Abstract
Nuclease-resistant, cytoplasmically resident molecular beacons were used to specifically label β-actin mRNA in living and motile chicken embryonic fibroblasts. β-actin mRNA signals were most abundant in active lamellipodia, which are protrusions that cells extend to adhere to surfaces. Time-lapse images show that the immediate sources of β-actin mRNA for nascent lamellipodia are adjacent older protrusions. During the development of this method, we observed that conventional molecular beacons are rapidly sequestered in cell nuclei, leaving little time for them to find and bind to their cytoplasmic mRNA targets. By linking molecular beacons to a protein that tends to stay within the cytoplasm, nuclear sequestration was prevented, enabling cytoplasmic mRNAs to be detected and imaged. Probing β-actin mRNA with these cytoplasmically resident molecular beacons did not affect the motility of the fibroblasts. Furthermore, mRNAs bound to these probes undergo translation within the cell. The use of cytoplasmically resident molecular beacons will enable further studies of the mechanism of β-actin mRNA localization, and will be useful for understanding the dynamics of mRNA distribution in other living cells.
INTRODUCTION
Fibroblasts growing on flat surfaces display dramatic motility. They extend protrusions, called lamellipodia, at their leading edges, which adhere to surfaces, allowing the cells to be pulled forward. Within active lamellipodia, actin is polymerized into a branched network of filaments that pushes the cell membrane forward. During protrusion, actin polymerization at the forward edge of the network exceeds actin depolymerization, whereas a net depolymerization occurs at the rear edge. The cells control the rates of polymerization and depolymerization of actin monomers, and the degree of branching in the actin filaments, to accomplish the protrusion of lamellipodia (Mitchison and Cramer, 1996; Pollard and Borisy, 2003; Ridley et al., 2003).
Singer and his colleagues have found that in some fibroblasts the density of β-actin mRNA is higher in subcellular zones proximal to the leading edges of the lamellipodia than in the rest of the cell (Lawrence and Singer, 1986; Hill et al., 1994). β-actin mRNA localizes in this manner only under conditions in which cells are moving, suggesting that the concentration of β-actin mRNA at leading edges may provide a high concentration of β-actin monomer to drive polymerization (Lawrence and Singer, 1986; Kislauskis et al., 1994). However, the role of β-actin mRNA localization in the motility of the cells has remained enigmatic, since protein synthesis inhibitors do not effect protrusion in the short term (Kislauskis et al., 1997; Shestakova et al., 2001). In addition to understanding the role of β-actin mRNA localization in cell motility, the mechanisms by which the β-actin mRNA becomes localized are the subject of ongoing investigations. A particular sequence at the 3′ end of β-actin mRNA binds to “zipcode-binding protein 1” (ZBP1) and the resulting complex is actively transported to the growing lamellipodia (Ross et al., 1997; Oleynikov and Singer, 2003). To further dissect this transport mechanism, and to clarify the role of mRNA localization in cell motility, it is important to be able to visualize native β-actin mRNA in living cells.
A number of different methods have been developed to image the distribution of mRNAs in live cells. In one approach, the gene encoding a target mRNA is modified by the addition of sequences, which in the expressed mRNA serve as binding sites for the coat protein of bacteriophage MS2. Cells expressing these modified mRNAs are also engineered to synthesize green fluorescent protein (GFP) that is fused to MS2 coat protein. Regions of the cell where the modified target mRNA is concentrated attract the fused GFP-MS2 coat protein and become more intensely fluorescent (Bertrand et al., 1998). Alternative approaches, designed to detect native mRNAs in unmodified cells, by using oligonucleotide probes tagged with fluorescent moieties whose emission is altered upon hybridization, have been used with varying degrees of success. Utilizing double-stranded quenched oligonucleotides in which the target can displace one of the strands, resulting in the restoration of fluorescence, Sixou et al. (1994) were able to detect a microinjected oligonucleotide target within live cells. Utilizing fluorescence correlation spectroscopy and oligodeoxythymidines as probes, Politz et al. (1998) were able to detect polyadenylated mRNAs in live cells. Finally, using binary hybridization probes that were labeled at their respective 5′ and 3′ ends with interactive fluorophores that induce fluorescence resonance energy transfer between the labels when the two probes are bound to the target RNA at adjacent sites, Tsuji et al. (2000) detected as few as 40,000 copies of target RNA expressed from transfected plasmids in living cells.
Concentrating on the detection of native mRNAs, we developed hairpin-shaped oligonucleotide probes, called “molecular beacons” (Tyagi and Kramer, 1996; Tyagi et al., 1998), that have been used to detect RNAs in living cells (Matsuo, 1998; Sokol et al., 1998; Dirks et al., 2001; Bratu et al., 2003). Molecular beacons are oligonucleotides that possess complementary sequences on either end of a probe sequence, enabling the molecules to assume a hairpin configuration in which the fluorophore and the quencher are held in close proximity. The formation of a probe-target hybrid disrupts the hairpin stem, removing the fluorophore from the vicinity of the quencher, restoring the probe's fluorescence. To prevent degradation of the probes by cellular nucleases, and to prevent destruction of the target mRNA by cellular ribonuclease H, the molecular beacons were synthesized from 2′-O-methylribonucleotides (Tsourkas et al., 2003), which do not occur naturally. Utilizing these probes, we were able to image the distribution and trafficking of oskar mRNA in live Drosophila oocytes (Bratu et al., 2003). In these studies, when the localization pattern of oskar mRNA was altered by genetic manipulation of the mRNA's 3′-untranslated region, or by chemical perturbation of the intracellular tubulin network in the oocyte (which plays a central role in the transport of oskar mRNA), the intracellular distribution of the fluorescence signals changed accordingly, demonstrating that the signals are target specific.
When we attempted to image the distribution of β-actin mRNA in chicken embryonic fibroblasts with these conventional molecular beacons, we found that the molecular beacons were rapidly sequestered in the nuclei of the cells. Sequestration was so rapid and extensive that the probes could not find the cytoplasmic β-actin mRNA before they were shunted to the nucleus. The same phenomenon was observed in other cells in culture and for probes that were specific for other mRNAs. In this report, we describe features of this sequestration process, demonstrate that the attachment of a nonkaryophilic protein (a protein that has no tendency to enter the nucleus) to the molecular beacons prevents nuclear sequestration of the probes, and utilize these modified probes to image the distribution and transport of β-actin mRNA in live fibroblasts. Our results show that it is possible to track β-actin mRNA within lamellipodia as they are extended and retracted.
MATERIALS AND METHODS
Molecular beacons
Molecular beacons possessing 2′-O-methylribonucleotide backbones were synthesized on an Applied Biosystems (Foster City, CA) 394 DNA synthesizer using 2′-O-methylribonucleotide β-cyanoethyl phosphoramidites. They contained a fluorophore at their 5′ ends and a biotin moiety at their 3′ ends (Table 1). The quencher moiety, dabcyl, was internally located at a thymidine residue within the 3′-stem sequence. The biotin moiety was introduced by using a controlled-pore glass column bearing triethyleneglycol-biotin; the internal dabcyl moiety was introduced via a deoxythymidine-dabcyl phosphoramidite; and a fluorophore was introduced at the 5′ end in a postsynthetic reaction. To enable the addition of the fluorophore, the last residue added during automated synthesis possessed a 5′-terminal sulfhydryl group protected by a trityl moiety. Each oligonucleotide was then purified by high-pressure liquid chromatography through a C-18 reverse-phase column, with the trityl moiety still attached. An iodoacetamide derivative of tetramethylrhodamine or Texas red was then coupled to the 5′-sulfhydryl group. DNA synthesis reagents were obtained from Glen Research (Sterling, VA) and activated fluorophores were obtained from Molecular Probes (Eugene, OR). The completed oligonucleotides were purified by high-pressure liquid chromatography, and their signal/background ratio was determined as described previously (Tyagi et al., 1998). A detailed protocol for molecular beacon synthesis is available at http://www.molecular-beacons.org.
TABLE 1.
Molecular beacons used in this study
| Molecular beacon | Sequence |
|---|---|
| Molecular beacon analogue | TMR-CCGCGAUUGUUGUUUUGGAGCACGGAAAGAUCGCGG-TEG-biotin |
| CEF-β-actin-38-61 | TMR-ACCACGGAGUAACGCGGUCAGTCAGGUGG(dT-dabcyl)-TEG-biotin |
| CEF-β-actin-66-83 | TMR-CAAUAUCAUCAUCCAUGGCAUUG(dT-dabcyl)-TEG-biotin |
| CEF-β-actin-240-262 | TMR-ACGACAGGAUACCUCUUUUGCUCUGGGUCG(dT-dabcyl)-TEG-biotin |
| Control molecular beacon | Texas red-ACGCGUUUGUAGAUCAGGUGGCCGGUCGCG(dT-dabcyl)-TEG-biotin |
| GFP-216-234 | TMR-GCUUCGUGGUCGGGGUAGCGGCUGAAGC(dT-dabcyl)-TEG-biotin |
| GFP-546-565 | TMR-CCGCGGGGGUGUUCUGCUGGUAGUGGUGCGG(dT-dabcyl)-TEG-biotin |
| GFP-624-642 | TMR-ACGCCUUCUCGUUGGGGUCUUUGCUCGGCG(dT-dabcyl)-TEG-biotin |
The numbers in the names of the CEF β-actin molecular beacons refer to the β-actin mRNA sequence (accession number L08165), with numbering beginning at the start of the sequence. The numbers in the names of the GFP molecular beacons refer to the GFP cDNA sequence in plasmid pTRE-d2EGFP (Clontech), with numbering beginning with the first nucleotide of the start codon. The molecular beacon analogue was designed to bind to mRNA encoding firefly luciferase, and the control molecular beacon was designed to bind to Chinese hamster elongation factor 1 mRNA; neither of their target sequences is likely to be present in CEF mRNAs. TEG refers to the triethylene glycol spacer.
Transcripts
An RNA whose nucleotide sequence corresponded to positions 29–1725 of embryonic chicken β-actin mRNA (accession number L08165) was synthesized by transcription in vitro from a DNA template. The DNA template was generated in a reverse-transcription polymerase chain reaction (PCR) initiated with isolated chicken embryonic fibroblast RNA. One of the PCR primers possessed a tail sequence containing a bacteriophage T7 RNA polymerase promoter, enabling the resulting amplicons to serve as templates in subsequent in vitro RNA synthesis reactions. An mRNA encoding green fluorescent protein was similarly generated, using plasmid pTRE-d2EGFP (Clontech, Palo Alto, CA) as the template in a PCR reaction. The DNA template was then destroyed by digestion with deoxyribonuclease. Real-time hybridization reactions (150 μL) were performed at 37°C in a spectrofluorometer (Photon Technology International, Lawrenceville, NJ). The reaction mixtures contained 30 nM of each of the three molecular beacons, 20 nM embryonic chicken β-actin mRNA, 20 mM Tris-HCl (pH 8.0), 7.5 mM MgCl2, and either no or 2 μM streptavidin.
Chicken embryonic fibroblasts
Chicken embryonic fibroblast (CEF) cells were obtained from Charles River Laboratories (Wilmington, MA) and stored in liquid nitrogen. Aliquots of the cell suspension were thawed, mixed with Dulbecco's modified Eagle's medium and 10% fetal bovine serum (Invitrogen, Carlsbad, CA), and then plated onto glass-bottom culture dishes containing a thin layer of gelatin. The gelatin was deposited onto the glass surface to promote adherence of the cells to the substratum. For this purpose, a solution of 0.2% gelatin was added to the plates and removed after a 2-min incubation. Cells were plated after the glass surface dried completely. The fibroblasts were cultured for 48 h. In control experiments, serum starvation was accomplished by incubating the cells with serum-free medium for 16 h.
Microinjection
Cell culture dishes were transferred from the CO2 incubator to the microscope stage, where they were maintained at 37°C. Microinjection was performed using a Femtojet microinjection apparatus (Brinkman, Westbury, NY). The concentrations of the microinjected reagents were: molecular beacon analog, 0.5 μM; CEF β-actin mRNA-specific molecular beacons, 0.5 μM each; control molecular beacon, 1.5 μM; and wheat germ agglutinin, 50 μg/ml. A 20-fold molar excess of streptavidin was mixed with the molecular beacons at least 10 min before microinjection. In the studies of protein synthesis from GFP mRNA-molecular beacon hybrids in Chinese hamster ovary cells, the completion of hybridization was ensured by using a high concentration of each molecular beacon (1.5 μM) relative to the concentration of GFP mRNA transcript (0.1 μM) and by continuing the hybridization for a relatively long time (4 h). The reagents used for microinjection were dissolved in water so that no extraneous salt was introduced into the cell. An exception to this was the injection of hybrids of GFP mRNA with its molecular beacons, which were dissolved in phosphate-buffered saline supplemented with 1 mM MgCl2 to ensure that the hybrids remained stable. The volume of microinjected materials was estimated to be ∼1 × 10−12 L. Antibodies against the nuclear protein fibrillarin were obtained from Abcam (Cambridge, MA) and the manufacturer-recommended protocol was followed for indirect immunofluorescence. The secondary antibodies in this procedure were labeled with Cascade blue.
Imaging
For live cell imaging, we used black T4 culture dishes (Bioptechs, Butler, PA) that possessed a 0.17-mm coverglass with a coating of conductive material on the underside to permit controlled heating. The temperature of the T4 culture dishes and the microscope objective was maintained at 37°C, using two Bioptechs controllers. During microinjection and imaging, the cells were maintained in Leibovitz's L-15 medium (purchased from Invitrogen and supplemented with 10% fetal bovine serum) that lacked the dye phenol red and was thus more transmissive and had less autofluorescence. An Axiovert 200M inverted fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a 100× oil objective and a CoolSNAP HQ camera (Photometrics, Pleasanton, CA), cooled to −30°C, was used to obtain the images. The fluorescence of tetramethylrhodamine (TMR) was imaged using excitation filter 546DF10, dichroic mirror 555DRLP, and emission filter 580DF30. The fluorescence of Texas red was imaged using excitation filter 590DF10, dichroic mirror 610DRLP, and emission filter 630DF30. All filters were purchased from Omega Optical (Brattleboro, VT). The images were acquired and analyzed using Openlab software (Improvision, Sheffield, UK). For quantitative measurements of the perimeter of a region of interest (such as a nucleus), the region was manually identified and then the average pixel intensity in the region was determined for each image in a time series. Background fluorescence was separately determined for each image, using an area just outside of the cell.
RESULTS
Sequestration of molecular beacons within nuclei
The characteristics of a good probe for an intracellular target are that it should be stable inside the cell, it should not destroy or otherwise perturb the target, it should be distributed homogeneously within the cell, and it should only give a signal when and where the target is present. To satisfy the first and second criteria, we used nuclease-resistant molecular beacons (synthesized from 2′-O-methylribonucleotides). They are stable for at least two days in fibroblasts and the target mRNA in the hybrids that they form is not digested by cellular ribonuclease H (Bratu et al., 2003). However, when we used these molecular beacons to image the distribution of mRNAs in cells in culture, we noticed that fluorescence from the molecular beacons was always most intense in the nucleus, even though the target RNA was expected to be present in the cytoplasm. To distinguish between unprocessed nuclear mRNAs and processed cytoplasmic mRNAs, we utilized molecular beacons that bind to an exon-exon junction within the mRNA sequence, yet we obtained the same result. Although there is a pool of mature mRNAs in the nucleus (Weil et al., 2000), a significant fraction of the fluorescence signal must have been nonspecific, because control molecular beacons, having no target inside the cell, also exhibited nuclear fluorescence, though at lower intensities.
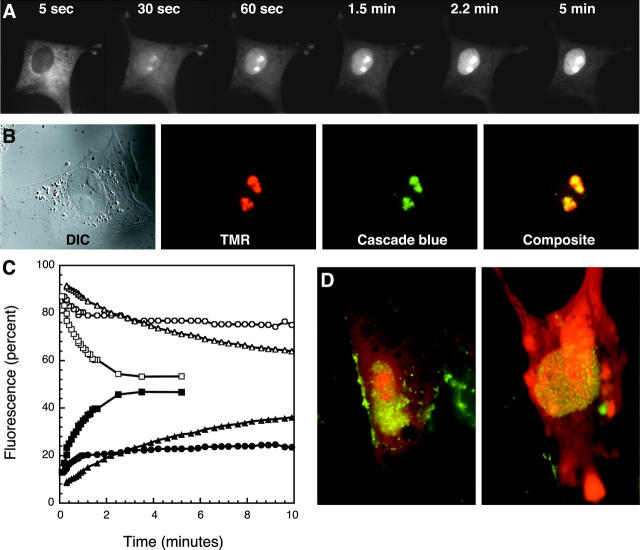
To systematically investigate this phenomenon, we prepared a molecular beacon analog that did not possess a quencher, and was thus fluorescent even when not bound to an mRNA. Its sequence was derived from the firefly luciferase gene and chosen so that it would not find a complementary RNA in the chicken fibroblasts (Table 1). A blast search did not detect a significant complementarity with any metazoan sequence (including chicken sequences) thus far available. The intracellular distribution of this molecular beacon analog in fibroblasts was imaged over time, beginning immediately after its injection into the cytoplasm. Strikingly, we found that within 2 min the nuclei became intensely fluorescent, whereas the cytoplasm lost most of its fluorescence (Fig. 1 A). The distribution of molecular beacons was not homogenous within the nuclei; several small regions within the interior of the nuclei were always more intensely fluorescent (Fig. 1 A). These regions were shown to be nucleoli, as they were denser in diffraction interference contrast images and they were stained by antibodies against the RNA-binding protein fibrillarin, which is found only within nucleoli (Kill, 1996) (Fig. 1 B). Measurements of the relative proportion of nuclear and cytoplasmic fluorescence as a function of time derived from the time-lapse images in Fig. 1 A are shown in Fig. 1 C. Even though the nuclei of these cells occupy only ∼5% of the cell volume, they acquired half of the fluorescence. The same phenomenon was observed with all of the cells that we have explored, including HeLa cells, Chinese hamster ovary cells, and the nurse cells of Drosophila egg chambers (Bratu et al., 2003). The only exception to this phenomenon was seen in Drosophila oocytes, where the nuclei do not sequester molecular beacons, perhaps due to their transcriptional dormancy. Nuclear sequestration occurred with all oligonucleotides tested, irrespective of whether their backbone was composed of 2′-O-methylribonucleotides, deoxyribonucleotides, or peptide nucleic acids. Moreover, nuclear sequestration occurred irrespective of whether or not the oligonucleotide could form a hairpin structure and irrespective of the identity of their attached fluorophore.
FIGURE 1.
Sequestration of molecular beacons in the nuclei of chicken embryonic fibroblasts. (A) Changes in the intracellular distribution of a molecular beacon analog that lacks a quencher, and is designed to not have a target in the cell, as a function of time. (B) Identification of the sites within the nucleus where molecular beacons preferentially localize. TMR-labeled molecular beacon analog was injected into the cytoplasm of a fibroblast. Ten minutes after injection, the cells were fixed, permeabilized, and stained with fibrillarin-specific antibodies. Two bean-shaped regions within the nucleus in the diffraction interference contrast (DIC) image are the nucleoli, which acquire molecular beacon analog (TMR) and are stained by fibrillarin-specific antibodies (Cascade blue). (C) Kinetics of nuclear sequestration of the molecular beacon analog under different conditions. Solid symbols show the increase in nuclear fluorescence and open symbols show the decrease in cytoplasmic fluorescence. Square symbols represent free molecular beacon analog at 37°C, triangular symbols represent free molecular beacon analog at 25°C, and circular symbols represent molecular beacon analog complexed with streptavidin at 37°C. (D) Effect of prior cytoplasmic injection of wheat germ agglutinin (green) on the distribution of the molecular beacon analog (red).
Active transport accounts for nuclear sequestration
These observations suggested that many constituents of nuclei have affinity for molecular beacons. However, it was not clear why molecular beacons enter so rapidly into nuclei. Nuclear pores allow <40 kD molecules to diffuse passively into nuclei, but prevent the entry of larger molecules unless they are attached to karyopherins, whose transport into nuclei is greatly facilitated (Ribbeck and Gorlich, 2001). Molecular beacons might traverse the nuclear pores by passive diffusion or by facilitated transport. To distinguish between these alternatives, the kinetics of nuclear sequestration were studied as a function of temperature (Zasloff, 1983; Lorenz et al., 2000; Ribbeck and Gorlich, 2001). We measured the kinetics of accumulation of fluorescence in fibroblast nuclei at 25°C and compared it with the kinetics of accumulation of fluorescence at 37°C. At 25°C, the initial rate of accumulation of fluorescence in the nuclei was 6% of the total fluorescence per minute, whereas it was 36% per minute at 37°C (Fig. 1 C). If passive diffusion were the primary mechanism of transport, only a 4% difference in the rate of transport at these two temperatures would have been seen, assuming a linear relationship between the rate of transport and the diffusion constant, which is proportional to the absolute temperature expressed in degrees Kelvin (Ribbeck and Gorlich, 2001). It has been shown that although the speed of nucleocytoplasmic transport drops by decreasing the temperature, its fundamental characteristics remain unchanged (Ribbeck and Gorlich, 2001).
The involvement of active transport became more apparent from the results of a second experiment in which we inhibited nuclear pore-mediated facilitated transport by the addition of wheat germ agglutinin (WGA), which is a small protein that binds specifically to O-linked N-acetylglucosaminitol, a sugar moiety found on a number of proteins that line the lumen of nuclear pores and aid in the transport process. The introduction of WGA into the cytoplasm leads to the inhibition of facilitated transport, without affecting passive diffusion through the nuclear pores (Hanover et al., 1987; Dabauvalle et al., 1988). Since WGA is membrane impermeable, we injected WGA into the cytoplasm of fibroblasts, incubated the cells for 30 min at 37°C, and then injected the quencherless molecular beacon analog. The cells were imaged after an additional incubation of 30 min. To mark the cells that received WGA, we used WGA that was labeled with fluorescein. Two representative cells are shown in Fig. 1 D, in which the green color indicates the presence of WGA and the red color represents the fluorescence of the coinjected molecular beacon analog. The nuclei of these cells possessed 24% of the total cellular molecular beacon analog fluorescence, compared to 45% in control cells that did not receive WGA, indicating that WGA inhibits the entry of molecular beacons into the nuclei. The specificity of WGA binding was apparent from the fluorescent staining of nuclear pores by WGA. Taken together, the results of these two experiments suggest that molecular beacons enter nuclei via facilitated transport.
This phenomenon is reminiscent of what occurs when antisense oligonucleotides are used to suppress gene expression by binding them to specific cytoplasmic mRNAs. Antisense oligonucleotides are also rapidly sequestered within nuclei (thus markedly decreasing the suppression of gene expression). Early studies suggested that antisense oligonucleotides enter the nuclei passively (Leonetti et al., 1991), whereas more recent studies indicate that antisense oligonucleotides enter the nuclei via facilitated transport (Hartig et al., 1998). However, irrespective of the mechanism by which oligonucleotides enter nuclei, it is clear that nuclear sequestration of molecular beacons poses a serious problem for the detection and imaging of mRNAs within cytoplasm.
Overcoming nuclear sequestration
We adopted a strategy described by Tsuji et al. (2000) to retain the molecular beacons within the cytoplasm. In this technique, a bulky protein, streptavidin, is attached to the molecular beacons. Streptavidin, being of prokaryotic origin, has no natural nuclear localization signal, and since it is a 75-kD protein that exists as a tetramer, it cannot diffuse passively through nuclear pores. It is simple to incorporate biotin, the natural ligand of streptavidin, into molecular beacons during automated synthesis of the probes. To ensure that the streptavidin does not adversely effect hybridization of the molecular beacon to its target RNA, the biotin moiety was linked to the 3′ end of the molecular beacon, and was separated from the body of the probe by a triethyleneglycol linker.
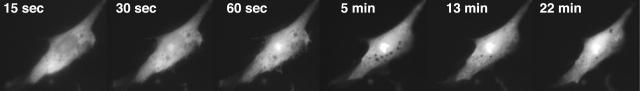
To find out if the presence of streptavidin prevents molecular beacons from entering nuclei, we performed additional experiments with the quencherless molecular beacon analog. When this analog was linked to streptavidin and microinjected into fibroblasts, we observed that its entry into nuclei was severely inhibited. Micrographic images showing the distribution of fluorescence between the nucleus and the cytoplasm of a representative cell at different times after microinjection are presented in Fig. 2, and measurements made from these micrographs are plotted in Fig. 1 C. These results show that ∼20% of the injected molecular beacons can still enter the nucleus, but the majority remain within the cytoplasm for a long time. The fraction that enters the nucleus does so with kinetics that are similar to the kinetics seen with free molecular beacons (Fig. 1 C), suggesting that this fraction was not bound to streptavidin. However, enough molecular beacons remained in the cytoplasm to bind to their target mRNAs.
FIGURE 2.
Intracellular distribution of molecular beacon analogs complexed with streptavidin, as a function of time after cytoplasmic injection. The measurements of nuclear and cytoplasmic fractions as a function of time are presented in Fig. 1 C (solid and open circles).
Hybridization of molecular beacons linked to streptavidin
We synthesized molecular beacons that were designed to detect β-actin mRNA in chicken embryonic fibroblasts. These probes contained a fluorophore, TMR, at their 5′ ends and a triethyleneglycol spacer linked to a biotin moiety at their 3′ ends. The quencher moiety, dabcyl, was covalently linked to a thymidine residue immediately adjacent to the triethyleneglycol spacer. The stem sequences were designed so that in the hairpin state of the molecular beacon the quencher moiety is directly opposite the fluorophore.
To improve the strength of the hybridization signals, we prepared three different streptavidin-linked molecular beacons, each designed to bind to β-actin mRNA at separate sites (Table 1). The target sites for these molecular beacons were selected with the aid of computer programs that identify regions of the target RNA that are likely to be accessible to hybridization probes (Bratu et al., 2003). The efficacy of these sites was confirmed via hybridization experiments, in which each molecular beacon was bound to β-actin transcripts in vitro. In addition, the results of the in vitro experiments confirmed that when all three probes are used together, the strength of the fluorescence signal is cumulatively enhanced.
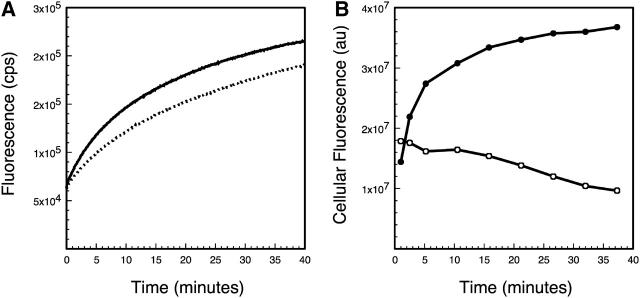
It was possible that the presence of streptavidin would adversely affect the ability of the probe to bind to its target, and might interfere with the quenching of the fluorophore. We therefore measured the kinetics of hybridization of free molecular beacons to full-length β-actin transcripts and compared those results to the kinetics of hybridization of molecular beacon-streptavidin complexes to the same transcripts. Both the rate of hybridization and the extent of hybridization of the two reactions were similar to each other (Fig. 3 A), indicating that streptavidin does not significantly interfere with hybridization. Furthermore, the common intercept of the two curves on the fluorescence axis in Fig. 3 A indicates that the presence of streptavidin does not adversely affect the quenching of TMR by dabcyl.
FIGURE 3.
Kinetics of hybridization of molecular beacons to chicken embryonic fibroblast β-actin mRNA. (A) Comparison of the kinetics of in vitro hybridization of free molecular beacons (continuous line) with molecular beacons linked to streptavidin (broken line). Fluorescence intensity is plotted as a function of time after the addition of a synthetic β-actin mRNA transcript. (B) Changes in total cellular fluorescence after injection of a mixture of streptavidin and biotinylated molecular beacons. Three of the molecular beacons in the mixture were labeled with TMR and were specific for β-actin mRNA (solid circles), and one was labeled with Texas red and had no target mRNA in the cells (open circles).
We then hybridized molecular beacons linked to streptavidin to native β-actin mRNA in live fibroblasts. For these experiments, we also prepared a Texas red-labeled control molecular beacon that could not bind to β-actin mRNA. The control molecular beacon contained the same quencher and biotin moieties. To see whether β-actin mRNA-specific molecular beacons are able to hybridize to their targets within fibroblasts, a mixture containing 0.5 μM of each of the three β-actin-specific molecular beacons and 1.5 μM of the control molecular beacon in the presence of a 22-fold molar excess of streptavidin was injected into these cells. The large excess of streptavidin was used to ensure that only one of the four biotin binding sites on each of the tetrameric streptavidin molecules would be occupied by a (biotinylated) molecular beacon. In preliminary experiments, we determined that ∼100,000 molecules of each molecular beacon were delivered into each cell by microinjection. The cells were imaged with respect to both TMR and Texas red as a function of time. The change in fluorescence intensity of these two differently colored fluorophores observed in a typical microinjected cell is shown in Fig. 3 B. The fluorescence of TMR increased with kinetics that indicate that the signal was due to the hybridization of the molecular beacons to β-actin mRNA, whereas the fluorescence of Texas red remained the same or decreased slightly (perhaps due to photobleaching).
Imaging the distribution of native β-actin mRNA in chicken embryonic fibroblasts
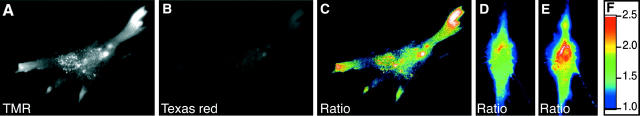
To image the distribution of β-actin mRNA in living fibroblasts, the culture dishes were returned to the CO2 incubator after injection. After 1 h of incubation, the cells were imaged with respect to their TMR and Texas red fluorescence, switching the optical filters for the two fluorophores in rapid succession. As shown for a representative cell (Fig. 4, A and B), TMR fluorescence was most concentrated in the largest and most active lamellipodia. Although the intensity of Texas red fluorescence was generally lower, its distribution did not appear to be very distinct from the distribution of the TMR fluorescence. Both images, in part, reflected the depth of the cytoplasm in various regions of the cell. In the lamellipodia, the depth of the cytoplasm is greater than in the intervening region between the lamellipodia and the perinuclear area (Hill et al., 1994). Thus, to extract the hybridization-dependent signals from the background fluorescence, which differs from region to region, we utilized an image-processing program to calculate a “ratio image,” by dividing the intensity of the TMR florescence at each pixel by the intensity of the Texas red fluorescence at the same pixel (Fig. 4 C). This ratio-imaging approach compensates for geographically variable levels of background fluorescence, and also compensates for geographical variations in the thickness of the cell (Dunn and Maxfield, 1998). In the ratio image, the magnitude of the ratio at each pixel is indicated by the color of the pixel, with warmer colors identifying higher ratios and cooler colors identifying lower ratios (Fig. 4 F). The preponderance of warmer colors in the most prominent lamellipodia indicates that β-actin mRNA was very abundant in those regions. Although one probe could yield detectable signals in these experiments, the signals were generally more reliable when all three probes were used together.
FIGURE 4.
Imaging the distribution of β-actin mRNA in living fibroblasts. A mixture of three TMR-labeled β-actin mRNA-specific molecular beacons and a Texas red-labeled control molecular beacon, each complexed with streptavidin, was microinjected into each cell. After 1 h of incubation, a typical cell was imaged in quick succession with respect to (A) TMR and (B) Texas red. A ratio image (C) was obtained by dividing the fluorescence intensity of TMR by the fluorescence intensity of Texas red at every pixel in the image. The color of each pixel reflects the value of the ratio, with warmer colors representing higher ratios and cooler colors representing lower ratios. (D and E) Two ratio images of a typical cell that was grown in the absence of serum. Image D shows the steady state in the absence of serum, and image E shows the changes that occurred 2 min after the addition of fresh serum to the medium. (F) Table of colors used to represent the ratio at each pixel in images C, D, and E.
To provide a biological control that would confirm the specificity of the signals that we obtained, the experiment was repeated with fibroblasts that were starved for serum. Under these conditions, the fibroblasts are very sluggish in their movement and their lamellipodia are poorly formed. In such cells, β-actin mRNA is symmetrically distributed in the perinuclear region of the cytoplasm, rather than being concentrated in the lamellipodia (Hill et al., 1994). When such quiescent cells were injected with the same mixture of biotinylated molecular beacons and streptavidin, and ratio images were obtained in the manner described above, β-actin mRNA-dependent fluorescence occurred mainly in the perinuclear domain (Fig. 4 D). However, within 2 min of adding fresh serum to media containing a serum-starved fibroblast, the distribution of the high-ratio signals shifted to one side of the nucleus and began to flow into nascent lamellipodia (Fig. 4 E). There is higher “integrated fluorescence” in Fig. 4 E compared to Fig. 4 D because the cell flattens as it prepares to move and as a result more fluorescent materials come into focus. These results demonstrate the ability of this technique to visualize rapid changes in the distribution of β-actin mRNA in living cells in response to environmental stimuli.
To image changes in the distribution of β-actin mRNA as the cells move and reorganize their lamellipodia, we obtained a series of time-lapse images with respect to TMR fluorescence (Supplementary Movie 1). Fluorescence was concentrated in the most active lamellipodia. The highly dynamic protrusive edges of the lamellipodia were more intensely labeled. As new lamellipodia formed adjacent to older lamellipodia, fluorescent material appeared to flow from older lamellipodia to new lamellipodia. Some of the fluorescence appeared as particulate objects that moved either from the nucleus toward a lamellipodium, or moved in the opposite direction. These dynamics are very similar to the dynamics displayed by the β-actin mRNA-binding protein, ZBP1 (Oleynikov and Singer, 2003). To confirm that these dynamic changes in fluorescence distribution reflect changes in the distribution of β-actin mRNA, we repeated the experiment with another motile fibroblast, this time collecting time-lapse ratio images by rapidly switching the TMR and Texas red filters at every time point (Supplementary Movie 2). Although some degradation of spatial resolution was apparent, the movement of the high-ratio signals was similar to the movement seen when imaging TMR fluorescence alone, confirming the specificity of dynamic fluorescence imaging with molecular beacons. To understand the sensitivity of this approach, it is important to note that the number of β-actin mRNA in each cell is ∼2500 molecules (Kislauskis et al., 1997) and each lamellipodium harbors only a fraction of the total population.
Effect of molecular beacon binding on translation of the mRNA
The introduction of a relatively small number of oligonucleotide probes should not have a deleterious effect on the life of a cell, unless the probe specifically affects the synthesis of the protein encoded by its target mRNA. Since the molecular beacons that we injected possessed a 2′-O-methylribonucleotide backbone, the target mRNA could not be destroyed by cellular ribonuclease H (Bratu et al., 2003). However, if the probes bind tightly to the coding regions of target mRNAs, the passage of ribosomes during translation might be blocked, or if the probes bind to the region containing the start codon (as molecular beacon CEF β-actin-66-83 does), the initiation of protein synthesis might be blocked (Dias and Stein, 2002). However, we did not observe any deleterious effects in fibroblasts caused by the presence of molecular beacons. The majority of the cells that were injected with molecular beacons remained attached to the substratum and continued to move about.
To address this issue more broadly, we explored the effect of molecular beacon binding on the translation of a synthetic mRNA encoding green fluorescent protein in live cells. Mimicking the experiments with β-actin mRNA, we prepared three molecular beacons that were specific for the coding region of GFP mRNA (Table 1) and hybridized them in vitro to a synthetic transcript of GFP mRNA. These molecular beacons were not bound to streptavidin. In preparing these hybrids, we utilized an excess of the three molecular beacons (each molecular beacon was 1.5 μM and GFP mRNA was 0.1 μM), and we confirmed that hybridization reached completion by real-time fluorescence monitoring. These hybrids were then injected into live Chinese hamster ovary cells. The development of green fluorescence in these cells could only occur if the injected GFP mRNA-molecular beacon hybrids were successfully translated. The injected cells were examined after an overnight incubation. We found that about half of the injected cells (identified by the fluorescence of the molecular beacons) developed green fluorescence (Fig. 5), indicating that some of the injected mRNA was capable of undergoing translation and produced functional green fluorescent protein. Either some of the mRNA molecules were not bound to molecular beacons (despite our efforts to maximize hybridization) and were therefore successfully translated, or the ribosomes were able to displace the molecular beacons as they moved along the mRNAs. Our prior results with oskar mRNA (Bratu et al., 2003), which indicate that a fraction of mRNA often fails to bind to molecular beacons, support the earlier possibility.
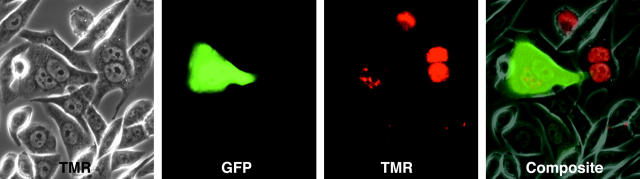
FIGURE 5.
Translation of GFP mRNA hybridized to molecular beacons. A synthetic GFP mRNA hybridized within its coding region to three molecular beacons was injected into the cytoplasm of Chinese hamster ovary cells, and the cells were imaged for GFP and molecular beacons (TMR) after an overnight incubation. Injected cells are identified by the red fluorescence of the excess molecular beacons that accumulated in their nuclei. Two of the four injected cells show production of green fluorescent protein.
Although these results indicate that mRNA can be imaged in living cells without seriously affecting their translation, there are several steps that can be taken to further reduce the possibility of deleterious effects occurring as a result of the binding of molecular beacons. One approach is to use a 3′-untranslated region of the mRNA as a target for the probes, and a second approach is to use molecular beacons that are designed so that they bind weakly to their targets, thus rapidly binding and detaching from the mRNA under physiological conditions (Bonnet et al., 1999).
DISCUSSION
We showed that when conventional molecular beacons are introduced into cells to image the distribution of specific mRNAs, they are rapidly sequestered within the nuclei, leaving little time for molecular beacons to hybridize to their cytoplasmic mRNA targets. We prevented nuclear sequestration by the attachment of a nonkaryophilic protein, streptavidin, to the molecular beacons. The use of these modified molecular beacons enabled us to image the distribution of β-actin mRNA in live chicken embryonic fibroblasts. In particular, we imaged dynamic changes in β-actin mRNA distribution that occurred as the fibroblasts extended and withdrew their lamellipodia. As a possible drawback of this approach, the binding of excess streptavidin to cellular biotin may adversely affect cellular physiology. However, that can be overcome by prior purification of the molecular beacon-streptavidin complex or by employing another protein.
The ability to visualize and track β-actin mRNA in motile cells will aid studies of the role that β-actin mRNA localization plays in fibroblast motility. In the absence of serum, fibroblasts are quiescent and hardly move (Mitchison and Cramer, 1996; Pollard and Borisy, 2003; Ridley et al., 2003). However, within 2 min of the addition of serum, vigorous lamellipodial movements begin. Components of serum initiate a series of signaling events that rapidly and directly modify actin polymerization and depolymerization dynamics (Ridley and Hall, 1992; Mitchison and Cramer, 1996; Pollard and Borisy, 2003; Ridley et al., 2003). The rapidity of the onset of motility, and its insensitivity to protein-synthesis inhibitors, suggest that localized β-actin mRNAs may not play a direct role in the process (Kislauskis et al., 1997; Shestakova et al., 2001). However, since localized β-actin mRNA is present in the lamellipodia of steadily moving cells, the localization of this mRNA must be important for motility at some stage. Molecular beacons modified for use in living cells will enable studies that elucidate differences between the kinetics of the onset of motility and the recruitment of β-actin mRNA to the lamellipodia. This may shed light on the role of β-actin mRNA localization in the mechanism of cellular motility.
The ability to track regions of cytoplasm possessing a high β-actin mRNA concentration will also help in the study of processes that are responsible for mRNA localization. For example, it has been postulated that ZBP1 binds to nascent β-actin mRNA in the nucleus, escorting it through the nuclear pores into the cytoplasm. Once in the cytoplasm, this complex attaches to motor proteins, which transport the mRNA to the lamellipodia, utilizing elements of the cytoskeleton as guides (Oleynikov and Singer, 2003). We will be able to test these postulates by visualizing the association of ZBP1 to β-actin mRNA in living fibroblasts. The β-actin mRNA can be followed with TMR-labeled molecular beacons and the ZBP1 can be fused to green fluorescent protein. It will be possible to detect the association of the mRNA and the protein by the coincidence of their fluorescence, or more specifically, by imaging fluorescence resonance energy transfer between them.
Although the experiments that we carried out utilized molecular beacons for the detection of β-actin mRNA in chicken embryonic fibroblasts, the significance of using the modified molecular beacons that we describe lies in their ability to specifically detect and image any desired mRNA in living cells. Of course, the scope of molecular beacon-based mRNA imaging, as for all fluorescence-based methods, is limited by the number of target mRNA molecules that are present in a cell. β-actin mRNA is one of the more abundant messages in the cell, with an estimated copy number of 2500 molecules per cell (Kislauskis et al., 1997), yet the intensity of the fluorescence signals resulting from the hybridization of the molecular beacons was only 2.5-fold higher than the background fluorescence. Moreover, the β-actin mRNA was concentrated in small regions of the cell. This indicates that detection of most mRNAs that may be spread throughout the cell will be challenging. Technical improvements that produce a 10- to 100-fold increase in the intensity of the signals (or a similar decrease in background levels), will be necessary to visualize most mRNAs by this method.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Diana P. Bratu, Cindy Fung, Fred Russell Kramer, Salvatore A. E. Marras, Musa M. Mhlanga, Arjun Raj, and Diana Y. Vargas for their contributions.
This work was supported by National Institutes of Health grants GM-070357 and EB-000277.
Osama Alsmadi's present address is King Faisal Specialist Hospital and Research Centre, MBC-03, PO Box 3354, Riyadh 11211, Kingdom of Saudi Arabia
References
- Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 2:437–445. [DOI] [PubMed] [Google Scholar]
- Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA. 96:6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu, D. P., B.-J. Cha, M. M. Mhlanga, F. R. Kramer, and S. Tyagi. 2003. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. USA. 100:13308–13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabauvalle, M. C., B. Schulz, U. Scheer, and R. Peters. 1988. Inhibition of nuclear accumulation of karyophilic proteins in living cells by microinjection of the lectin wheat germ agglutinin. Exp. Cell Res. 174:291–296. [DOI] [PubMed] [Google Scholar]
- Dias, N., and C. A. Stein. 2002. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 1:347–355. [PubMed] [Google Scholar]
- Dirks, R. W., C. Molenaar, and H. J. Tanke. 2001. Methods for visualizing RNA processing and transport pathways in living cells. Histochem. Cell Biol. 115:3–11. [DOI] [PubMed] [Google Scholar]
- Dunn, K., and F. R. Maxfield. 1998. Ratio imaging instrumentation. Methods Cell Biol. 56:217–236. [PubMed] [Google Scholar]
- Hanover, J. A., C. K. Cohen, M. C. Willingham, and M. K. Park. 1987. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 262:9887–9894. [PubMed] [Google Scholar]
- Hartig, R., R. L. Shoeman, A. Janetzko, S. Grub, and P. Traub. 1998. Active nuclear import of single-stranded oligonucleotides and their complexes with non-karyophilic macromolecules. Biol. Cell. 90:407–426. [PubMed] [Google Scholar]
- Hill, M. A., L. Schedlich, and P. Gunning. 1994. Serum-induced signal transduction determines the peripheral location of β-actin mRNA within the cell. J. Cell Biol. 126:1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kill, I. R. 1996. Localisation of the Ki-67 antigen within the nucleolus. Evidence for a fibrillarin-deficient region of the dense fibrillar component. J. Cell Sci. 109:1253–1263. [DOI] [PubMed] [Google Scholar]
- Kislauskis, E. H., X. Zhu, and R. H. Singer. 1994. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis, E. H., X. Zhu, and R. H. Singer. 1997. β-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 136:1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, J. B., and R. H. Singer. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 45:407–415. [DOI] [PubMed] [Google Scholar]
- Leonetti, J. P., N. Mechti, G. Degols, C. Gagnor, and B. Lebleu. 1991. Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 88:2702–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, P., T. Misteli, B. F. Baker, C. F. Bennett, and D. L. Spector. 2000. Nucleocytoplasmic shuttling: a novel in vivo property of antisense phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 28:582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, T. 1998. In situ visualization of mRNA for basic fibroblast growth factor in living cells. Biochim. Biophys. Acta. 1379:178–184. [DOI] [PubMed] [Google Scholar]
- Mitchison, T. J., and L. P. Cramer. 1996. Actin-based cell motility and cell locomotion. Cell. 84:371–379. [DOI] [PubMed] [Google Scholar]
- Oleynikov, Y., and R. H. Singer. 2003. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 13:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J. C., E. S. Browne, D. E. Wolfe, and T. Pederson. 1998. Intranuclear diffusion and hybridization state of oligonucleotides measured by flurescence correlation spectroscopy in living cells. Proc. Natl. Acad. Sci. USA. 95:6043–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 70:389–399. [DOI] [PubMed] [Google Scholar]
- Ridley, A. J., M. A. Schwartz, K. Burridge, R. A. Firtel, M. H. Ginsberg, G. Borisy, J. T. Parsons, and A. R. Horwitz. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. [DOI] [PubMed] [Google Scholar]
- Ross, A. F., Y. Oleynikov, E. H. Kislauskis, K. L. Taneja, and R. H. Singer. 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17:2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova, E. A., R. H. Singer, and J. Condeelis. 2001. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA. 98:7045–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixou, S., F. C. Szoka Jr., G. A. Green, B. Giusti, G. Zon, and D. J. Chin. 1994. Intracellular oligonucleotide hybridization detected by fluorescence resonance energy transfer. Nucleic Acids. Res. 22:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol, D. L., X. Zhang, P. Lu, and A. M. Gewirtz. 1998. Real time detection of DNA.RNA hybridization in living cells. Proc. Natl. Acad. Sci. USA. 95:11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsourkas, A., M. A. Behlke, and G. Bao. 2003. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 30:5168–5174. [PMC free article] [PubMed] [Google Scholar]
- Tsuji, A., H. Koshimoto, Y. Sato, M. Hirano, Y. Sei-Iida, S. Kondo, and K. Ishibashi. 2000. Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys. J. 78:3260–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49–53. [DOI] [PubMed] [Google Scholar]
- Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303–308. [DOI] [PubMed] [Google Scholar]
- Weil, D., S. Boutain, A. Audibert, and F. Dautry. 2000. Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA. 6:962–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff, M. 1983. tRNA transport from the nucleus in a eukaryotic cell: carrier-mediated translocation process. Proc. Natl. Acad. Sci. USA. 80:6436–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.