Abstract
DNA ejection from bacteriophage T5 can be passively driven in vitro by the interaction with its specific host receptor. Light scattering was used to determine the physical parameters associated with this process. By studying the ejection kinetics at different temperatures, we demonstrate that an activation energy of the order of 70 kBT must be overcome to allow the complete DNA ejection. A complex shape of the kinetics was found whatever the temperature. This shape may be actually understood using a phenomenological model based on a multistep process. Passing from one stage to another requires the mentioned thermal activation of pressurized DNA inside the capsids. Both effects contribute to shorten or to lengthen the pause time between the different stages explaining why the T5 DNA ejection is so slow compared to other types of phage.
INTRODUCTION
Nearly 20 years ago, it was hypothesized that forces driving DNA ejection from bacteriophage into the host cell could come from the strong repulsions that the neighboring DNA portions experience due to being locally confined in their capsids (Riemer and Bloomfield, 1978; Earnshaw and Casjens, 1980). Many recent theories, simulations, and experiments support this hypothesis (Odijk, 1998; Kindt et al., 2001; Evilevitch et al., 2003). The strong confinement and bending of the entire genome inside the viral capsid impose force values as high as 50 pN (Smith et al., 2001). Consequently, one can liken these phage particles to DNA cannons that expel their genetic materials once their specific membrane receptors trigger the opening of the proteinaceous gatekeeper. This strategy has been observed on λ-phage in vitro (Evilevitch et al., 2003) but may vary from phage to phage. Interestingly, for T5 phage, two steps in the in vivo DNA transfer have been reported (Lanni, 1968; Mc Corquodale and Warner, 1988). How does such a multistep process occur and what is the mechanism able to interrupt the expulsion or to block the high pressurized DNA during its transfer? To address the T5 DNA ejection strategy, we measured here the kinetics of in vitro DNA ejection.
In vitro T5 DNA ejection may be simply triggered by the addition of its Escherichia coli receptor FhuA (Boulanger et al., 1996). The phage tail tip binds to the receptor, leading to conformational changes that are transmitted to the head-tail connector triggering its opening and the release of the DNA. This release was monitored earlier by measuring the increase of the fluorescence intensity of a DNA intercalating dye (Boulanger et al., 1996). In this study, we determined for the first time the ejection averaged kinetics by light scattering, thus avoiding all the drawbacks that might be associated with fluorescence staining. For instance, the detection of the slow ejection kinetics may be impeded by the photobleaching effect and the possible diffusion of the probe through the permeable capsids. The light-scattering method is well known for many decades and is commonly used to characterize and quantify the mass and size of macromolecules dispersed into solution. We observed that the ejection kinetics was not a simple order process. Its complex shape could be explained using a phenomenological model based on rate equations and including a multistep description. Moreover temperature and ionic conditions were varied to clarify which kind of barrier blocks the DNA expulsion.
MATERIALS AND METHODS
Preparation of phage T5 and purification of its E. coli membrane protein receptor FhuA
In this study, we used T5st(0) (114 kbp), a T5 heat stable mutant deleted of 7.2% of its genome (Scheible et al., 1977). Roughly one-third of the phage mass comes from the proteins (Mproteins = 3 × 107 Da) and two-thirds from DNA (MDNA = 7.1 × 107 Da). Phage particles were produced on E. coli Fsuβ+ and purified as described by Bonhivers et al. (1996). They were stored in the working buffer: 100mM NaCl, 1mM MgSO4, 1mM CaCl2, and 10mM Tris-HCl pH 7.6. The final titer was 1013 phage/ml. The membrane protein receptor FhuA was overexpressed in E. coli HO830fhuA transformed with plasmid pHX405 and purified following the protocol described by Boulanger et al., (1996).
For light-scattering measurements, phage T5 and FhuA were diluted in the working buffer containing 0.03% LDAO (N, N-dimethyldodecylamine-N-oxide), the detergent used to solubilize FhuA. A typical sample contained 3 × 1010 T5 particles in a final volume of 0.3 ml, leading to a phage concentration (DNA + proteins) equal to 16.8 mg/l. The concentration of the membrane receptor FhuA was varied from 2 to 20 mg/l, which corresponds to a number of FhuA molecules per phage particle varying from 100 to 1000.
Light scattering
A homemade light-scattering apparatus was used to measure the DNA ejection from capsids. Most of the practical aspects and explanations required to set up and to use this type of apparatus are detailed in Huglin (1972), Berne and Pecora (1976), and Chu (1991). We used a He-Ne laser polarized light source of wavelength λ0 = 632.8 nm and of power 75 mW. This incident light was attenuated by a factor 30. The scattered light was detected by a HAMAMATSU photon counting head (H7421 series) and recorded using a RACAL-DANA counter (Universal Counter 1991) in a frequency mode of counting interval 0.1 s. Phage samples were placed into a thermostated cell, at the center of a goniometer that allowed us to collect the scattered intensity at different angles θ from the incident light. The scattering vector q = (4π ns/λ0) sin(θ /2) with ns the buffer refractive index corresponds to the inverse of the observation length scale. In this analysis of the initial and final states, θ angles were varied from 40° to 140° leading to a length scale q−1 from 110 to 40 nm. Regarding the kinetics, most of the data were recorded at θ = 90°. For a solution sufficiently diluted to neglect the interactions between isolated phage, the intensity I(θ) or I(q) scattered by phage may be expressed by I(q) = α C M P(q), C and M denoting the phage concentration and molar mass, respectively. To obtain the intensity I(q) only scattered by phage, the intensity scattered by buffer should be in principle simply subtracted from the total detected intensity. However because of its low scattering level, the buffer contribution to the signal was neglected here. The numerical prefactor α includes the Rayleigh's scattering law in 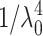 and the contrast (∂n/∂C)2 between phage particles and buffer −(∂n/∂C) being the refractive index increment. Using toluene to calibrate the experimental set up, this prefactor may be written as
and the contrast (∂n/∂C)2 between phage particles and buffer −(∂n/∂C) being the refractive index increment. Using toluene to calibrate the experimental set up, this prefactor may be written as 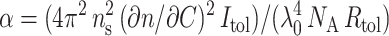 with NA the Avogadro's number and Rtol = 1.4 × 10−5 cm−1 the toluene Rayleigh's ratio. In our standard conditions, the scattering intensity of toluene being equal to Itol = 2.9 × 103, the prefactor α simply reduced to α = 1478 × (∂n/∂C)2. Now the angular dependence of I(q) is expressed via the form factor P(q), which is lower or equal to unity depending on the product qRg, where Rg corresponds to the phage radius of gyration. In our q range, the phage form factor P(q) may be approximated by a Guinier expansion law: 1/P(q) = 1 + (qRg)2/3. In the limit of nil angle, P(q) becomes equal to unity and the scattered intensity expression divided by the concentration C reduces to I(q→0)/C = αM and therefore the relative intensity becomes proportional to the molar mass of phage. For most of our samples, the working phage concentration was equal to C = 16.8 × 10−3 g/l and the corresponding scattered intensity extrapolated at zero angle reached the value I(q→0) = 1.66 × 105. Since the phage molar mass M is known, one may evaluate α or more precisely their contrast when they are filled up with DNA: (∂n/∂C)tot = 0.253 cm3/g.
with NA the Avogadro's number and Rtol = 1.4 × 10−5 cm−1 the toluene Rayleigh's ratio. In our standard conditions, the scattering intensity of toluene being equal to Itol = 2.9 × 103, the prefactor α simply reduced to α = 1478 × (∂n/∂C)2. Now the angular dependence of I(q) is expressed via the form factor P(q), which is lower or equal to unity depending on the product qRg, where Rg corresponds to the phage radius of gyration. In our q range, the phage form factor P(q) may be approximated by a Guinier expansion law: 1/P(q) = 1 + (qRg)2/3. In the limit of nil angle, P(q) becomes equal to unity and the scattered intensity expression divided by the concentration C reduces to I(q→0)/C = αM and therefore the relative intensity becomes proportional to the molar mass of phage. For most of our samples, the working phage concentration was equal to C = 16.8 × 10−3 g/l and the corresponding scattered intensity extrapolated at zero angle reached the value I(q→0) = 1.66 × 105. Since the phage molar mass M is known, one may evaluate α or more precisely their contrast when they are filled up with DNA: (∂n/∂C)tot = 0.253 cm3/g.
RESULTS
Before and after DNA ejection: the initial and final states
Solutions of phage T5 diluted in buffer without FhuA were characterized by measuring the time-averaged scattered intensity at different angles θ or different q values. A typical signal recorded at a temperature of T = 23°C is presented (see solid circles) in Fig. 1. This signal decreases with the detection angle, as expected from the intensity expression I(q) = αCMP(q)(cf. the Materials and Methods section for more details). The angular dependence provides information on the phage dimensions through the analysis of the form factor P(q). A simple fit by Guinier's law approximation leads to the phage radius of gyration Rg = 39 nm. Another size characteristic, such as the hydrodynamic radius RH, may also be extracted from an analysis of the time-dependent fluctuations of the signal scattered at each angle (Berne and Pecora, 1976). An averaged value of 3.7 × 10−12 m2/s was found for the phage diffusion coefficient D, leading to a hydrodynamic radius RH of 58 nm. Both radii are in very good agreement with the capsid radius (45 nm) (Mc Corquodale and Warner, 1988). The contrast between phage and buffer, which is actually incorporated in the prefactor α, was extracted from the signal magnitude. This contrast, namely the refractive index increment, was found equal to (∂n/∂C)tot = 0.253 cm3/g. Such a high value is consistent with the fact that DNA is tightly compacted into the capsid and that a concentrated DNA state enhances the refractive index increment (Wissemburg et al., 1995).
FIGURE 1.
Static spectra of the light scattered by phage T5 before and after FhuA addition. The detected scattered intensity is plotted as a function of the angle θ. After addition of FhuA, the intensity (open circles) at the final state (i.e., once DNA ejection is achieved), becomes ∼15 times lower than the initial intensity measured without FhuA (solid circles).
Addition of FhuA to the phage sample resulted in a strong decrease of the signal. Without phage, we checked that such a FhuA amount doesn't contribute to the detected signal. Since this FhuA concentration allows all phage to eject their DNA (Boulanger et al., 1996) the signal decrease is associated with DNA ejection from capsids and its release into the surrounding medium. After a certain period, the signal did not vary anymore suggesting that the DNA ejection process was achieved and that the final state was reached. The corresponding scattered intensity as a function of θ is illustrated (see open circle) in Fig. 1. Two observations may be noticed: i), its angular dependence is similar to the dependence observed in the previous initial state; and ii), its magnitude is ∼10 times less than that of the initial state. More precisely if Iinit and Ifinal denote the intensities measured before and after the DNA ejection, respectively, the ratio Ifinal/Iinit averaged over all the experiments is found equal to Ifinal/Iinit = 0.14 ± 0.02. Now how may such observations be interpreted remembering that I(q) = αCMP(q)? In this final state, the solution is mainly composed of empty capsids and ejected DNA but may also contain some residual phage that present some aberrations prohibiting partially or totally their DNA release. Therefore these three components could contribute to the final signal in the following way:
The usual fraction of residual phage is known to be a few percent of the total phage concentration (Labedan and Legault-Demare, 1974). So their contribution to the final signal should be only a few percent of the initial signal, which is not high enough to explain the 0.14 ratio.
Concerning the contribution of the ejected DNA chains to the final signal, DNase was added once ejection was achieved and as a result, no change was observed! Such an unchanged signal may be actually related to the conformational state of the ejected DNA that adopts a coil conformation of the order of one micron size, i.e., a size much larger than our observation length scale q−1. In this configuration, its form factor becomes much lower than unity in our q range and the mass of scattering base pairs M × P(q) becomes typically the base pairs mass, contained in the length scale q−1, of the order of a few hundred of base pairs (see Huglin, 1972 for more details). Therefore the DNA contribution to the final signal is expected to be of the order of a few thousand less than its contribution when it is tightly packaged inside the capsid, explaining why the signal remains unchanged in the presence of DNase.
The last contribution to the final state is concerned with the empty proteic capsids. First because their dimension is comparable to the dimension of the fully filled DNA capsids, the angular dependence of their signal and of the initial signal should be comparable in our q range, in accordance with the experimental curves. Secondly the magnitude of their signal should only differ from the initial signal via the product αproteins × Mproteins. Their contrast is not known but may be reasonably likened to the typical protein value reported in the literature (∂n/∂C)proteins = 0.185 cm3/g (Huglin, 1972). Combining this number with the estimated mass value Mproteins = 3 × 107 Da, one gets a ratio between the expected protein signal and the initial signal Iproteins/Iinit = 0.15, which exactly corresponds to the measured ratio 0.14 ± 0.02.
To sum up, the quantitative analysis of the signal in the initial and final states confirms the receptor's efficiency to trigger the DNA ejection process of almost all phage particles. Since the ejected DNA doesn't contribute to the detected signal, the decreasing signal once FhuA is added reflects directly the progressive loss of DNA mass confined in the capsids. Both initial and final states being clearly defined, the ejection process was studied by a temporal detection of the scattering intensity. Measurements were performed only at θ = 90° since the detected kinetics were shown not to depend on the angle within error bars.
Temperature effect on the kinetics
In the following study, t = 0 defines the time at which receptors were added to the thermostated phage sample. The receptor concentration was chosen so that the binding rate of the phage to FhuA was not a limiting step in the ejection in agreement with previous observations (Boulanger et al., 1996). After a brief and vigorous shake of the sample, the intensity I(t) was recorded as a function of the time t and at different temperatures ranging from 5°C to 41°C. Since the critical temperature required to denature the phage T5 st(0) is ∼50°C (Abelson and Thomas, 1966), phage particles and empty capsids are stable in the explored range. The normalised function F(t) = (I(t) − Ifinal)/(Iinit − Ifinal) is reported in Fig. 2 for all temperatures. In the log-lin representation, the curves present a complex shape and seem parallel. For T = 23°C, the signal had decreased by ∼90% after 1 h whereas 16 h were required to detect the reminding 10%. As a result, the complete process of DNA ejection is found to be extremely slow.
FIGURE 2.
Temperature effect on F(t) = (I(t) − Ifinal)/(Iinit − Ifinal) plotted as a function of time. All experiments were done with a large excess of receptors. By simply decreasing the temperature from 41 to 5°C, the time required to achieve DNA ejection from phages shifts from a few tens of minutes to several days.
If the temperature doesn't affect the shape of the curves, it greatly extends or shortens the temporal range during which phage eject their genome. For the two extreme temperatures, T = 41°C and 5°C, half F(t) was reached at 10 s and 10 h after FhuA addition, respectively. Even 1 week wasn't sufficient to complete the DNA ejection at T = 5°C! It may also be noticed that the curves, measured below 23°C, remain for a long time at their initial values before ejection started. We have verified that this delay didn't depend on the receptor concentration in our conditions and therefore was not due to a binding rate effect. The rate limiting step may be the opening of the head-tail connector and/or conformational changes in the tail that are required for DNA to be ejected.
Ionic condition effects on the kinetics of DNA ejection
To detect how a significant change in the ionic conditions affects DNA ejection, polyamines were added to the samples. These polyvalent cations that are well-known to condense DNA (Raspaud et al., 1998) permeate the capsids and are able to reduce significantly the pressure within them (Rau and Parsegian, 1992; Evilevitch et al., 2003). Experiments were performed at 30°C using the trivalent cation spermidine (10 mM); DNase was also added to digest any DNA condensate that the ejected DNA formed outside the capsid and which could otherwise contribute to the detected signal. DNase, spermidine, and phage were preincubated for 2 h before the addition of FhuA. The typical decreasing response due to the DNA ejection are illustrated in Fig. 3—the intensity I(t) being plotted relatively to its initial value Iinit . When the ionic concentration (100 mM NaCl) was too high for spermidine to condense DNA, the decreasing signal (empty circles) was similar to the previous measurement performed without polyamines; the entire genome was ejected as indicated by the final ratio Ifinal/Iinit = 0.14. For a salt concentration low enough to allow DNA to be condensed (10 mM NaCl), the signal decreased but deviated significantly from the earlier measurements. The final signal remained at an upper ratio Ifinal/Iinit = 0.61 meaning that only part of the genome was released and that the other part remained condensed inside the capsid. Such an inhibition is in agreement with earlier reports on in vivo injection from bacteriophage λ (see for instance Harrison and Bode, 1975).
FIGURE 3.
Ionic condition effect on the detected signal I(t) relative to its initial value Iinit., plotted as a function of time. Spermidine 10mM and DNase were added in two incubating samples. These two samples differed in their NaCl content. When the ionic condition was not sufficient to condense DNA (open circles), the NaCl content being 100 mM, the ejection kinetics were similar to the curve measured without polyamines (solid line without symbols). In the opposite case, when the ionic condition was sufficient to condense DNA (solid symbol), the NaCl content being reduced to 10 mM, the relative signal remained at the final ratio value Ifinal/Iinit = 0.61—measured the day after the kinetics experiment. In such an ionic condition, we observed a strong inhibition of the DNA release.
DISCUSSION
In vitro DNA ejection from phage particles was studied for the first time using light scattering. This technique, contrary to fluorescence that measured DNA released from the particles, provides averaged information on the amount of DNA remaining inside the capsids. Whatever the experimental conditions all the kinetics curves are found continuous suggestive of an uninterrupted DNA release. However in vivo experiments showed the existence of at least two ejection steps (Lanni, 1968; Mc Corquodale and Warner, 1988). In addition, in a recent report, images recorded by fluorescence microscopy on individual phage clearly indicate that in vitro DNA release also proceeds by stages (Mangenot et al., 2005). The apparent inconsistence between the two in vitro studies comes from the fact that measurements by light scattering are averaged over a large number of phage particles. Data from both techniques can be reconciled in a unique model: at a given time t, the ejection is complete for some phage, partial for others and unstarted for the rest, with all states contributing to the detected signal and leading to the special shape of the kinetics. Rate equations were introduced to describe the kinetics in terms of a multistep process. To simplify the calculation several approximations were made: i), DNA release between two successive steps was considered as instantaneous (Mangenot et al., 2005); ii), the receptors being in a large excess with regard to the phage particles, their binding was also considered as instantaneous; and iii), the number of intermediate states was reduced to two, corresponding to 50% and 10% of unejected DNA that remains in the capsid. These values are not crucial for this model, the most important notion being the succession of steps, as will be discussed later. The multistep process was made into a simplified chain of successive first-order reactions. This chain may be described as follows:
τ1, τ2, and τ3 describe the reverse of the decay rates of the phage fraction in each stage. To pass from a stage to another, phage must be activated or reactivated. This implies the following set of four differential equations: 1), dX1/dt = −(1/τ1) X1; 2), dX2/dt = (1/τ1) X1 − (1/τ2) X2; 3), dX3/dt = (1/τ2) X2 − (1/τ3) X3; and 4), dX4/dt = (1/τ3) X3, Xi denoting the phage fraction being in the stage i. At t = 0, although bound to the receptors, all phage are fully filled with DNA implying the initial condition: X1(0)=1 and Xi≠1(0) = 0, whereas, in the final stage t → + ∞, the DNA release being complete, only empty capsids are present: X4(t)=1 and Xi≠4(t)= 0. This set of rate equations was solved numerically allowing for the determination of the different fractions of phage in each stage as a function of time and then the computation of the normalized function F(t). Combining the 50% and 10% of unejected DNA mass together with their corresponding fraction, this function may be written as F(t) = X1(t)+ 0.50 × X2(t)+ 0.10 × X3(t). From this expression, it is seen that if other values of the DNA mass were considered, the prefactors would be different. However the fractions Xi blocked in each step can be adjusted to counterbalance this difference. Only the values of the intermediate DNA length have to be sufficiently different for the data to be fitted over the whole time range.
Fig. 4, left panel, shows the very good fit between the kinetics measured at 23°C and the kinetics obtained using the above model. By adjusting the three temporal parameters τ1, τ2, and τ3, we were able to reproduce accurately the special shape of the experimental curves. The good agreement between the experimental data and the rate equation results clearly demonstrates that the multistep behavior is an underlying factor in the continuous variation of the signal averaged on all phage particles. It should be added that without or with only one intermediate step, it was not possible to describe accurately the measurements. Nevertheless the data could also be fitted by the rate equations corresponding to two phage populations, each one having one distinct intermediate step at 50% and 10% of the DNA mass, respectively. This is actually a reasonable hypothesis given the known heterogeneities in the phage populations (Labedan, 1976; Mc Corquodale and Warner, 1988).
FIGURE 4.
Interpretation of the continuous kinetics using a phenomenological model of successive events. Two intermediate lengths have been considered here corresponding to 50% and 10% of the genome length. The typical fraction of unejected DNA length for one phage is illustrated as a function of time in Fig. 4, right panel. Each transition from one stage to another is described by a first-order law. Between two stages, the DNA ejection itself is considered as instantaneous. In our experiments and at a given time, different fractions of phage having different unejected DNA lengths coexist and contribute to the detected signal. By fitting the data with this model, the fraction of each and its temporal dependence may be evaluated. An example is given in Fig. 4, left and middle panels, the temperature being 23°C. By adjusting the characteristic times describing the different decay rates, it is possible to reproduce accurately the experimental data by this model.
Assuming a homogeneous phage population and two stopping places, the kinetics model allows the reproduction of the different curves in the whole temporal range and in particular the long tail off ending the kinetics. As shown in Fig. 4, middle panel, at the beginning of the tail off, X3(t) becomes maximal indicating that most of the phage particles still contain the last 10% of the DNA mass. This is due to the fact that the characteristic time τ3 required to pass through the last step is very long compared to the others τ1 and τ2—these characteristic times being directly related to the pause timings. From the analysis of all curves, we found that on average and regardless of the temperature value, τ2/τ1 = 9.9 and τ3/τ1 = 90. These ratio indicate that the probability to activate phage depends on the DNA length remaining unejected: longer time being required to transfer the last DNA fraction. This suggests that the DNA pressurization inside the capsid plays a role in the pause timing, lower pressurization due to lower unejected length leading to longer pause timings. The pressure role is even more manifest in the polyamine experiments. In our experimental conditions, we found that part of the genome remained unejected where polyamines are able to condense DNA. By inducing an attraction between neighboring DNA portions, polyamines create a “negative” pressure inside the capsid, thereby stopping its ejection. Therefore this result demonstrates the predominant role of the DNA pressurization in the multistep process. In this way, phage T5 seems to behave like other phage such as λ (Evilevitch et al., 2003), but how a progressive variation of the osmotic pressure—for instance, exerted by stressing polymers—acts on the multistep process, seems to be unpredictable. This effect will be analyzed in a future article (Tavares, personal communication).
How temperature affects the multistep process remains a key point to understand its origin. The single-molecule study (Mangenot et al., 2005) suggests that these stages or pauses are correlated to genetically defined single-strand interruptions (nicks) along the T5 DNA (Scheible et al., 1977). If the stopping places are only related to the nicks, why is the ejection kinetics dependent on the temperature? Previous studies have already shown that temperature affects DNA ejection from phage (Labedan, 1976; Boulanger and Letellier, 1992; Boulanger et al., 1996). Here we show that a decrease in temperature doesn't change the kinetics shape but greatly slows it but without stopping it even at 4°C (Fig. 2). The minutes required to achieve complete ejection at 41°C became days at 5°C. Such a huge effect cannot be associated to simple DNA motions during its release because the involved factors such as viscosity, diffusion coefficient, or friction vary only linearly with reciprocal temperature (1/T) (Gabashvili and Grosberg, 1992). Another mechanism must be responsible for the strong temperature dependence of the kinetics. When plotted in log-lin scales, kinetics measured at different temperatures look parallel (see Fig. 2). In other words the curves are shifted by a simple additive factor that depends in itself on the temperature. If ω(T) denotes this additive factor, then the kinetics may be replotted as a function of log t + ω(T) or log [t × exp ω(T)]. Therefore the temperature dependence of ω(T) acts exponentially on the temporal axis. Such a rescaling is generally used in mechanisms driven by activation energy. The factor ω(T) may then be assimilated to a simple energetic ratio ω(T) = −ΔH/kBT. where ΔH denotes the enthalpy required to activate the DNA ejection and where ΔH is compared to kBT the thermal energy. To superimpose the experimental curves measured at different temperatures and reported in Fig. 2, a constant energy ΔH = (2.9 ± 0.1) × 10−19 J (41.6 kcal/mol) is needed. The agreement achieved induced by such a scaling is illustrated in Fig. 5. At 5°C, the activation enthalpy corresponds to 67 kBT and at 41°C to 76 kBT. This variation of 9 kBT is sufficient to explain the temporal shift of the ejection kinetics because the probability of activating the phage at 41°C becomes larger than the probability at 5°C by a factor e9. What kind of transition is the activation energy related to? This discrete transition from one state to another could be related to a conformational change of DNA and/or proteins. In the first hypothesis, the energy required to induce a conformational change of DNA should depend on its pressurization state and therefore would depend on the unejected length. Here we observe that only one activation energy is sufficient to superimpose all curves in the whole temporal range. Equally the ratios τ2/τ1 and τ3/τ1 do not depend on the temperature because the three characteristic times vary with the temperature in the same manner. Since the obtained energy doesn't depend on the DNA unejected length, we believe that a discrete change in protein conformation is more likely. As the same energetic barrier of activation must be overcome thermally for each ejection step, the same proteic valve seems to block the ejection and must be thermally reactivated and reopened to continue DNA transfer until complete ejection.
FIGURE 5.
Interpretation of the temperature dependence of the DNA ejection kinetics. Since the curves seem parallel when plotted in log-lin scale (Fig. 2), a simple rescaling of the time t by an exponential allows us to obtain a good superimposition of the different data. Such a behavior is expected for an activation process, where the enthalpy ΔH required to activate the DNA ejection is compared to the thermal energy kB T.
The activation energy value 41.6 kcal/mol is relatively high to cause a change in protein conformation—for example five times larger than the energy required to open a large membrane protein pore (Hamill and Martinac, 2001). It could however reflect some important conformational changes in the large proteic complexes that compose the phage. Indeed, to initiate the DNA ejection, the receptor binding induces conformational changes that propagate from the tail tip to the 200-nm distant tail-head connector. From our experiments, we cannot determine which proteins regulate the channel aperture and the consequent DNA ejection. A modification of this proteic regulator conformation would probably alter the value of the activation energy and of the pause time at each stage, some of which in the extreme case could even be suppressed. Interestingly we may postulate whether a change in this proteic regulator could be related to the fact that only one intermediate stage was observed in vivo whereas four intermediate stages are reported by Mangenot et al., 2005. Indeed after the first step transfer of the viral genome into the bacterial host, it is already known that proteins encoded by the ejected part are synthesized and then partially bind to the membrane and to the phage (Mc Corquodale and Warner, 1988); the absence of these proteins blocks the ejection indefinitely.
Why these conformational changes occur at some specific places along the DNA chain still remains unclear. Surprisingly they occur and are able to block the DNA expulsion although it is highly pressurized inside the capsid. Comparison with other phage would be informative, in particular, with the phage λ, which appears to eject its genome in one step (Novick and Baldeschwieler, 1988; Evilevitch et al., 2003). It would also be informative to determine the activation energy required to trigger the DNA release once its protein receptor LamB is bound.
Figure s6.
Acknowledgments
E.R. thanks the Laboratoire de Physique des Solides for helping him financially to set up the light-scattering apparatus, and the technical, mechanical, and electronic engineers without whom this homemade apparatus would never be. We also thank Linda Roubieu for her participation in the preliminary experiments. We are grateful to Pascale Boulanger, Françoise Livolant, Amélie Leforestier, Paulo Tavares, Sandrine Brasiles, and Stéphanie Mangenot for valuable discussions. We thank Pascale Boulanger for providing the FhuA protein.
This work was supported in part by the Centre National de la Recherche Scientifique program “Dynamique et Réactivité des Assemblages Biologiques”.
References
- Abelson, J., and C. A. J. Thomas. 1966. The anatomy of the T5 bacteriophage DNA molecule. J. Mol. Biol. 18:262–291. [Google Scholar]
- Berne, B. J., and R. Pecora. 1976. Dynamic Light Scattering. John Wiley, New York.
- Bonhivers, M., A. Ghazi, P. Boulanger, and L. Letellier. 1996. FhuA, a transporter of the Escherichia coli outer membrane, is converted into a channel upon binding of bacteriophage T5. EMBO J. 15:1850–1856. [PMC free article] [PubMed] [Google Scholar]
- Boulanger, P., and L. Letellier. 1992. Ion channels are likely to be involved in the two steps of phage T5 penetration into Escherichia coli cells. J. Biol. Chem. 267:3168–3172. [PubMed] [Google Scholar]
- Boulanger, P., M. Le Maire, M. Bonhivers, S. Dubois, M. Desmadril, and L. Letellier. 1996. Purification and structural and functional characterization of FhuA, a transporter of the Escherichia coli outer membrane. Biochemistry. 35:14216–14224. [DOI] [PubMed] [Google Scholar]
- Chu, B. 1991. Laser Light Scattering, 2nd ed. Academic Press, San Diego, CA.
- Earnshaw, W. C., and S. R. Casjens. 1980. DNA packaging by the double-stranded DNA bacteriophages. Cell. 21:319–331. [DOI] [PubMed] [Google Scholar]
- Evilevitch, A., L. Lavelle, C. M. Knobler, E. Raspaud, and W. M. Gelbart. 2003. Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl. Acad. Sci. USA. 100:9292–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili, I. S., and A. Grosberg. 1992. Dynamics of double stranded DNA reptation from bacteriophage. J. Biomol. Struct. Dyn. 9:911–920. [DOI] [PubMed] [Google Scholar]
- Hamill, O., and B. Martinac. 2001. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 81:685–740. [DOI] [PubMed] [Google Scholar]
- Harrison, D. P., and V. C. Bode. 1975. Putrescine and certain polyamines can inhibit DNA injection from bacteriophage Lambda. J. Mol. Biol. 96:461–470. [DOI] [PubMed] [Google Scholar]
- Huglin, M. B. 1972. Light Scattering from Polymer Solutions. Academic Press, London.
- Kindt, J., S. Tzlil, A. Ben-Shaul, and W. M. Gelbart. 2001. DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. USA. 98:13671–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labedan, B., and J. Legault-Demare. 1974. Evidence for heterogeneity in populations of T5 bacteriophage. J. Virol. 13:1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labedan, B. 1976. A very early step in the T5 DNA ejection process. Virology. 75:368–375. [DOI] [PubMed] [Google Scholar]
- Lanni, Y. T. 1968. First-step-transfer deoxyribonucleic acid of bacteriophage T5. Bacteriol. Rev. 32:227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangenot, S., M. Hochrein, J. Raedler, and L. Letellier. 2005. Real time imaging of DNA ejection from single phage particles. Current Biology. In Press. [DOI] [PubMed]
- McCorquodale, J. D., and H. R. Warner. 1988. Bacteriophage T5 and related phages. In The Viruses. R. Calendar, editor. Plenum Press, New York. 1:439–476.
- Novick, S. L., and J. D. Baldeschwieler. 1988. Fluorescence measurement of the kinetics of DNA injection by bacteriophage lambda into liposomes. Biochemistry. 27:7919–7924. [DOI] [PubMed] [Google Scholar]
- Odijk, T. 1998. Hexagonally packed DNA within bacteriophage T7 stabilized by curvature stress. Biophys. J. 75:1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspaud, E., M. Olvera de la Cruz, J. L. Sikorav, and F. Livolant. 1998. Precipitation of DNA by polyamines: a polyelectrolyte behavior. Biophys. J. 74:381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, D. C., and V. A. Parsegian. 1992. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys. J. 61:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer, S. C., and V. A. Bloomfield. 1978. Packaging of DNA in bacteriophage heads: some considerations on energetics. Biopolymers. 17:785–794. [DOI] [PubMed] [Google Scholar]
- Scheible, P. P., E. A. Rhoades, and M. Rhoades. 1977. Localization of single-chain interruptions in bacteriophage T5 DNA. I. Electron microscopic studies. J. Virol. 23:725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature. 413:748–752. [DOI] [PubMed] [Google Scholar]
- Wissemburg, P., T. Odjik, P. Citkel, and M. Mandel. 1995. Multimolecular aggregation of mononucleosomal DNA in concentrated isotropic solutions. Macromolecules. 28:2315–2328. [Google Scholar]








