Abstract
A molecular dynamics simulation of the active unfolding of denatured rhodanese by the chaperone GroEL is presented. The compact denatured protein is bound initially to the cis cavity and forms stable contacts with several of the subunits. As the cis ring apical domains of GroEL undergo the transition from the closed to the more open (ATP-bound) state, they exert a force on rhodanese that leads to the increased unfolding of certain loops. The contacts between GroEL and rhodanese are analyzed and their variation during the GroEL transition is shown. The major contacts, which give rise to the stretching force, are found to be similar to those observed in crystal structures of peptides bound to the apical domains. The results of the simulation show that multidomain interactions play an essential role, in accord with experiments. Implications of the results for mutation experiments and for the action of GroEL are discussed.
INTRODUCTION
Although many proteins fold spontaneously in dilute solution (Anfinsen, 1973), the folding process is complicated in the cellular medium due to the high concentration of other molecules (Ellis and Hartl, 1999). They can interfere with the folding to the native state and may cause misfolding or aggregation of denatured proteins, leading to disease and death in some cases (Dobson, 2002). One mechanism to prevent such problems in the cell is the protection of the newly synthesized protein chains by chaperones (Hartl, 1996; Saibil, 2000). The best studied chaperone is the bacterial chaperonin, GroEL, a large protein shaped like a double ring with dyad symmetry under certain conditions (Boisvert et al., 1996; Xu et al., 1997; Grallert and Buchner, 2001). Each ring is composed of seven identical subunits, which enclose a central cavity. Each subunit consists of three domains. The interactions between the two rings arise from the equatorial domains, which contain an adenosine triphosphate (ATP) binding site, and the smaller intermediate domains that connect the equatorial and the apical domains. The apical domains form the entrance of the rings and are important for substrate binding (Fenton et al., 1994). Experiments have shown that the two seven-membered rings have large conformational changes, which alternate in the GroEL cycle involved in the folding of protein substrates. The denatured protein substrate is first bound to one of the rings (the cis ring) (Weissman et al., 1996; Rye et al., 1997, 1999) in the closed (or t) state; the binding of a protein to GroEL in this state appears to be dominated by interactions with the exposed H and I helices of the apical domains (Fenton et al., 1994; Buckle et al., 1997; Chen and Sigler, 1999). Binding of ATP to the equatorial domains of the cis ring initiates a downward motion of the intermediate domain, which triggers the subsequent steps of the GroEL cycle. The cis cavity expands primarily through motion of the apical domains to form the partly open (r′) state (Chen et al., 1994; Roseman et al., 1996; Llorca et al., 1997; Ma and Karplus, 1998; Ranson et al., 2001; Ma et al., 2000). The cochaperone GroES then binds (Chen et al., 1994; Roseman et al., 1996; Llorca et al., 1997) to close the top of the cis cavity while inducing an additional displacement of the apical domains that further enlarges the cavity in the open (r″) state (Chen et al., 1994; Roseman et al., 1996; Llorca et al., 1997; Xu et al., 1997). This leads to the release of the protein into the cavity, since GroES competes for the substrate-binding H and I helices of the apical domains (Xu et al., 1997). Hydrolysis of ATP and binding of denatured protein and ATP to the other (trans) ring subsequently leads to the release of GroES, adenosine diphosphate (ADP), and the protein from the cis cavity by a cooperative mechanism (Rye et al., 1997, 1999). It is believed that several binding-release cycles are required in many cases to yield fully folded and functional protein (M. J. Todd, 1994; Weissman et al., 1994). Each cycle takes ∼15 s (M. J. Todd, 1994) and consumes seven ATP molecules (M. J. Todd, 1994; Rye et al., 1999).
The GroEL transition is initiated by ATP binding to the cis ring (Inobe et al., 2001). It has been shown by simulations (Ma and Karplus, 1998; Ma et al., 2000) and confirmed by cryoelectron microscopy (Ranson et al., 2001) that binding of ATP results in a downward twisting motion of the intermediate domain that is the trigger for the major conformational changes. The intermediate domain displacement closes the ATP binding pocket, releases the apical domain to permit its upward motion, and pushes downward on the equatorial domain (Ma and Karplus, 1998; Ma et al., 2000). The r′ state is reached by a small upward motion and, looking down from the top of the cis ring, a small clockwise rotation of the apical domains. This is accompanied by a counterclockwise twist of the equatorial domains of the cis ring. The r′-to-r″ transition consists mainly of a further clockwise rotation and upward tilt of the apical domains. The motion has been shown to be highly cooperative within the cis ring (Thirumalai and Lorimer, 2001), due to steric and electrostatic effects (Ma et al., 2000; Ranson et al., 2001). The steric effects are due to van der Waals repulsions, which can be avoided only by a concerted motion of the seven subunits in the cis ring. The electrostatic effects involve an intraring, intersubunit salt bridge between Glu-386 and Arg-197 (Ma et al., 2000; Ranson et al., 2001), which is broken by the intermediate domain motion. Anticooperativity between the rings is primarily due to steric effects. The twisting of the equatorial domains upon ATP binding would result in severe van der Waals clashes if binding occurred in both rings (Ma et al., 2000). Overall, the observed allosteric pathway is the result of coupled tertiary structure changes, rather than quaternary structural effects (Ma and Karplus, 1998).
Although GroEL has an essential role in the folding of many proteins in Escherichia coli (Houry et al., 1999), it is still unclear what the chaperone system does. One hypothesis is that the major function of GroEL is to prevent aggregation by providing a shielded environment (“Anfinsen cage”) for folding. This “passive” mechanism is supported by structural data, which shows that the lining of the cis cavity changes from hydrophobic in the closed state to hydrophilic in the open state (Xu et al., 1997). Unfolded or misfolded proteins, which have exposed hydrophobic patches, bind to the closed state. Opening of the cis cavity releases the protein into a more hydrophilic environment, where the protein can fold spontaneously, without the possibility of aggregation. In addition, it has been suggested that the confinement of a protein substrate like ribulose-1,5-biphosphate carboxylase-oxygenase (RuBisCo) could speed up the folding rate by eliminating kinetically trapped intermediates (Brinker et al., 2001). It has also been proposed that misfolded conformations of the protein substrate are partly unfolded by GroEL (Zahn et al., 1996a,b; M. Todd et al., 1996; Walter et al., 1996; Shtilerman et al., 1999; Thirumalai and Lorimer, 2001; Hammarström et al., 2000, 2001). The unfolding could originate from the preferential binding of the denatured state to GroEL (Zahn et al., 1996a,b; M. Todd et al., 1996; Walter et al., 1996). Alternatively, the mechanical force generated by the interactions between the protein substrate and GroEL during the opening motion in the t-to-r′ transition could “pull” the protein into a more unfolded state (Thirumalai and Lorimer, 2001; Shtilerman et al., 1999; Wang and Boisvert, 2003). Transient, asymmetric states in which not all ATP binding sites of the cis ring are occupied by the protein could play a role (Wang and Boisvert, 2003). Clearly the prevention of aggregation does occur, but the contribution of active unfolding is still debated (Saibil et al., 2002). Hydrogen exchange experiments indicated that RuBisCo (Shtilerman et al., 1999) and barnase (Zahn et al., 1996b) are unfolded by GroEL, but corresponding studies of other proteins did not show such an effect (Groß et al., 1996; Chen et al., 2001). Also, lattice model calculations, although far from a realistic description of the system, have shown that a hydrophobic environment can pull an incorrectly folded protein model apart under certain conditions (Chan and Dill, 1996; Betancourt and Thirumalai, 1999), and that such unfolding can increase the yield of native protein in the chaperone cycle (Chan and Dill, 1996; Betancourt and Thirumalai, 1999; Sfatos et al., 1996).
To investigate whether active unfolding can occur as part of the GroEL cycle, we have performed molecular dynamics simulations with an atomic resolution model of the opening transition of the cis ring in the presence of denatured rhodanese as the substrate. The study concentrated on the transition from the closed-to-r′ state, before the protein is released into the cis cavity, since this is the most likely phase for an unfolding interaction to occur in the GroEL cycle. Since the opening motion is on the submillisecond timescale (Inobe et al., 2003), while simulations are limited to nanoseconds (Inobe et al., 2003), a staging method was introduced. Each step in the opening motion of the apical domains was induced by targeted molecular dynamics (Schlitter et al., 1993), and followed by a ten times longer relaxation phase in which the protein substrate was allowed to respond to the change in the interactions. During the simulation of the GroEL transition from the t to the r′ state, significant unfolding of rhodanese occurred. The contacts between GroEL and rhodanese gave rise to a stretching force, which increased the unfolding of the substrate.
METHODS
There are two difficulties in simulating the opening transition of GroEL in the presence of a protein substrate. The first is that GroEL is a large system (∼66,000 atoms in a polar hydrogen model and a total of ∼250,000 atoms if a reasonable explicit water environment is included). Consequently, we used only the apical domains of the cis ring (in the t and r′ states, prior to the release into the cis cavity, the substrate interacts only with the apical domains), and employed an implicit solvent model (EEF1) (Lazaridis and Karplus, 1999). The quality of the implicit solvent model was verified by test simulations on a GroEL-peptide system and comparison of the root mean-square difference (RMSD), the calculated B-factors, and the residues involved in binding with explicit water simulations and experimental results.
The second problem is that the opening motion is likely to be on the millisecond timescale; small-angle x-ray scattering and fluorescence studies indicate that the timescale is on the submillisecond range (Inobe et al., 2003), whereas nanosecond-length simulations are the limit for large systems (Böckmann and Grubmüller, 2002). Consequently, a staging method was introduced in which each step in the opening motion of the apical domains was induced by targeted molecular dynamics (TMD) (Schlitter et al., 1993), and followed by a ten times longer relaxation phase in which the protein substrate was allowed to respond to the change in interactions. The TMD method generates a trajectory from a known initial structure to a known target structure by use of a force that decreases the RMSD with a preset value at each step (Schlitter et al., 1993). The force was applied only to the Cα atoms of GroEL; all other atoms were treated by unbiased molecular dynamics. During the relaxation phase, the system was propagated by 10 steps of unbiased molecular dynamics while keeping the GroEL Cα atoms fixed.
Rhodanese (Protein Data Bank code 1RHS) (Gliubich et al., 1996) was unfolded by a high-temperature (550 K) denaturing simulation of 2.1 ns. From this simulation, the structure with the largest radius of gyration was selected (28.8 Å; the radius of gyration of the native state is 18.8 Å), slowly cooled to 300 K, and further equilibrated for 2 ns. A complex of rhodanese and GroEL was constructed by randomly placing the unfolded rhodanese on top of the minimized closed-state cis ring (Protein Data Bank code 1DER) (Boisvert et al., 1996) (Fig. 1). Only the apical domains of GroEL were included and care was taken to avoid initial contacts between rhodanese and GroEL. The complex was slowly heated to 300 K and equilibrated for 1.2 ns, while harmonically restraining the GroEL Cα atoms.
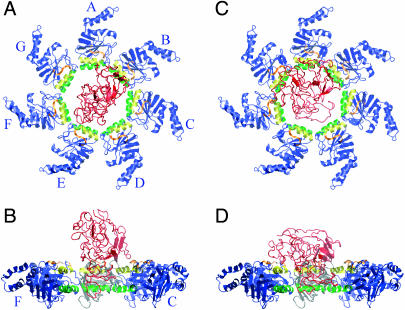
FIGURE 1.
Snapshots of the initial configurations of rhodanese. A and B show the starting configuration for the study of denatured rhodanese and closed-state GroEL; there are no contacts between GroEL and rhodanese in this configuration. C and D show the starting configurations for the TMD simulation; these are the endpoints of the closed-state simulation. The overall motion of rhodanese during the closed-state simulation is a translation by 10.7 Å and a 45.9° rotation. The H helices of GroEL are colored yellow, the I helices are green, the loop formed by residues 310–315 is orange, and the rest of the GroEL apical domains are blue. Rhodanese is shown in red. A and C show the top views, in which the A subunit is at the 12-o'clock position and the other subunits (B–G) follow in a clockwise fashion as indicated. B and D show the side views, obtained by a 90° rotation. For clarity, subunits D and E have been removed in B and D, and subunit A is colored gray. Figs. 1, 2, and 6–8 were prepared with VMD (Humphrey et al., 1996).
Normal mode analysis (Ma and Karplus, 1998) and a TMD simulation of the transition for a single GroEL subunit (Ma et al., 2000) had identified the r′ state as lying approximately halfway between the closed and open state. The r′state cis ring for this study was constructed by fitting the equatorial domain of the r′ state from the single-subunit TMD simulation to each equatorial domain of the closed-state cis ring of the crystal structure (Boisvert et al., 1996). After the fit, the equatorial and intermediate domains of the r′ state were removed, and the remaining apical domains were minimized. The trajectory from closed to r′ state was generated using four intermediate GroEL conformations; i.e., the new TMD simulation connected the chosen states (from the initial structure to the first intermediate, from the first intermediate to the second, and so on). These intermediate conformations were generated from the coordinate sets of the single-subunit TMD simulation (Ma et al., 2000), with the trajectory 10%, 20%, 30%, and 40% completed. Since the RMSD from the fully open (r″) state decreased linearly with time in the closed-to-open state single-subunit TMD simulation, we refer to these snapshots as the 10%, 20%, 30%, and 40% open states, respectively (the r′ state being the 50% open state). The total RMSD between the r′ and closed states along the intermediates is 23.1 Å. To avoid a problem arising from the collapse of the phase space (Schlitter et al., 1993), the transition was simulated only until a RMSD of 0.6 Å from each target was reached. Each TMD step decreased the RMSD by 0.0004 Å; no qualitative differences were observed for smaller steps.
The temperature was held within the 300 ± 3 K range by independent rescaling of the GroEL and rhodanese velocities after each TMD relaxation cycle, and a time step of 2 fs was used. The overall simulation time to go from the t to the r′ state was 1.2 ns. Bonds involving hydrogen atoms were fixed with the SHAKE algorithm (Ryckaert et al., 1977). All simulations were performed with the CHARMM program (Brooks et al., 1983); the solvent-accessible surface area was calculated by the Lee and Richards algorithm (Lee and Richards, 1971) with a 1.4 Å water probe. In total seven simulations, each with a different initial position and orientation of the rhodanese substrate, were performed.
RESULTS
In what follows, we describe one unfolding simulation in detail to illustrate the specific behavior and the types of interactions involved in the force generation; other simulations (not shown) are similar, though the interactions are not identical.
Binding of substrate to the closed (t)-state apical domains
After a denaturation simulation of rhodanese in isolation (see Methods), a starting structure for the GroEL opening simulation was generated by randomly placing the denatured rhodanese on top of the minimized closed-state cis ring and equilibrating the closed-state complex for 1.2 ns. During the first 900 ps of the closed-state simulation, the number of contacts between rhodanese and GroEL increased gradually; after that, the number and type of contacts remained roughly constant. The binding of rhodanese to closed-state GroEL was mostly through contacts with the H and I helices and with the 310-315 loops at the rim of the cis cavity (Fig. 1). Initial contacts with one apical domain quickly led to contacts with the neighboring domains, so that rhodanese closed off and partly penetrated into the cis cavity, although the molecule was too large to fit completely inside (∼22% of the rhodanese atoms were inside the cis cavity, where they occupied 14% of the apical cavity volume). All seven apical domains were involved in the binding, but most of the contacts were made between rhodanese and the A, D, E, and F apical domains (see Fig. 1). Residue-residue contacts within 4.0 Å between rhodanese and the individual subunits of GroEL during the last 100 ps of the closed-state simulation varied from 15.3 (A and E) to 5.7 (G) and 3.0 (C). The average number of hydrogen bonds between rhodanese and each of the A–G apical domains in this period was equal to 2.8, ranging from 1.5 (C) to 4.0 (D) for the different apical domains. The binding energy was provided by both van der Waals and electrostatic interactions, with the former 1.5-fold more negative than the latter. The majority of the van der Waals binding energy originated from interactions with the aliphatic portions of the Lys, Arg, and Glu side chains of the H and I helices of GroEL. These side chains also accounted for most of the hydrogen bonding between rhodanese and GroEL. Other hydrophobic interactions involved Ala, Ile, Leu, and Val residues of the H and I helices.
Opening transition
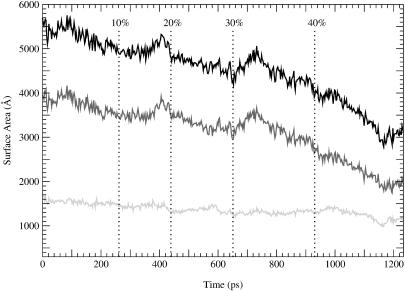
During the closed- to r′-state transition, rhodanese lost contact with two apical domains (Fig. 2). Contacts with the B apical domain were lost at the 20% open state and contacts with the E subunit were lost at the 46% open state. Contacts with the other five subunits persisted throughout the transition. Most of the contacts between rhodanese and GroEL continued to involve the H and I helices. Rhodanese followed the upward motion of these helices and slowly moved outward from the cavity during the transition. At the r′ state, rhodanese still penetrated into the cis cavity but with 25% fewer atoms than in the closed state. The loss of contacts between rhodanese and GroEL was due mostly to a decrease in nonpolar contacts. Fig. 3 shows the buried surface area, obtained from the difference in solvent-accessible surface area of GroEL and rhodanese in the uncomplexed and complexed state. The buried binding surface was divided into a polar and nonpolar contribution, based on the atomic surface area values. In the closed state the surface area of the binding site is 5695 Å2, which is large compared to the value for most protein-protein complexes (between 550 and 4900 Å2, with an average of ∼800 Å2 (Veselovsky et al., 2002; Brooijmans et al., 2002)). The binding free energy of protein complexes with buried binding surface areas over 2000 Å2 are in the range of −10.0 to −14.3 kcal/mol (Brooijmans et al., 2002). This suggests that the binding free energy of the rhodanese-GroEL complex is of the same order or larger, so that it is within the range of protein denaturation free energies. In the r′ state, the surface area of the binding site decreased to 3129 Å2; most of this decrease is due to the nonpolar contribution. During the transition, the nonpolar contribution decreased by 2014 Å2 (from 3948 to 1934 Å2), whereas the polar contribution decreased by 552 Å2 (from 1747 to 1195 Å2). This means that polar contacts become relatively more important during the transition: in the closed state the polar contribution to the surface area of the binding site is 30%, in the r′ state this has increased to 40%. Due to the loss of contacts, the interaction energy between rhodanese and GroEL was 45% less negative in the r′ than in the closed state, in accord with suggestions that the substrate binding is strongest in the closed state (Yifrach and Horovitz, 1996). The electrostatic component gained in relative importance during the transition, so that in the r′ state the electrostatic and van der Waals components were equal in strength.
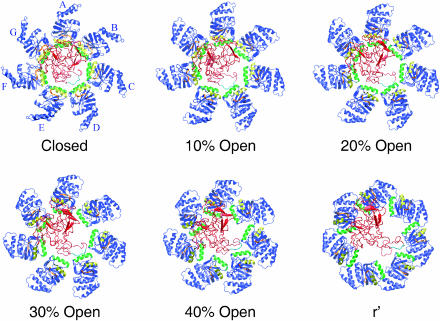
FIGURE 2.
Snapshots during the unfolding simulation; for the description of the different states, see Methods and text. The H helices of GroEL are colored yellow, the I helices are green, the loops formed by residues 310–315 are orange, and the rest of GroEL is blue. GroEL is viewed from the top, looking down into the cis cavity, as in Fig. 1. Subunit A is at the 12-o'clock position, the other subunits (B–G) follow in a clockwise fashion (see Fig. 1). Rhodanese is shown in red, except for the loop consisting of residues 45–50, which is in light blue.
FIGURE 3.
Contact area between GroEL and rhodanese during the GroEL closed-to-r′ transition. The black line corresponds to the total contact area, the dark gray line to the hydrophobic portion of the contact area, and the light gray line to the polar portion of the contact area.
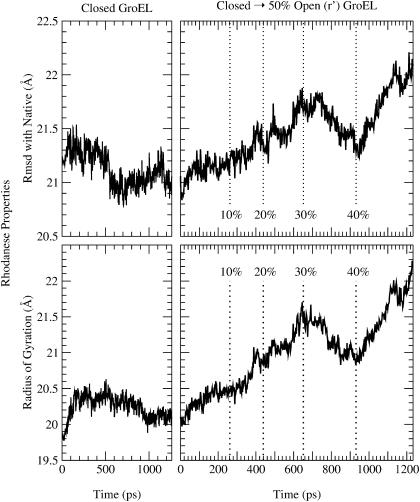
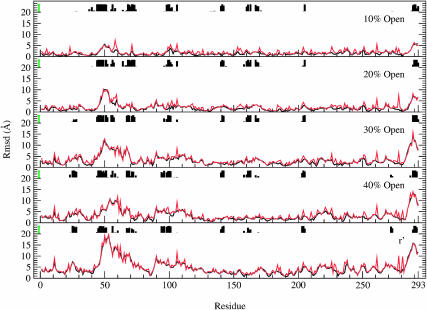
Although the size and shape of rhodanese is essentially unchanged from that of the free denatured molecule upon binding to the closed state, the closed-to-r′ state transition of GroEL had a significant effect on the conformation of rhodanese (Fig. 2). The total solvent-accessible surface of rhodanese (including the buried surface area) increased from 13,463 Å2 in the closed state, to 14,760 Å2 in the r′ state; the polar surface area increased by 407 Å2 (from 2890 to 3297 Å2), the nonpolar surface area increased by 890 Å2 (from 10,572 to 11,462 Å2). The transition caused a further unfolding of rhodanese, increasing the radius of gyration by 2.1 Å and the overall RMSD from the native rhodanese structure by 1.2 Å (Fig. 4); however, certain loops undergo very large changes (see below). The degree of unfolding is strongly correlated with the volume of the cis cavity. Rhodanese was most actively unfolded during the 40% open to r′ part of the transition, when the cis cavity expanded the most. During the 30% to 40% open state transition, the RMSD and radius of gyration of rhodanese actually decreased. This parallelled a decrease in cavity volume, which was due to the rotation of the apical domains in the plane normal to the symmetry axis. The unfolding force had the strongest impact on residues 42–70, 90–112, and 283–293 (Fig. 5). The RMSD per residue of rhodanese in the final (r′) state, compared to the closed-state bound structure at the start of the transition, is 18.5 Å for residues 42–70, to 9.0 Å for residues 90–112, and up to 15.8 Å for the 283–293 loop. Fig. 5 shows clearly that unfolding of rhodanese was localized in areas where the protein was in contact with GroEL. This suggests that GroEL actively unfolded rhodanese during the simulation, by exerting a force on the bound protein. Snapshots of the simulation show that the unfolding of rhodanese corresponds to stretching of the protein by pulling on certain loops (Fig. 2). In what follows we analyze the results, so as to provide an understanding of the mechanism involved. We note here that the qualitative features of the results are preserved in other unfolding trajectories (results not shown), although the details of the interactions vary somewhat from one trajectory to another. The specific results presented here correspond to one unfolding scenario.
FIGURE 4.
Rhodanese properties during the simulation. The top panels show the RMSD from the native state, and the bottom panels show the radius of gyration. The left panels correspond to rhodanese bound to the closed state of GroEL, whereas the right panels correspond to the closed-to-r′ state transition.
FIGURE 5.
The RMSD per residue for rhodanese during the closed-to-r′ state transition of GroEL. The RMSD is with respect to the closed-state bound structure. Black lines show the backbone RMSD, red lines the RMSD for the entire residue. Contacts between rhodanese and GroEL are indicated by the histograms. The height of the histograms represents the lifetime of these contacts during each stage of the transition; these lifetimes were measured between the closed and 10% open state for the top plot, between the 10% and 20% open state for the 20% open plot, etc. The maximum possible lifetimes are indicated by the green bars on the left; it is evident that many contacts are present during the entire transition (see text).
Figs. 6 and 8 show snapshots of the structures that illustrate the results, whereas Figs. 7 and 9 delineate the specific interactions between GroEL and rhodanese. In the latter, Fig. 7 summarizes the interactions that are observed, and Fig. 9 shows when they are present as a function of simulation time. This provides statistics as to their stability and functional role during the entire simulation.
FIGURE 6.
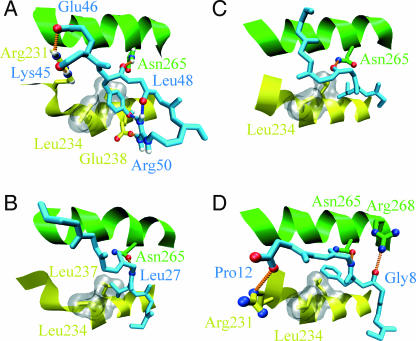
Binding to closed-state GroEL in simulation and experiments. The peptides are shown in light blue, the GroEL H helix is yellow, and the GroEL I helix is shown in green. The hydrophobic pocket formed by Leu-234 and Leu-237 of the H helix is shown by the gray surface. Hydrogen bonds between the peptide and GroEL are indicated by the orange dotted lines; intramolecular hydrogen bonds in the peptides are indicated by blue dotted lines. (A) The starting TMD configuration for rhodanese residues 45–50. Hydrogen bonds are formed between rhodanese Lys-45 and GroEL Arg-231, rhodanese Glu-46 and GroEL Arg-231, rhodanese Tyr-47 and Asn-265, and rhodanese Arg-50 and GroEL Glu-238. Intramolecular hydrogen bonds exist between rhodanese Tyr-47 and Arg-50 and between rhodanese Leu-48 and Arg-50. For clarity, the hydrogen bond between Glu-46 and Arg-268 of the E subunit is excluded from the figure. The hydrophobic pocket is occupied by Tyr-47. (B) GroES residues 24–30 binding to GroEL (Xu et al., 1997). There is a hydrogen bond between GroES Leu-27 and GroEL Asn-265; the hydrophobic pocket is occupied by Val-26. (C) Residues 184-189 of the N-terminal extension binding to GroEL (Buckle et al., 1997). There is a hydrogen bond between Val-186 of the N-terminal extension and Asn-265 of GroEL. The hydrophobic pocket is occupied by Leu-185. (D) Residues 6–12 of the SBP peptide binding to GroEL, taken from chain F and B of the protein data bank structure 1DKD (Chen and Sigler, 1999). Hydrogen bonds are formed between SBP Gly8 and GroEL Arg-268, SBP Leu-10, and GroEL Asn-265, and SBP Pro-12 and GroEL Arg-231. The hydrophobic pocket is occupied by Phe-9.
FIGURE 8.
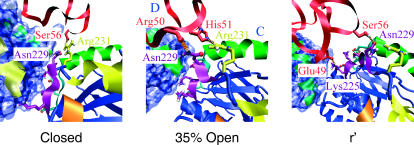
The interface between the apical domains of the C and D subunit (see text). The H helices of GroEL are colored yellow, the I helices are green, loop 310–315 is orange, loop 225–230 is light blue, and loop 252–256 is purple; the rest of GroEL is blue. Subunit C is shown as a ribbon diagram with the polar and charged side chains of loops 225–230 and 252–256 of subunit C as stick models; Arg-231 is shown as a yellow stick model to indicate the closeness of this residue to the H helix (analogous to Fig. 7). For subunit D the solvent-accessible surface is shown in blue. Rhodanese is shown in red, hydrogen bonds between GroEL residues 225–231 and rhodanese are indicated by the orange dotted lines. GroEL is viewed from the top; the viewing angle is identical to that of Fig. 2.
FIGURE 7.
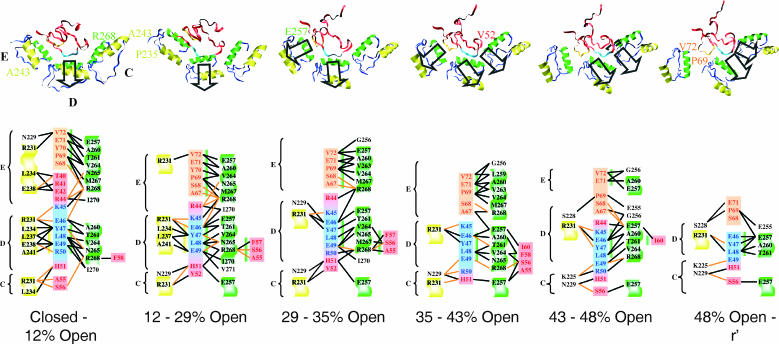
Stretching and interactions for the 42–70 loop of rhodanese. The H helices of GroEL (residues 234–243) are colored yellow, the I helices (residues 257–268) are green, and the rest of GroEL is blue. The symbols C, D, and E refer to the different GroEL subunits. GroEL is viewed from the top of subunit D, looking down into the cis cavity. The viewing angle is the same for all snapshots. Rhodanese residues 45–50 are light blue, residues 67–72 are orange; the other rhodanese residues are shown in red. The direction of the stretching force is indicated by the black arrows. The diagrams below show the interactions between these rhodanese residues and GroEL during part of the transition. Hydrogen bonding is indicated by the orange lines, heavy atom contacts within 4.0 Å, which are mainly van der Waals contacts, are shown by the black lines. The diagrams show all interactions that are present for 10 ps or more during the entire interval; not all contacts and hydrogen bonds are present at every instant of the interval. In the diagrams the H helix is shown on a yellow background, the I helix on a green background, and other GroEL residues are shown on a white background. Residue Arg-231 of GroEL is shown on a yellow background to indicate the closeness of this residue to the H helix. The coloring of rhodanese is identical to the structures above. Binding of rhodanese parallel to the H helix is indicated by a yellow bar (left), binding of rhodanese parallel to the I helix by the green bar (right and left). The presence of both bars for certain residues (e.g., Glu-46, Tyr-47) indicate that this residue was bound in the cleft formed by the H and I helices; this happens in the closed to 12% open and the 12% to 29% open intervals.
FIGURE 9.
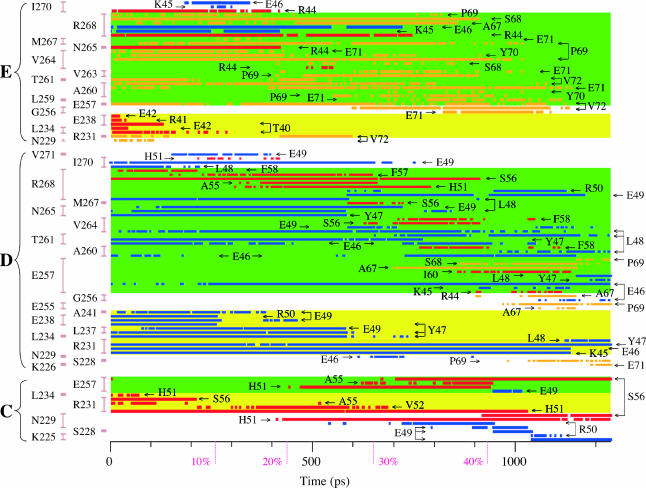
Heavy atom contacts within 4.0 Å between rhodanese residues 42–72 and GroEL; the conventions (labels and colors) are the same as in the other figures. This figure should be used in conjunction with Fig. 7. GroEL residues of subunits C, D, and E are shown on the left; the vertical bars group contacts with a given residue. The H helices and Arg-231 are indicated by the yellow background, the I helices by the green background; other GroEL residues are shown on a white background. Rhodanese residues are indicated by the arrows; residues 45–50 are shown in blue, residues 67–72 in orange, and the other residues in red.
Residues 42–70
The binding of this region of rhodanese to closed state GroEL reveals some striking similarities with the binding in the GroES-GroEL complex (Xu et al., 1997), and two peptides for which the crystal structures have been solved (these are a covalently attached N-terminal extension of an apical domain bound to a neighboring apical domain (Buckle et al., 1997) and a 12-residue peptide bound to an apical domain (Chen and Sigler, 1999); see Fig. 6). Rhodanese residues 46-49 are bound in an extended conformation in the cleft between the H and I helices of the D apical domain in the closed state. The binding of these residues is very tight, due to favorable hydrophobic interactions and a number of strong hydrogen bonds (Fig. 6 A). The phenyl group of Tyr-47 fits neatly in the hydrophobic pocket formed by Leu-234 and Leu-237 of the H helix. Glu-46 makes hydrogen bonds to Arg-231 of the H helix of the D subunit (energy of −0.5 kcal/mol) and to Arg-268 of the I helix of the E subunit (−1.6 kcal/mol). The carbonyl oxygen of Tyr-47 hydrogen bonds to Asn-265 of the I helix (−2.8 kcal/mol), the carbonyl oxygen of Lys-45 binds to Arg-231 of the H helix (−2.5 kcal/mol), and another hydrogen bond is formed between Arg-50 and Glu-238 of the H helix (−0.8 kcal/mol). Rhodanese also has two intramolecular hydrogen bonds in this region, between Arg-50 and the carbonyl oxygen of Leu-48, and between Arg-50 and the hydroxyl group of Tyr-47. In all structures (simulation and crystal structures, see Fig. 6) the substrate peptide is bound in an extended conformation in the cleft between the H and I helix. Also, a large hydrophobic group (Val-26 of GroES, Leu-185 of the N-terminal extension, Phe-9 of the peptide, and the phenyl group of rhodanese Tyr-47 in the simulation) binds in the hydrophobic pocket formed by residues Leu-234 and Leu-237. Finally, Asn-265 of the I helix forms a hydrogen bond with the peptide backbone. The importance of these Leu and Asn residues has been established by mutation studies (Fenton et al., 1994). Arg-231 forms a hydrogen bond with the peptide in the 12-mer peptide complex and in the simulation structure, but not in the N-terminal extension complex. In the GroEL-GroES structure the side chain of Arg-231 is not resolved, but the distance between the Cα of Arg-231 and the peptide backbone is similar to the 12-mer peptide complex and simulation structures.
The 42–70 loop, which is the only part of rhodanese that is in contact with both the H and the I helices of the closed state, is the region where the stretching is most evident. Between the closed and the 12% open state, and between the 12–29% open state the 42–70 loop is stretched radially outwards (top panel of Fig. 7). The unfolding force originates from contacts with rhodanese residues 46–49 (see lower panels of Fig. 7). The strength of these contacts force the rhodanese loop to follow the rotation of the D apical domain, causing the stretching and partial unfolding of the loop. At the 29% open state, an additional unfolding force begins to contribute. Residues 67–69, which are bound parallel to the I helix of the E subunit, are pulled radially outwards toward the E subunit. The binding involves hydrophobic contacts between rhodanese Pro-69 and Val-263–Val-264 of the I helix, and hydrogen bonds between rhodanese Ser-68 and Ala-67, and Arg-268 of the I helix. Stretching also continues at residues 46–49, which are now bound parallel to the I helix of the D subunit. The upward rotation had gradually made the H helix of the D subunit less available for binding, causing Tyr-47 to move out of the hydrophobic pocket. The only contacts with this H helix now involve a hydrogen bond between Arg-231 and rhodanese Lys-45 and van der Waals contacts between Arg-231 and rhodanese Glu-46. Another hydrogen bond is made between Glu-49 and Arg-268 of the I helix; the other contacts are nonpolar. The 35% open state represents an important stage in the transition. At this point, the highly polar residues of the 225–230 loop of the apical domains start to become available for binding to rhodanese (the highly polar residues of the 252–256 loop become available too, but no binding to these residues was observed in this simulation). In the closed state, these regions point toward the neighboring apical domain and are inaccessible to the protein substrate inside the cavity (Fig. 8). During the transition they are rotated inwards, and become part of the lining of the cis cavity. Residues 50–51 of rhodanese start to hydrogen bond with Asn-229 and Arg-231, resulting in a stretching force toward the C subunit (Fig. 7). At the same time, the stretching of residues 46–49 and 67–69 continues. Contacts between subunit D and loop 46–49 are mainly formed by hydrogen bonds between Thr-261 and rhodanese Glu-46, and Arg-268 and rhodanese Glu-49. Contacts between loop 67–69 of rhodanese and subunit D consist mainly of hydrophobic contacts, also extending to rhodanese residues 71 and 72. The stretching of residues 46–49 is now more toward the C apical domain, resulting in a decrease of the radius of gyration. At the 43% open state, contacts between the E subunit and rhodanese start to break. The stretching of residues 46–49 continues, and subunit C binds and stretches Ser-56 by a hydrogen bond to Asn-229. At the 48% open state, all the stretching is due to the C subunit, involving hydrogen bonds to Lys-225 and Asn-229.
Fig. 9 shows that there are a number of contacts between GroEL and rhodanese that lasted throughout (most of) the transition. Contacts between Arg-231 of the C subunit and rhodanese Ser-56, and Arg-231 of the D subunit and rhodanese Tyr-47 were present during the entire transition. Contacts between Arg-231 of the D subunit and rhodanese Glu-46 and Lys-45 were present from the closed to the 47% open state, contacts between Glu-257 of the D subunit and rhodanese Glu-46 were present from the 8% open state to the r′ state. Other contacts form clusters of contacts that start and break as a group. For example, contacts between Glu-238 and Ala-241 of the D subunit with rhodanese Tyr-47, Glu-49, and Arg-50 exist between the closed and 12% open state; contacts between Leu-234 and Leu-237 of the D subunit with rhodanese Tyr-47 and Glu-49, and contacts between Asn-265 of the D subunit with rhodanese Tyr-47 and Leu-48 exist between the closed and 29% open state (Figs. 7 and 9). The formation and breaking of the contacts often corresponds to a change in the direction of the unfolding force, as can be seen from Fig. 7.
Residues 283–293
Contacts between GroEL and rhodanese residues 283–293 mainly involve hydrogen bonds between Ser-289, Gln-290, and Gly-291 and the H helix of subunit A. Residues 283–293 form the carboxylate tail of rhodanese, which makes them susceptible to a stretching force. The residues bind parallel to the H helix and the rotational motion of the H helix leads to stretching of this loop. Partial unfolding of residues 90–112 does not occur until after the 30% open state is reached. Initially the rhodanese loop has two hydrogen bonds with the B subunit and one with the A subunit. The hydrogen bond with the A apical domain is between Gly-100 of rhodanese and Glu-232 of the H helix; this hydrogen bond persists throughout the simulation. The contacts with the B subunit, which involve a β-turn conformation of rhodanese without stable hydrophobic interactions, are broken at the 20% open state and the loop binds exclusively to subunit A. At the 31% open state a second hydrogen bond is formed between His-94 of rhodanese and Arg-231 of the A subunit. Together, these hydrogen bonds attach the 90–112 loop firmly to the A subunit, causing the local stretching of this portion of rhodanese.
Other regions of rhodanese
Not all regions of rhodanese that are in contact with GroEL are stretched and unfolded. Residues 140–142, 158–169, and 203–205 bind to the rim of the cis cavity by contacts with the top of the H helices and the 310–315 loop of the F and G apical domains. Residues 141–142 and 168–170 bind perpendicular to the H helix of subunits F and G, respectively. Residues 158–163 and 203–204 bind to the sides of the H helices, in between the F and G subunit, and form hydrogen bonds only to Glu-232 and Arg-231. During the transition, contacts between these residues and GroEL continue to involve a few hydrogen bonds with the H helices, but no stretching occurs. It is interesting to note that this region of rhodanese showed the lowest backbone fluctuation and had the largest number of intramolecular hydrogen bonds in the simulation of free denatured rhodanese, which suggests a high stability for these loops. The unfolding of these loops may require very tight binding to GroEL (parallel to the H and I helices, rather than perpendicular to them), which was not observed during the transition studied in detail.
Sampling
To ensure that the observed unfolding of rhodanese was not a computational artifact due to insufficient sampling, we repeated one of the simulations using a ten times longer relaxation phase. The starting configuration for the transition was identical to the simulation described in detail above. Analysis showed very similar behavior in the two simulations; rhodanese was unfolded in the same areas, and the same interactions with GroEL were involved. The longer simulation showed a larger degree of unfolding: the radius of gyration increased by 3.0 Å (compared to 2.1 Å), and the RMSD per residue of rhodanese in the r′ state compared to the closed-state bound structure was up to 23.6 Å for the 42–70 loop (compared to 18.5 Å). This difference was due to the increased flexibility of rhodanese in the longer simulation. Although the lifetime of intramolecular hydrogen bonds in rhodanese was identical in the two simulations, these hydrogen bonds were broken more often in the longer simulation. This resulted in greater flexibility of the 42–70 loop of rhodanese, which allowed it to be more tightly bound with more extensive unfolding.
Comparison with experiments
The importance of the H and I helices for the binding and unfolding of rhodanese found in the simulations agrees with the results of mutation experiments (Fenton et al., 1994). Mutation of Leu-234 and Leu-237 of the H helix, and Val-263, Val-264, and Asn-265 of the I helix have been shown to eliminate peptide and GroES binding and chaperone assisted protein folding. Mutation of Glu-238 eliminated GroES binding and decreased protein folding. Also, the chemical type (hydrophobic, polar, and negatively charged) of these residues is strongly conserved (Stan et al., 2003). All of these residues were important for the binding and unfolding of rhodanese in the simulations. Mutation studies (Fenton et al., 1994) also indicated that the loop 199–204, Leu-259, Leu-309, and Asp-361 were involved in substrate and GroES binding and protein folding. No contacts between rhodanese and these residues were observed in any of the simulations. Looking down into the cis cavity, loop 199–204 is located underneath the I helix at the bottom of the apical domains, where it remained during the closed-to-r′ state transition. Most of these residues become accessible only in the r′-to-r″ transition to the fully open GroES bound state. Thus, it is not surprising that they do not contribute during the closed-to-r′ transition. Experiments showed a decrease in the folding activity of GroEL upon mutation of Leu-314 (Fenton et al., 1994), but no unfolding force was generated by this residue in any of the simulations, although contacts between Leu-314 and rhodanese did occur. This would suggest that Leu-314 has an indirect effect (e.g., in terms of preserving the structural integrity of GroEL) on the folding efficiency of GroEL. Hydrogen bonds to the polar and charged residues of loop 225–230 aided the unfolding of rhodanese in the simulation, but mutation experiments on residues in this loop (Lys-225→Glu, Ile-227→Ser, Ile-230→Ser) did not show a decrease in peptide binding or folding (Fenton et al., 1994). The disagreement is not surprising, since the mutations retain or introduce polar residues. Mutation of the polar groups in this loop (Lys-225, Lys-226, Ser-228, Asn-229) into nonpolar groups would be more revealing. No experimental data are available for Pro-235, Ala-241, and Lys-242 of the H helix, Glu-257, Ala-260, Thr-261, Met-267, and Arg-268 of the I helix, Arg-231, Glu-232, and loop 252–256, which were involved in the binding and unfolding of rhodanese in the simulations. Such studies would be of great interest for testing the details of the unfolding mechanism proposed here.
DISCUSSION
Molecular dynamics simulations have shown that interactions between the apical domains of GroEL and the bound rhodanese substrate during the closed-to-r′ state transition of GroEL exert a force on rhodanese that leads to partial unfolding. The contacts between GroEL and rhodanese and their variation as a function of the GroEL transition were analyzed. Many contacts were parts of clusters of contacts that formed and broke as a group. The formation and breaking of the contacts often corresponded to a change in the direction of the stretching force. Two factors are found to be important in the generation of the stretching force. The first is the presence of strong contacts between GroEL and the bound protein that continue to exist for a considerable portion of the transition. The second factor is the interaction of the protein substrate with multiple apical domains; it is the relative motion of several apical domains that pulls apart the rhodanese substrate.
The H and I helices are shown to play the primary role, in accord with the suggestions based on mutation studies (Fenton et al., 1994) and peptide-bound x-ray structures (Xu et al., 1997; Buckle et al., 1997; Chen and Sigler, 1999). During the first part of the transition (closed to 29% open state) the stretching force is mostly generated by the residues in the center of the H and I helices. Residues involved are Leu-234 and Leu-237, which form a hydrophobic pocket in which a phenyl ring of rhodanese is bound, and Asn-265, which forms hydrogen bonds with the backbone of the protein substrate. The binding of rhodanese to these residues closely resembles the binding of GroES (Xu et al., 1997) and peptides (Buckle et al., 1997; Chen and Sigler, 1999) to the apical domains in the available crystal structures. Analysis of the interaction energy and the buried binding surface reveal the importance of hydrophobic contacts. During the second part of the transition, hydrogen bonding becomes more important for the unfolding force, in agreement with suggestions based on the comparison of the cavity lining in the closed and open state (Xu et al., 1997). In this part of the transition the GroEL loops 225–230 and 252–256 become available for binding to the substrate. These loops contain charged and polar residues, which can hydrogen bond to the protein substrate; i.e., at the 48% open state the pulling forces on rhodanese loop 42–70 come from hydrogen bonds to Lys-225 and Asn-229 of the C subunit (Fig. 7). The importance of these GroEL loops for the unfolding force is in accord with a suggestion based on a bioinformatics study (Stan et al., 2003). The charged residues at the end of the H and I helices (especially Arg-231, but also Glu-257) play an important role in the transfer of the pulling force from the residues in the center of the H and I helix to the highly polar 225–230 loop. The aliphatic portions of the Arg and Glu side chains compensate for the loss of the van der Waals interaction with the hydrophobic pocket, whereas the position of Arg-231 and Glu-257 next to the 225–230 and 252–256 loops, respectively, facilitate the transfer of hydrogen bonds to these loops. This transfer makes possible the continous stretching of the protein during the transition and frees the hydrophobic pockets of the H and I loops for their interaction with the incoming GroES. It would be interesting to verify the importance of the Arg-231 and Glu-257 residues for the unfolding of the substrate by mutation experiments.
The importance of multidomain contacts for substrate binding is in agreement with a study of the binding of rhodanese to mutated GroEL molecules (Farr et al., 2000). The GroEL mutants in that study consisted of covalently linked GroEL subunits with various arrangements of apical domains that were either wild-type (available for substrate binding), or mutants that were rendered inactive for substrate binding (Farr et al., 2000). The experimental study revealed that the binding of rhodanese to GroEL generally increases with an increasing number of available apical domains. As already mentioned, the simulations show that contacts with several apical domains are crucial for the additional unfolding of the bound substrate. In some of the simulations, the unfolding diminished or stopped when contacts with nonneighboring subunits were broken and contacts with only two or three neighboring subunits were present (data not shown). Loss of some contacts in the r′ state may also be important for freeing regions required for interaction with GroES (Thirumalai and Lorimer, 2001). To unfold the rhodanese substrate, it has to be “attached” to one part of GroEL and be pulled by another part. This is done most effectively by spatially separated (nonneighboring) subunits. When rhodanese interacts with only two or three neighboring subunits, it is merely displaced by the forces involved and does not unfold (data not shown). The importance of contacts to multiple nonneighboring subunits for the unfolding force explains the strong correlation between the volume of the cis cavity and the degree of unfolding. When the volume increases, the separation between nonneighboring rhodanese binding subunits increases, resulting in a stretching force. This force is maximized when the increase in volume is largest. The importance of the arrangement of substrate contacts to GroEL for the unfolding force could be examined experimentally, by performing hydrogen exchange studies on substrates bound to the GroEL mutants of Farr et. al. (2000). These experiments could provide information concerning the minimum number and arrangement of apical domains required for an unfolding force to be active. In addition, some mutations of residues that are involved in the unfolding interaction but have not been studied experimentally would be of interest (loop 225–230, Arg-231, Glu-232, Pro-235, Ala-241, Lys-242, Glu-257, Ala-260, Thr-261, Met-267, Arg-268, and loop 252–256).
It is important to note that the unfolding force could play a role also for systems that do not enter the cis cavity (Chaudhuri et al., 2001; Hammarström et al., 2000). The simulations showed that the protein does not need to be inside the cavity to be subjected to the stretching force. Binding to the top of the apical domains and only partial penetration into the cis cavity suffices.
The biological role of the additional partial unfolding may be the resetting of the starting conformation for spontanous folding, as has been suggested previously (Shtilerman et al., 1999; Thirumalai and Lorimer, 2001). A mechanism of such partial unfolding has been demonstrated here for the first time in a realistic simulation. Stretching of the protein could eliminate structure elements that are trapped in a particularly misfolded conformation. The removal of intraprotein contacts and the increase of solvent-accessible surface area could facilitate the spontaneous refolding process after release into the cavity or in solution. The present study illustrates what has been suggested to be a general property of molecular “motors” (Yang et al., 2003), in that binding energy (rather than hydrolysis) is converted into mechanical work by highly coordinated conformational changes.
Acknowledgments
We thank Ioan Andricioaei and Wei Yang for stimulating discussions, and Robert Yelle, Martin Spichty and Andrei Golosov for assistance with the computing environment.
The work done at Harvard was supported in part by the National Institutes of Health. Some of the simulations were performed at the National Energy Research Scientific Computing Center. A.vdV. was supported by a Marie Curie Individual Fellowship.
References
- Anfinsen, C. 1973. Principles that govern the folding of protein chains. Science. 181:223–230. [DOI] [PubMed] [Google Scholar]
- Betancourt, M., and D. Thirumalai. 1999. Exploring the kinetic requirements for enhancement of protein folding rates in the GroEL cavity. J. Mol. Biol. 287:627–644. [DOI] [PubMed] [Google Scholar]
- Böckmann, R., and H. Grubmüller. 2002. Nanoseconds molecular dynamics simulation of primary mechanical energy transfer steps in F1-ATP synthase. Nat. Struct. Biol. 9:198–202. [DOI] [PubMed] [Google Scholar]
- Boisvert, D., J. Wang, Z. Otwinowski, A. Horwich, and P. Sigler. 1996. The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATPγS. Nat. Struct. Biol. 3:170–177. [DOI] [PubMed] [Google Scholar]
- Brinker, A., G. Pfeifer, M. Kerner, D. Naylor, F. Hartl, and M. Hayer-Hartl. 2001. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 107:223–233. [DOI] [PubMed] [Google Scholar]
- Brooijmans, N., K. Sharp, and I. Kuntz. 2002. Stability of macromolecular complexes. Proteins. 48:645–653. [DOI] [PubMed] [Google Scholar]
- Brooks, B., R. Bruccoleri, B. Olafson, D. States, S. Swaninathan, and M. Karplus. 1983. CHARMM: A program for macromolecular energy minimization and dynamics calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- Buckle, A., R. Zahn, and A. Fersht. 1997. A structural model for GroEL-polypeptide recognition. Proc. Natl. Acad. Sci. USA. 94:3571–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, H., and K. Dill. 1996. A simple model of chaperonin-mediated protein folding. Proteins. 24:345–351. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, T., G. Farr, W. Fenton, S. Rospert, and A. Horwich. 2001. GroEL/GroES-mediated folding of a protein too large to be encapsulated. Cell. 107:235–246. [DOI] [PubMed] [Google Scholar]
- Chen, J., S. Walter, A. Horwich, and D. Smith. 2001. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat. Struct. Biol. 8:721–728. [DOI] [PubMed] [Google Scholar]
- Chen, L., and P. Sigler. 1999. The crystal structure of a GroEL/peptide complex: Plasticity as a basis for substrate diversity. Cell. 99:757–768. [DOI] [PubMed] [Google Scholar]
- Chen, S., A. Roseman, A. Hunter, S. Wood, S. Burston, N. Ranson, A. Clarke, and H. Saibil. 1994. Location of a folding protein and shape changes in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature. 371:261–264. [DOI] [PubMed] [Google Scholar]
- Dobson, C. 2002. Getting out of shape. Nature. 418:729–730. [DOI] [PubMed] [Google Scholar]
- Ellis, R., and F. Hartl. 1999. Principles of protein folding in the cellular environment. Curr. Opin. Struct. Biol. 9:102–110. [DOI] [PubMed] [Google Scholar]
- Farr, G., K. Furtak, M. Rowland, N. Ranson, H. Saibil, T. Kirchhausen, and A. Horwich. 2000. Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell. 100:561–573. [DOI] [PubMed] [Google Scholar]
- Fenton, W., Y. Kashi, K. Furtak, and A. Horwich. 1994. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 371:614–619. [DOI] [PubMed] [Google Scholar]
- Gliubich, F., M. Gazerro, G. Zanotti, S. Delbono, G. Bombieri, and R. Berni. 1996. Active site structural features for chemically modified forms of rhodanese. J. Biol. Chem. 271:21054–21061. [DOI] [PubMed] [Google Scholar]
- Grallert, H., and J. Buchner. 2001. Review: A structural view of the GroE chaperone cycle. J. Struct. Biol. 135:95–103. [DOI] [PubMed] [Google Scholar]
- Groβ, M., C. Robinson, M. Mayhew, F. Hartl, and S. Radford. 1996. Significant hydrogen exchange protection in GroEL-bound DHFR is maintained during iterative rounds of substrate cycling. Prot. Science. 5:2506–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström, P., M. Persson, and U. Carlsson. 2001. Protein compactness measured by fluorescence resonance energy transfer. J. Biol. Chem. 276:21765–21775. [DOI] [PubMed] [Google Scholar]
- Hammarström, P., M. Persson, R. Owenius, M. Lindgren, and U. Carlsson. 2000. Protein substrate binding induces conformational changes in the chaperonin GroEL. J. Biol. Chem. 275:22832–22838. [DOI] [PubMed] [Google Scholar]
- Hartl, F. 1996. Molecular chaperones in cellular protein folding. Nature. 381:571–580. [DOI] [PubMed] [Google Scholar]
- Houry, W., D. Frishman, C. Eckerskorn, F. Lottspeich, and F. Hartl. 1999. Identification of in vivo substrates of the chaperonin GroEL. Nature. 402:147–154. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD - Visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- Inobe, T., M. Arai, M. Nakao, K. Ito, K. Kamagata, T. Makio, Y. Amemiya, H. Kihara, and K. Kuwajima. 2003. Equilibrium and kinetics of the allosteric transition of GroEL studied by solution X-ray scattering and fluorescence spectroscopy. J. Mol. Biol. 327:183–191. [DOI] [PubMed] [Google Scholar]
- Inobe, T., T. Makio, E. Takasu-Ishikawa, T. Terada, and K. Kuwajima. 2001. Nucleotide binding to the chaperonin GroEL: non-cooperative binding of ATP analogs and ADP, and cooperative effect of ATP. Biochim. Biophys. Acta. 1545:160–173. [DOI] [PubMed] [Google Scholar]
- Lazaridis, T., and M. Karplus. 1999. Effective energy function for proteins in solution. Proteins. 35:133–152. [DOI] [PubMed] [Google Scholar]
- Lee, B., and F. Richards. 1971. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55:379–400. [DOI] [PubMed] [Google Scholar]
- Llorca, O., S. Marco, J. Carrascosa, and J. Valpuesta. 1997. Conformational changes in the GroEL oligomer during the functional cycle. J. Struct. Biol. 118:31–42. [DOI] [PubMed] [Google Scholar]
- Ma, J., and M. Karplus. 1998. The allosteric mechanism of the chaperonin GroEL: A dynamic analysis. Proc. Natl. Acad. Sci. USA. 95:8502–8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., P. B. Sigler, Z. Xu, and M. Karplus. 2000. A dynamic model for the allosteric mechanism of GroEL. J. Mol. Biol. 302:303–313. [DOI] [PubMed] [Google Scholar]
- Ranson, N., G. Farr, A. Roseman, B. Gowen, W. Fenton, A. Horwich, and H. Saibil. 2001. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 107:869–879. [DOI] [PubMed] [Google Scholar]
- Roseman, A., S. Chen, H. White, K. Braig, and H. Saibil. 1996. The chaperonin ATPase cycle: Mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell. 87:241–251. [DOI] [PubMed] [Google Scholar]
- Ryckaert, J., G. Ciccotti, and H. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- Rye, H., S. Burston, W. Fenton, J. Beechem, Z. Xu, P. Sigler, and A. Horwich. 1997. Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature. 388:792–798. [DOI] [PubMed] [Google Scholar]
- Rye, H., A. Roseman, S. Chen, K. Furtak, W. Fenton, H. Saibil, and A. Horwich. 1999. GroEL-GroES cycling: ATP and nonnative polypeptide direct alternation of folding-active rings. Cell. 97:325–338. [DOI] [PubMed] [Google Scholar]
- Saibil, H. 2000. Molecular chaperones: Containers and surfaces for folding, stabilising or unfolding proteins. Curr. Opin. Struct. Biol. 10:251–258. [DOI] [PubMed] [Google Scholar]
- Saibil, H., A. Horwich, and W. Fenton. 2002. Allostery and protein substrate conformational change during GroEL/GroES-mediated protein folding. Adv. Prot. Chem. 59:45–72. [DOI] [PubMed] [Google Scholar]
- Schlitter, J., M. Engels, P. Krüger, E. Jacoby, and A. Wollmer. 1993. Targeted molecular dynamics simulation of conformational change-application to the T↔R transition in insulin. Mol. Simulat. 10:291–308. [Google Scholar]
- Sfatos, C., A. Gutin, V. Abkevich, and E. Shakhnovich. 1996. Simulations of chaperone-assisted folding. Biochemistry. 35:334–339. [DOI] [PubMed] [Google Scholar]
- Shtilerman, M., G. Lorimer, and S. Englander. 1999. Chaperonin function: Folding by forced unfolding. Science. 284:822–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan, G., D. Thirumalai, G. Lorimer, and B. Brooks. 2003. Annealing function of GroEL: Structural and bioinformatic analysis. Biophys. Chem. 100:453–467. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D., and G. Lorimer. 2001. Chaperonin-mediated protein folding. Annu. Rev. Biophys. Biomol. Struct. 30:245–269. [DOI] [PubMed] [Google Scholar]
- Todd, M., G. Lorimer, and D. Thirumalai. 1996. Chaperonin-facilitated protein folding: Optimization of rate and yield by an iterative annealing mechanism. Proc. Natl. Acad. Sci. USA. 93:4030–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, M. J., P. V. Viitanen, and G. H. Lorimer. 1994. Dynamics of the chaperonin ATPase cycle: Implications for facilitated protein folding. Science. 265:659–666. [DOI] [PubMed] [Google Scholar]
- Veselovsky, A., Y. Ivanov, A. Ivanov, A. Archakov, P. Lewi, and P. Janssen. 2002. Protein-protein interactions: Mechanisms and modification by drugs. J. Mol. Recognit. 15:405–422. [DOI] [PubMed] [Google Scholar]
- Walter, S., G. Lorimer, and F. Schmid. 1996. A thermodynamic coupling mechanism for GroEL-mediated unfolding. Proc. Natl. Acad. Sci. USA. 93:9425–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., and D. Boisvert. 2003. Structural basis for GroEL-assisted protein folding from the crystal structure of (GroEL-KMgATP)14 at 2.0 Å resolution. J. Mol. Biol. 327:843–855. [DOI] [PubMed] [Google Scholar]
- Weissman, J., Y. Kashi, W. Fenton, and A. Horwich. 1994. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell. 78:693–702. [DOI] [PubMed] [Google Scholar]
- Weissman, J., H. Rye, W. Fenton, J. Beechem, and A. Horwich. 1996. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 84:481–490. [DOI] [PubMed] [Google Scholar]
- Xu, Z., A. Horwich, and P. Sigler. 1997. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 388:741–750. [DOI] [PubMed] [Google Scholar]
- Yang, W., Y. W. Gao, Q. Cui, J. Ma, and M. Karplus. 2003. The missing link between thermodynamics and structure in F1-ATPase. Proc. Natl. Acad. Sci. USA. 100:874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yifrach, O., and A. Horovitz. 1996. Allosteric control by ATP of non-folded protein binding to GroEL. J. Mol. Biol. 255:356–361. [DOI] [PubMed] [Google Scholar]
- Zahn, R., S. Perrett, and A. Fersht. 1996a. Conformational states bound by the molecular chaperones GroEL and SecB: A hidden unfolding (annealing) activity. J. Mol. Biol. 261:43–61. [DOI] [PubMed] [Google Scholar]
- Zahn, R., S. Perrett, G. Stenberg, and A. Fersht. 1996b. Catalysis of amide proton exchange by the molecular chaperones GroEL and SecB. Science. 271:642–644. [DOI] [PubMed] [Google Scholar]