and the binding patterns observed in the force spectroscopy experiments on these molecules.
By using cantilevers, it is possible to pull individual subunits out of collagen fibers from tendon. However, the low noise performance of a small cantilever (length <20 μm) gives more details than the conventional cantilever. The force versus extension curves are complex and show different features, e.g., an exponential increase in force that cannot be fitted by common models, like the worm-like chain model. The fact that we typically did not see reformation of bonds if we waited with the cantilever above the surface, is in marked contrast to the situation for titin and other modular proteins with compact domains (Clausen-Schaumann et al., 2000; Zlatanova et al., 2000; Carrion-Vazquez et al., 2000; Oberhauser et al., 2001). It is also in contrast to the situation for lustrin (Smith et al., 1999) and disordered collagen (Thompson et al., 2001).
Two types of repeating rupture events occur with periodicities of 78 nm and 22 nm. It is therefore likely that there are bonds, which break at forces >100 pN, between subunits in fibrils every 78 nm (stretched length) as well as very weak bonds, which break at forces <10 pN, every 22 nm. Both types of bonds, however, break with forces less than we would expect for breaking a collagen backbone, which should take forces larger than 1000 pN. The histogram (Fig. 3 C) shows a more distinct peak at 22 nm than at 78 nm. This is a statistical disparity, because the 23-nm banding appeared in <3% of the images of collagen fibrils. However, the forces needed to break the bonds with the 22-nm distance were significantly smaller as compared to the forces needed to break the 78-nm bonds. Thus, we propose that both periodicities are present in all fibrils. In most cases the strong bonds with the 78-nm periodicity are dominant in the formation of the fibrils, which leads to the more common 67-nm banding in the images. Whether or not the bonds result solely from the collagen proteins or from other molecules of the matrix is not clear yet. Most likely we investigated the structural properties of the shell of the collagen fibrils in our experiments. The structural properties of the softer core could therefore be different. This concept of bonds with different strength and periodicities that lead to an optimization of mechanical properties could be the basis for new materials.
Acknowledgments
We thank Herbert Waite and Herman Gaub for helpful discussions and Jacqueline Cutroni and Sanje Woodsorrel for helpful assistance. We are grateful to Diane McClure and Amber Griffin of the campus animal resource center for saving rat tails for us from rats sacrificed for other experiments.
This work was supported by the National Science Foundation, through the MRSEC program (award number DMR 9988640) and the MRL program (award number DMR00-80034), the National Institutes of Health (award number GM65354), NASA/URETI on Bio Inspired Materials (award number NCC-1-02037), the University of Bologna, the Erzherzog-Johann-University Graz, and the Deutsche Forschungsgemeinschaft (project GU 568/2-1).
Thomas Gutsmann's present address is Research Center Borstel, Leibniz Center for Medicine and Biosciences, Dept. of Immunochemistry and Biochemical Microbiology, Div. of Biophysics, Parkallee 10, D-23845 Borstel, Germany.
References
- Baselt, D. R., J. P. Revel, and J. D. Baldeschwieler. 1993. Subfibrillar structure of type I collagen observed by atomic force microscopy. Biophys. J. 65:2644–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, N., E. Oroudjev, S. Mutz, J. P. Cleveland, P. K. Hansma, C. Y. Hayashi, D. E. Makarov, and H. G. Hansma. 2003. Molecular nanosprings in spider capture-silk threads. Nat Mater. 2:278–283. [DOI] [PubMed] [Google Scholar]
- Bustamante, C., C. Rivetti, and D. J. Keller. 1997. Scanning force microscopy under aqueous solutions. Curr. Opin. Struct. Biol. 7:709–716. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez, M., A. F. Oberhauser, T. E. Fisher, P. E. Marszalek, H. Li, and J. M. Fernandez. 2000. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 74:63–91. [DOI] [PubMed] [Google Scholar]
- Chapman, J. A., and D. J. S. Hulmes. 1984. Electron microscopy of the collagen fibril. In Ultrastructure of the Connective Tissue Matrix. P. M. Motta and A. Ruggeri, editors. Kluwer Academic Publishers, Boston, MA. 1–33.
- Clausen-Schaumann, H., M. Seitz, R. Krautbauer, and H. E. Gaub. 2000. Force spectroscopy with single bio-molecules. Curr. Opin. Chem. Biol. 4:524–530. [DOI] [PubMed] [Google Scholar]
- Cribb, A. M., and J. E. Scott. 1995. Tendon response to tensile stress: an ultrastructural investigation of collagen:proteoglycan interactions in stressed tendon. J. Anat. 187:423–428. [PMC free article] [PubMed] [Google Scholar]
- Diamant, J., A. Keller, E. Baer, M. Litt, and R. G. Arridge. 1972. Collagen; ultrastructure and its relation to mechanical properties as a function of ageing. Proc. R. Soc. Lond. B Biol. Sci. 180:293–315. [DOI] [PubMed] [Google Scholar]
- Fisher, T. E., A. F. Oberhauser, M. Carrion-Vazquez, P. E. Marszalek, and J. M. Fernandez. 1999. The study of protein mechanics with the atomic force microscope. Trends Biochem. Sci. 24:379–384. [DOI] [PubMed] [Google Scholar]
- Fratzl, P., K. Misof, I. Zizak, G. Rapp, H. Amenitsch, and S. Bernstorff. 1998. Fibrillar structure and mechanical properties of collagen. J. Struct. Biol. 122:119–122. [DOI] [PubMed] [Google Scholar]
- Gosline, J., M. Lillie, E. Carrington, P. Guerette, C. Ortlepp, and K. Savage. 2002. Elastic proteins: biological roles and mechanical properties. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357:121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsmann, T., G. E. Fantner, M. Venturoni, A. Ekani-Nkodo, J. B. Thompson, J. H. Kindt, D. E. Morse, D. K. Fygenson, and P. K. Hansma. 2003. Evidence that collagen fibrils in tendons are inhomogeneously structured in a tubelike manner. Biophys. J. 84:2593–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma, H. G., K. J. Kim, D. E. Laney, R. A. Garcia, M. Argaman, M. J. Allen, and S. M. Parsons. 1997. Properties of biomolecules measured from atomic force microscope images: a review. J. Struct. Biol. 119:99–108. [DOI] [PubMed] [Google Scholar]
- Katsura, N., and T. Ono. 1998. A noval helical filament model of hen tendon collagen fibril. In Extracellular Matrix-Cellular Interactions: Molecules to Diseases. Y. Ninomiya, editor. Japan Sci. Soc. Press/S. Karger, Tokyo, Japan/Basel, Switzerland. 185–202.
- Lin, A. C., and M. C. Goh. 2002. Investigating the ultrastructure of fibrous long spacing collagen by parallel atomic force and transmission electron microscopy. Proteins. 49:378–384. [DOI] [PubMed] [Google Scholar]
- Marszalek, P. E., H. Li, A. F. Oberhauser, and J. M. Fernandez. 2002. Chair-boat transitions in single polysaccharide molecules observed with force-ramp AFM. Proc. Natl. Acad. Sci. USA. 99:4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, D. J., W. Baumeister, and A. Engel. 1999. Controlled unzipping of a bacterial surface layer with atomic force microscopy. Proc. Natl. Acad. Sci. USA. 96:13170–13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhauser, A. F., P. K. Hansma, M. Carrion-Vazquez, and J. M. Fernandez. 2001. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA. 98:468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhauser, A. F., P. E. Marszalek, H. P. Erickson, and J. M. Fernandez. 1998. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 393:181–185. [DOI] [PubMed] [Google Scholar]
- Oroudjev, E., J. Soares, S. Arcdiacono, J. B. Thompson, S. A. Fossey, and H. G. Hansma. 2002. Segmented nanofibers of spider dragline silk: atomic force microscopy and single-molecule force spectroscopy. Proc. Natl. Acad. Sci. USA. 99(Suppl 2):6460–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruska, J. A., and A. J. Hodge. 1964. A subunit model for the tropocollagen macromolecule. Proc. Natl. Acad. Sci. USA. 51:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puxkandl, R., I. Zizak, O. Paris, J. Keckes, W. Tesch, S. Bernstorff, P. Purslow, and P. Fratzl. 2002. Viscoelastic properties of collagen: synchrotron radiation investigations and structural model. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandra, G. N., and G. Karthan. 1955. Structure of collagen. Nature. 176:593–595. [DOI] [PubMed] [Google Scholar]
- Revenko, I., F. Sommer, D. T. Minh, R. Garrone, and J. M. Franc. 1994. Atomic force microscopy study of the collagen fibre structure. Biol. Cell. 80:67–69. [DOI] [PubMed] [Google Scholar]
- Rief, M., F. Oesterhelt, B. Heymann, and H. E. Gaub. 1997. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science. 275:1295–1297. [DOI] [PubMed] [Google Scholar]
- Rief, M., J. Pascual, M. Saraste, and H. E. Gaub. 1999. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286:553–561. [DOI] [PubMed] [Google Scholar]
- Schmitt, F. D., C. E. Hall, and M. A. Jakns. 1942. Electron microscopy investigations of the structure of collagens. J. Cell. Comp. Physiol. 20:11–33. [Google Scholar]
- Scott, J. E. 1991. Proteoglycan: collagen interactions and corneal ultrastructure. Biochem. Soc. Trans. 19:877–881. [DOI] [PubMed] [Google Scholar]
- Seifert, U. 2000. Rupture of multiple parallel molecular bonds under dynamic loading. Phys. Rev. Lett. 84:2750–2753. [DOI] [PubMed] [Google Scholar]
- Shadwick, R. E. 1990. Elastic energy storage in tendons: mechanical differences related to function and age. J. Appl. Physiol. 68:1033–1040. [DOI] [PubMed] [Google Scholar]
- Smith, B. L., T. E. Schäffer, M. Viani, J. B. Thompson, N. A. Frederick, J. Kindt, A. Belcher, G. D. Stucky, D. E. Morse, and P. K. Hansma. 1999. Molecular mechanistic origin of the toughness of natural adhesives, fibres and composites. Nature. 399:761–763. [Google Scholar]
- Strunz, T., K. Oroszlan, R. Schafer, and H. J. Guntherodt. 1999. Dynamic force spectroscopy of single DNA molecules. Proc. Natl. Acad. Sci. USA. 96:11277–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. L., Z. P. Luo, A. Fertala, and K. N. An. 2002. Direct quantification of the flexibility of type I collagen monomer. Biochem. Biophys. Res. Commun. 295:382–386. [DOI] [PubMed] [Google Scholar]
- Tanzer, M. L. 1976. Cross-Linking. In Biochemistry of Collagen. G. N. Ramachandra and H. Redl, editors. Plenum Press, New York. 137–62.
- Thompson, J. B., J. H. Kindt, B. Drake, H. G. Hansma, D. E. Morse, and P. K. Hansma. 2001. Bone indentation recovery time correlates with bond reforming time. Nature. 414:773–776. [DOI] [PubMed] [Google Scholar]
- Venturoni, M., T. Gutsmann, G. E. Fantner, J. H. Kindt, and P. K. Hansma. 2003. Investigations into the polymorphism of rat tail tendon fibrils using atomic force microscopy. Biochem. Biophys. Res. Commun. 303:508–513. [DOI] [PubMed] [Google Scholar]
- Viani, M. B., T. E. Schaeffer, A. Chand, M. Rief, H. E. Gaub, and P. K. Hansma. 1999. Small cantilevers for force spectroscopy of single molecules. J. Appl. Phys. 86:2258–2262. [Google Scholar]
- Zlatanova, J., S. M. Lindsay, and S. H. Leuba. 2000. Single molecule force spectroscopy in biology using the atomic force microscope. Prog. Biophys. Mol. Biol. 74:37–61. [DOI] [PubMed] [Google Scholar]