Abstract
The effect of external osmotic pressure on the extent of DNA ejection from bacteriophage-λ was recently investigated (Evilevitch et al., 2003). The total length of DNA ejected was measured via the 260-nm absorption by free nucleotides, after opening of the capsids in the presence of varying amounts of polyethylene glycol 8000 and DNase I. As a function of osmolyte concentration, this absorption was shown to decrease progressively, ultimately vanishing completely for a sufficiently high external osmotic pressure. In this work we report the results of both sedimentation and gel analysis of the length of DNA remaining inside the capsids, as a function of osmolyte concentration. It is confirmed in this way that the progressive inhibition of DNA ejection corresponds to partial ejection from all of the capsids.
INTRODUCTION
Recently an in vitro experiment was reported in which it was demonstrated that the ejection of double-stranded DNA from a phage capsid can be suppressed by external osmotic pressure (Evilevitch et al., 2003). Basically, the force driving the ejection, feject, is balanced by a force, fresist, resisting entry of the DNA into solution. The DNA packaged in the capsid is highly stressed because it has been strongly bent and crowded on itself (Kindt et al., 2001; Purohit et al., 2003; Tzlil et al., 2003). This stored energy gives rise to the force ejecting the DNA along its length through the viral tail, upon binding of the tail to a receptor in the host cell (bacterium) membrane. The force decreases monotonically as ejection proceeds, whereas fresist is an increasing function of the concentration of osmolyte in the external solution (Castelnovo et al., 2003; Evilevitch et al., 2004). For a given external osmolyte concentration, then, the ejection will stop as soon as feject drops to the corresponding value of fresist. In this way, the extent of ejection can be continuously controlled simply by increasing the external osmolyte concentration. In the case of λ-phage in TM buffer, for example, it was shown that an external osmotic pressure of 30 atm is sufficient to completely suppress ejection, whereas at a pressure of 5 atm as much as half of the genome is ejected (Evilevitch et al., 2003). These results are consistent with earlier studies (Serwer et al., 1983) that showed packaging and ejection efficiencies can be influenced by the presence of an osmolyte.
Our earlier measurements were carried out by digesting all of the ejected DNA and, after spinning down the system, using 260-nm absorption to determine the concentration of nucleotides in the supernatant for each of successively higher concentrations of osmolyte. These measured concentrations were converted to fractions of genome ejected by assuming that ejection was complete at 0% osmolyte concentration and comparing the ultraviolet (UV) absorptions at different osmolyte concentrations with that at 0%. For example (see above) the absorbance at 5 atm was found to be half that at zero external osmotic pressure (0% osmolyte). There are two very different ways to interpret this result, however, and the experiment could not distinguish between them. In the first scenario, all of the capsids eject half of their genomes in the presence of a concentration of osmolyte corresponding to an osmotic pressure of 5 atm in the host external solution. Alternatively, the effect of 5-atm pressure might have been to inhibit ejection from half of the capsids completely whereas the other half ejected all of their genome.
We report here the results of experiments that clearly distinguish between the two mechanisms. They consist of direct measurements of the distribution of lengths of DNA remaining in the capsids after ejection rather than the amount of ejected DNA. Once again, polyethylene glycol (PEG) is used to control the ejection of the DNA but now after ejection we recover the capsids, lyse them, and examine the exposed DNA by pulse-field electrophoresis and sedimentation velocity. We find that at each PEG concentration there is a single length of DNA that remains inside the capsid and that this length varies with concentration in a way that is consistent with the earlier measurements of the ejected length. Moreover, we find that in the absence of PEG (and hence for fresist = 0), no DNA remains inside the capsids.
EXPERIMENTAL METHODS
Details of the sample preparation procedure are presented elsewhere (Evilevitch et al., 2003). Bacteriophage-λ EMBL3 (Goldschmidt-Clermont and Rahire, 1986) with genome length 41.5 kb (isolated from an infected culture of Escherichia coli NM539; Moseley et al., 2002) was mixed with its receptor protein, LamB (membrane protein from Shigella sonneii strain 3070, expressed in and purified from pop 154, a strain of E. coli K12), to allow phage to eject its DNA in vitro (Graff et al., 2002; Randall-Hazelbauer and Schwartz, 1973). DNA ejection was complete within seconds (Novick and Baldeschwieler, 1988). The ejection was studied at 37°C in PEG8000 solutions with concentrations between 0 and 20% by weight. The ionic strength was set by the TM buffer (10 mM MgSO4 and 50 mM Tris-HCl, pH 7.4). All the phage-receptor mixtures contained DNase I, which cut the ejected DNA into nucleotides. After ejection, the samples were incubated for 1 h to assure that the enzyme had sufficient time to act.
After DNase I digestion the phage capsids were opened by denaturation in phenol/chloroform mixture (50% phenol, 48% chloroform, 2% isoamyl alcohol) saturated with TE buffer (10 mM Tris-HCl, pH 8.0, 1.0 mM EDTA). The DNA remaining in the aqueous phase was subjected to a second extraction with chloroform/isoamyl alcohol solution (24:1) that removed the remaining traces of PEG and protein. The DNA was then precipitated by addition of sodium acetate to a final concentration of 0.3 M and 70% alcohol. After centrifugation, the DNA pellet was purified by an additional wash in 70% ethanol, dried at 50°C, and then dissolved in TE buffer (Silhavy, 1984). Because intact λ-DNA has cohesive ends that can circularize, samples were heated to 65°C for 5 min and then cooled in ice to eliminate this possibility before measurements of length distributions were made.
The length of the extracted DNA fragments was determined by pulsed-field electrophoresis. Measurements were carried out with a CHEF-DRII electrophoresis system from Bio-Rad (Hercules, CA). Separations were performed on a 1.0% agarose gel in 0.5× TBE buffer (0.5× 45 mM Tris-borate, 1 mM EDTA, pH 8) at 14°C. The switch time was ramped from 0.2 to 3.4 s, with a field of 15 V/cm, and run time was 15 h. For size determination of the DNA fragments, we used CHEF DNA size standards for pulsed-field electrophoresis from Bio-Rad. The bands were visualized with ethidium bromide staining.
The extracted DNA in 150 mM NaCl, 10 mM Tris, 1 mM EDTA, pH 7.4, was also examined by sedimentation velocity analysis at 20°C in a Beckman Optima XL-A analytical ultracentrifuge (Fullerton, CA) at speeds of 20,000 rpm using absorption optics at 260 nm and a 12-mm pathlength double-sector cell. Apparent sedimentation coefficient (s) distributions, uncorrected for diffusion, were determined as g(s) plots using the Beckman ORIGIN-based software (Version 3.01). The values of g(s), which are proportional to the weight fractions of material with a given sedimentation coefficient, s = (1/ω2t)(ln(rm/r)), were calculated from g(s) = (1/co)t(dc/dt)(r/rm)2/s, where ω is the angular velocity of the rotor, co the initial concentration, r the radius, rm the radius of the meniscus, and t the time (Stafford, 1992). They were corrected for density and viscosity to an s20,w value using 0.55 ml/g for the partial specific volume at 20°C (Laue, 1992). To minimize the effects of the concentration dependence of the sedimentation coefficient, concentrations of the samples were low (6 μg/ml or less). At these low concentrations, sedimenting boundaries are sometimes destabilized by mechanical and thermal convection in the cell. As a precaution, a crude sucrose gradient sufficient to stabilize the sedimenting boundaries was prepared. A 0.6% solution of sucrose in half the sample was used as an underlayer for the other half. The layers were gently mixed by tilting the cell, which generates a crude sucrose gradient that is sufficient to stabilize the sedimenting boundaries.
The molecular weight M of the DNA fragments can be the determined from the experimental sedimentation coefficients by using semiempirical relations (Eisenberg and Crothers, 1979). The relation for a flexible DNA chain is 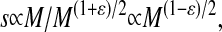 where
where 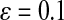 (Crothers and Zimm, 1965). Because M is proportional to the length of DNA, L, we can use this relation to obtain an estimate of the length of the DNA fragment relative to that of the full length, L0, λ-genome:
(Crothers and Zimm, 1965). Because M is proportional to the length of DNA, L, we can use this relation to obtain an estimate of the length of the DNA fragment relative to that of the full length, L0, λ-genome: 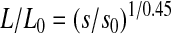 .
.
RESULTS AND DISCUSSION
For analysis of the DNA length distribution remaining in the capsid, DNA was extracted and purified from the phage after ejection in 0, 5, 10, and 20% PEG8000. As a calibration standard, the entire λ-genome was also extracted from a phage sample lacking both receptor (LamB) and PEG, i.e., the capsid was not opened and the entire DNA remained inside. We can compare the results from the two independent methods for determination of the length of DNA remaining in the capsid after ejection with the previously established complementary UV-analysis data on the ejected fraction of DNA.
A photograph of the pulsed-field gel is shown in Fig. 1. The right-most lane, labeled “DNA ladder,” is an 8–48.5-kb standard consisting of 13 bands of DNAs of known length. The next lane is the band corresponding to the DNA from an unopened λ-capsid, which is 41.5-kb long because we worked with a 41.5-kb EMBL3 mutant phage rather then the 48.5-kb wild type. The next lane to the left corresponds to the DNA extracted when LamB and 5% PEG were added to the phage solution; here we see the result of a partial ejection that left a length of ∼9 kb inside. It is evident from the other lanes that higher concentrations of PEG led to progressively longer DNA fragments, e.g., at 10% PEG ∼17 kb remains, whereas by 20% PEG the major part of the genome remains encapsidated; a band is most prominent at 30 kb. The length of the DNA fragments was determined by a least-squares fit to the DNA size standard “DNA ladder” sample.
FIGURE 1.

Results from pulsed-field gel electrophoresis. Analysis of the DNA length remaining in the capsid after phenol-extraction. (Right to left) “DNA ladder” is the 8–48-kb size standard; the 41.5-kb band is the entire EMBL3 λ-genome extracted from phage solution missing both LamB receptor and PEG; the 9-kb band is the DNA remaining in the capsid after ejection in 5% PEG; the 17-kb band is the DNA remaining in the capsid after ejection in 10% PEG; and the 30-kb band is the DNA remaining in the capsid after ejection in 20% PEG. Partial ejection and its suppression with increasing PEG concentration are evident. The 41.5-kb band present in all the samples results from a fraction of phage that remains unopened and is the same in all the samples within the same batch.
As is also evident from the gel, intact 41.5-kb DNA is also present in all the samples. This DNA is from a rather small fraction of phages (∼10% estimated from analytical ultracentrifugation analysis) that remain unopened even when there is an excess of LamB. This was shown in the earlier analytical ultracentrifugation analyses (Evilevitch et al., 2003) comparing DNA content from thermally ruptured phage (when all of the DNA comes out) with that from samples treated with LamB with and without PEG. The electron micrograph in Fig. 2 shows two phage particles sharing a single receptor molecule, with the consequence that only one of them has ejected its genome; this situation might account for the small fraction of unopened phage that is routinely observed.
FIGURE 2.
Cryo-EM images of phage in the presence of the LamB receptor. When two phages attach to the same receptor, only one of the phages ejects its DNA whereas the other remains filled.
Fig. 3 shows the sedimentation boundaries measured as 260-nm UV absorbance as a function of height in the cell at constant time intervals for two samples: (panel a) no PEG-no LamB (entire genome) sample and (panel b) 10% PEG sample. There is only a single boundary in the absence of PEG, whereas two boundaries that move at different rates are evident in Fig. 3 b; the slower moving one corresponds to the shorter DNA fragment (∼20 ± 5 kb) remaining in the phage and the other one to the entire λ-genome, which is always present because of a small fraction of intact capsids.
FIGURE 3.

Analytical ultracentrifugation analysis of phenol-extracted DNA from: (a) phage not treated with receptor (entire λ-genome), showing only one length distribution of DNA; (b) receptor-treated phage in 10% w/w PEG8000 solution showing two length distributions of DNA corresponding to a short DNA fragment remaining in the capsids after ejection and a full-length genome remaining in unopened phage. Samples were centrifuged at 20,000 rpm, 20°C, and the concentration of DNA length distribution populations was followed by scanning the cells at 260 nm. The moving boundary was monitored at 4.5-min time intervals in panel a and 16-min intervals in panel b. To minimize the concentration dependence of the sedimentation coefficient, samples were run at low concentrations of 6 μg/ml or less (corresponding OD at 260 nm was 0.05–0.12).
Sedimentation coefficients, s20,w, obtained from these data are given in Fig. 4, which shows g(s) plots for each DNA length distribution population in the sample. The samples are DNA fragments extracted from the phage in a no LamB-no PEG sample (entire λ-genome), and from those in 5, 10, and 20% PEG mixtures. Note that the relative areas associated with the two optical density (OD) peaks within the same sample do not show the correct relation between the numbers of intact and opened capsids, because they involve weight (versus number) averages, and hence the OD for shorter DNA fragments is substantially lower than that for the entire genome. The single peak in Fig. 4 a at s20,w = 32.9 S corresponds to the sedimentation coefficient for the 41.5-kb intact DNA. In the 5% PEG sample in Fig. 4 b the major peak at s20,w = 20 S corresponds to a length of 14 ± 4 kb. The peaks in Fig. 4, c and d, which are for the 10 and 20% PEG samples, respectively, are at s20,w = 24.2 S, corresponding to 20 ± 5 kb, and s20,w = 30 S, corresponding to 34 ± 6 kb. Fig. 4, b–d, also display a second peak at s20,w = 32.9 S, which is associated with the unopened capsids present in all samples. The two peaks are completely resolved only in Fig. 4 b. Higher resolution was not possible at the low signal/noise ratios that result from the very low DNA concentrations that have been employed to minimize the effects of concentration on s.
FIGURE 4.
Sedimentation coefficient (s) distributions of phenol-extracted DNA determined from g(s) plots, corrected for density and viscosity to an s20,w value. (a) Sample of phage lacking LamB receptor and PEG (entire genome); (b) DNA from receptor-treated phage in 5% PEG; (c) DNA from receptor-treated phage in 10% PEG; and (d) DNA from receptor-treated phage in 20% PEG. In 5, 10, and 20% w/w PEG samples there are two peaks in the sedimentation coefficient distributions, one from the shorter DNA fragment remaining in the phage after ejection and one from the full λ-genome from unopened phage.
Data from the pulsed-field electrophoresis and sedimentation velocity analysis are compared with the earlier UV absorbance measurements (Evilevitch et al., 2003) in Fig. 5, a plot of the fraction of DNA ejected against the external osmotic pressure. The horizontal error bars are standard deviations estimated by propagation of the uncertainties in the PEG concentration related to the difficulties in accurate pipetting of viscous solutions. The vertical error bars are standard deviations associated with uncertainties in the respective analytical approaches. For the pulsed-field gel electrophoresis data the vertical error bars were estimated from the widths of the DNA bands observed on the gel; here there is excellent agreement with the UV-absorbance measurements. The data from the sedimentation velocity analysis are systematically lower by a few percent, but display the same trend; even at the low concentrations employed in the sedimentation experiments, the sedimentation coefficients are affected by their concentration dependence and this effect will increase the values of s by a few percent.
FIGURE 5.
Combined data for the fraction of DNA ejection as a function of the osmotic pressure (calculated from the concentration of PEG8000). The results from three experimental methods are shown: pulsed-field electrophoresis (○), sedimentation velocity analysis (□), and previously measured UV absorbance data (▵) for three different batches of phage (Evilevitch et al., 2003).
The agreement between three independently measured data sets illustrates nicely that the previously determined fraction of ejected DNA does correspond directly to the length of the genome ejected versus external osmotic pressure. This result confirms the earlier physical interpretation of the experiment, namely, that the ejection of DNA in the presence of osmotic pressure is partial and the corresponding length of ejected genome decreases with increasing osmotic force until it becomes completely suppressed.
The experiments appear to be in excellent accord with a physical model in which the force of ejection derives from the energy stored in the bent and compressed DNA (Kindt et al., 2001; Tzlil et al., 2003; Purohit et al., 2003; Evilevitch et al., 2003). In the last stage of ejection, however, the DNA remaining in the capsid and the tail is no longer bent or compressed; the diameter of the capsid (64 nm) and the length of the tail (172 nm) (Hendrix, 1983) are sufficient to accommodate a 700-bp linear segment of DNA that would not be subject to an ejection force. Moreover, if there were a strong attractive interaction between the inner surface of the capsid and the DNA, an even greater fraction of the DNA might remain in the capsid when the ejection force had fallen to zero.
To investigate this possibility we have examined the length of DNA remaining in the capsid after ejection has taken place in a solution without added osmolyte. The experimental protocol was identical to that previously used for the solutions containing PEG. In this case, however, because the segments remaining were expected to be short the electrophoresis was carried out on a 1% agarose gel to resolve the fragments >1 kb in length, and an 8% acrylamide gel that can resolve fragments between 70 and 1000 bp. Complementary sedimentation velocity analysis was performed as well. No DNA was detected in any of these experiments, suggesting that the ejection is complete (e.g., no DNA remains due to attraction to the capsid).
It is possible that, in the absence of attraction to the inside of the capsid or tail, the final, unstressed, >700-bp segment simply diffuses out into solution on the timescale of its incubation with DNase. But there are at least a couple of possibilities for additional forces acting along the length of the viral genome, beyond the ejection and osmotic forces that we have considered here. First, although the DNA is cut into nucleotides by the DNase, the timescale for ejection is much shorter than that for complete digestion. Thus, entropic fluctuations in the longer ejected part of the genome may be sufficient to pull out the remaining DNA; this effect would be driven by a force of order kT/ξ, where kT is the thermal energy and ξ the DNA persistence length. Another possibility is that the pulling force arises from the binding of the DNase I (Zandi et al., 2003), i.e., from its adsorption onto the chain; here the force is of order U/d, where U is the binding energy and d the average distance between adsorption events. In either case, however, it seems clear that if there is any attraction between the capsid interior and the DNA, it cannot be strong, because it is overwhelmed by one or both of these forces. Preliminary cryoelectron microscopy (cryo-EM) investigations, carried out by one of us (A.E.) in the absence of DNase I, suggest that some DNA remains in the capsid; it is observed that the ejected DNA, which is not being digested, attaches to the hydrophilic grid boundaries and appears to remain connected to the phage tails.
Acknowledgments
We are pleased to acknowledge many helpful discussions with Lucienne Letellier, Paulo Tavares, Eric Raspaud, Françoise Livolant, Amélie Leforestier, Bengt Jönsson, and Ulf Olsson. Gunnel Karlsson is gratefully acknowledged for help with cryo-EM imaging performed at the Center for High Resolution Electron Microscopy at Lund University, Sweden.
The work has been financially supported by the National Science Foundation, through grant No. CHE00-7931 to C.M.K. and grant No. CHE99-88651 to W.M.G., and by the Swedish Foundation for Internationalization of Research and Higher Education through a grant to A.E.
Alex Evilevitch's present address is Lund University, Center for Chemistry and Chemical Engineering, Dept. of Molecular Biophysics, Box 124, SE-221 00 Lund, Sweden.
References
- Castelnovo, M., R. K. Bowles, H. Reiss, and W. M. Gelbart. 2003. Force resisting insertion in a colloidal suspension. Eur. Phys. J. E. Soft Matter. 10:191–197. [DOI] [PubMed] [Google Scholar]
- Crothers, D. M., and B. H. Zimm. 1965. Viscosity and sedimentation of the DNA from bacteriophages T2 and T7 and the relation to molecular weight. J. Mol. Biol. 12:525–536. [DOI] [PubMed] [Google Scholar]
- Eisenberg, D., and D. Crothers. 1979. Physical Chemistry with Applications to the Life Sciences. The Benjamin/Cummings Publishing Company, San Francisco, CA.
- Evilevitch, A., M. Catselnovo, C. M. Knobler, and W. M. Gelbart. 2004. Measuring the force ejecting DNA from phage. J. Phys. Chem. B. 108:6838–6843. [Google Scholar]
- Evilevitch, A., L. Lavelle, C. M. Knobler, E. Raspaud, and W. M. Gelbart. 2003. Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl. Acad. Sci. USA. 100:9292–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M., and M. Rahire. 1986. Sequence, evolution and differential expression of the 2 genes encoding variant small subunits of ribulose bisphosphate carboxylase-oxygenase in Chlamydomonas reinhardtii. J. Mol. Biol. 191:421–432. [DOI] [PubMed] [Google Scholar]
- Graff, A., M. Sauer, P. V. Gelder, and W. Meier. 2002. Virus-assisted loading of polymer nanocontainer. Proc. Natl. Acad. Sci. USA. 99:5064–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix, R. W. 1983. Lambda II. R. W. Hendrix, editor. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Kindt, J., S. Tzlil, A. Ben-Shaul, and W. M. Gelbart. 2001. DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. USA. 98:13671–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue, T. 1992. Analytical Ultracentrifugation in Biochemistry and Polymer Science. S. E. Harding, A. J. Rowe, and J. C. Horton, editors. Redwood Press, Cambridge, UK.
- Moseley, J. L., M. D. Page, N. P. Alder, M. Eriksson, J. Quinn, F. Soto, S. M. Theg, M. Hippler, and S. Merchant. 2002. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell. 14:673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, S. L., and J. D. Baldeschwieler. 1988. Fluorescence measurement of the kinetics of DNA injection by bacteriophage-lambda into liposomes. Biochemistry. 27:7919–7924. [DOI] [PubMed] [Google Scholar]
- Purohit, P. K., J. Kondev, and R. Phillips. 2003. Mechanics of DNA packaging in viruses. Proc. Natl. Acad. Sci. USA. 100:3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall-Hazelbauer, L., and M. Schwartz. 1973. Isolation of the Bacteriophage Lambda Receptor from Escherichia coli. J. Bacteriol. 116:1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer, P., W. E. Masker, and J. L. Allen. 1983. Stability and in vitro DNA packaging of bacteriophages: effects of dextrans, sugars, and polyols. J. Virol. 45:665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy, T. J. 1984. Experiments with Gene Fusions. T. J. Silhavy, M. L. Berman, L. W. Enquist, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Stafford, W. F. 1992. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 203:295–301. [DOI] [PubMed] [Google Scholar]
- Tzlil, S., J. T. Kindt, W. M. Gelbart, and A. Ben-Shaul. 2003. Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 84:1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi, R., D. Reguera, J. Rudnick, and W. M. Gelbart. 2003. What drives the translocation of stiff chains? Proc. Natl. Acad. Sci. USA. 100:8649–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]





