Abstract
Intrinsically disordered proteins such as FlgM play important roles in biology, but little is known about their structure in cells. We use NMR to show that FlgM gains structure inside living Escherichia coli cells and under physiologically relevant conditions in vitro, i.e., in solutions containing high concentrations (≥400 g/liter) of glucose, BSA, or ovalbumin. Structure formation represents solute-induced changes in the equilibrium between the structured and disordered forms of FlgM. The results provide insight into how the environment of intrinsically disordered proteins could dictate their structure and, in turn, emphasize the relevance of studying proteins in living cells and in vitro under physiologically realistic conditions.
Most proteins require a defined three-dimensional structure to perform their function. Intrinsically disordered proteins seem paradoxical because they lack stable structure, yet they play key roles in diverse biological processes including signal transduction, transcription, and neurodegenerative diseases (1, 2). Here, we report results from studies of the so-called intrinsically disordered protein, FlgM, a 97-residue polypeptide from Salmonella typhimurium that regulates flagellar synthesis by binding the transcription factor σ28. The FlgM-σ28 complex inhibits transcription of the genes encoding the late flagellar proteins, but inhibition is relieved when FlgM exits the cell via the basal hook body of the flagella (3). Free FlgM is mostly unstructured in dilute solution, but its C-terminal half forms a transient helix (4). One signature of protein structure can be the absence of crosspeaks in 1H-15N heteronuclear single quantum correlation (HSQC) NMR spectra because of conformational exchange (5–7). NMR studies in dilute solution indicate that the C-terminal half of FlgM becomes structured on binding σ28, as shown by the disappearance of crosspeaks from residues in the C-terminal half of FlgM in the FlgM-σ28 complex (8). This bipartite behavior (i.e., disappearance of crosspeaks from the C-terminal half with retention of crosspeaks from the N-terminal half) provides a valuable built-in control for studying the response of FlgM to different solution conditions. We discuss our results in terms of two types of intrinsically disordered proteins: those that gain structure under crowded conditions and those that do not. FlgM is an example of both types in a single protein.
Materials and Methods
FlgM was overexpressed and purified as described (4, 8). In vitro NMR data were acquired by using a 0.4 mM uniformly 15N-enriched sample in 10 mM sodium acetate, pH 5.0/10 mM sodium chloride/0.02% sodium azide/10.0% (vol/vol) D2O at 25°C. For the crowding experiments, glucose, BSA, or ovalbumin were incorporated into the sample. The sample used for live-cell NMR spectroscopy was obtained by overexpressing 15N-labeled FlgM in BL21 Gold Escherichia coli bacteria and by preparing as described (9). Two-dimensional gradient-enhanced 1H-15N HSQC spectra (10, 11) were acquired at 600 MHz. Except for the in-cell NMR experiment, sweep widths of 5,199.87 Hz and 1,337.55 Hz were used in the 1H and 15N dimensions, respectively. The 1H acquisition dimension consisted of 1,024 complex data points, whereas the 15N dimension consisted of 256 complex increments. For the in-cell NMR experiment, sweep widths of 6,994.74 Hz and 1,337.55 Hz were used in the 1H and 15N dimensions, respectively, and 1,024 complex data points and 128 complex increments were collected.
Results
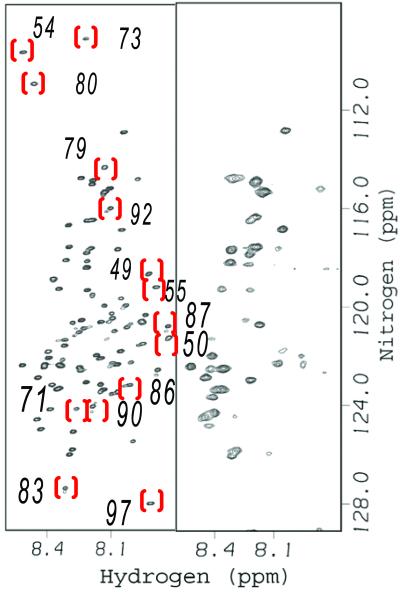
The HSQC spectrum of 15N-enriched FlgM in dilute solution is contrasted with the spectrum in living E. coli cells in Fig. 1. In-cell NMR experiments are possible because FlgM is overexpressed upon induction (≈100 mg of FlgM can be purified from 1 liter of saturated culture). The overexpression allows the FlgM spectrum to be observed on top of signals arising from other 15N-enriched proteins in the cell, which contribute a uniform background (9). As shown in Fig. 1, about half the crosspeaks disappear. The crosspeaks that disappear are the same ones that disappear on σ28 binding in simple buffer solution (8). The crosspeaks that persist are the same ones that persist on σ28 binding (8). The retention of crosspeaks from the N-terminal half of FlgM provides an internal control by showing that the loss of crosspeaks from the C-terminal half is not caused by nonspecific line broadening brought about by the intracellular environment.
Figure 1.
FlgM is structured in E. coli. The HSQC spectrum of FlgM in dilute solution (Left) and in living E. coli (Right). Red brackets surround some of the crosspeaks from the C-terminal half in FlgM that disappear in E. coli. The number near each bracket is the residue number (8). Dilute solution sample: 400 μM FlgM/10 mM sodium acetate, pH 5.0/10 mM NaCl/0.02% NaN3/10% (vol/vol) D2O at 298 K. The E. coli spectrum was acquired as described by Serber et al. (9).
These data suggest that the C-terminal portion of FlgM is structured in cells, but the N-terminal portion remains unstructured. We also performed important controls. The cells could be plated and grown after the experiment, proving that they were alive during the experiment. To show that the FlgM was intracellular, the cells were pelleted and the supernatant was subjected to SDS/PAGE with Coomassie staining. A band at the molecular mass of FlgM is not observed, proving that the spectrum in Fig. 1 comes from intracellular FlgM. However, an FlgM band could be found in cells overexpressing FlgM, proving that the loss of crosspeaks is not caused by proteolysis. The results are not explained by overexpression of FlgM because we used the Bradford assay (12) to show that both induced and uninduced cells have the same amount of total protein per g of wet cells. Most importantly, the results cannot be explained by the binding of FlgM to the E. coli homolog of σ28 because the homolog is not overexpressed, and, as described below, the same pattern of crosspeaks is observed in vitro in the absence of σ28 when the spectrum is acquired at high solute concentrations. These data prove that FlgM gains structure in cells and show that important information can be gained by studying proteins in cells rather than in dilute solution.
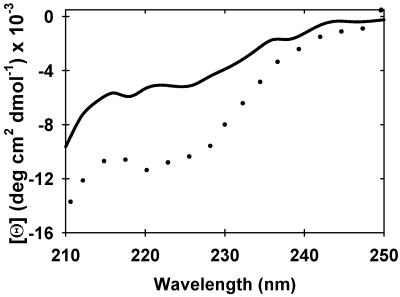
Why is FlgM unstructured in dilute solution but structured in cells? One obvious difference is the solute concentration, which can reach hundreds of grams per liter in cells (13, 14). We explored this variable by using sugar to mimic the intracellular environment. The HSQC and circular dichroic (CD) spectra of FlgM in the presence and absence of 2.5 M (450 g/liter) glucose is shown in Figs. 2 and 3, respectively. The HSQC spectrum shows that the same crosspeaks that are lost in cells are also lost in the presence of 2.5 M glucose. The glucose-induced ellipticity decrease at 222 nm (Fig. 3) indicates FlgM gains α-helical structure at high sugar concentration, consistent with earlier NMR data suggesting the C-terminal end of the protein forms nascent helical structure in dilute solution (4). These results are also consistent with previous studies showing that high concentrations of sugars and other osmolytes induce structure in other intrinsically disordered proteins (15–18). The dilute solution spectrum reappears on diluting the sugar, proving that the reaction is reversible. Two observations show that the effect of glucose on α-helix formation is nonspecific. First, other sugars induce comparable CD spectra (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Second, sugar concentrations of <200 g/liter have no effect, suggesting that the dissociation constant for glucose is much greater than 1 M.
Figure 2.
Glucose induces structure in FlgM. Overlaid HSQC spectra of FlgM in dilute solution (red) and in 450 g/liter 2.5 M glucose (black). Conditions: 400 μM FlgM/10 mM sodium acetate, pH 5.0/10 mM NaCl/0.02% NaN3/10% (vol/vol) D2O at 298 K.
Figure 3.
Glucose induces α-helix in FlgM. Far-UV CD spectra of 15 μM FlgM in dilute solution (solid line) and in 2.5 M (450 g/liter) glucose (dotted line). Both samples contain 10 mM sodium acetate (pH 5.0), 0.02% NaN3.
How is structure formation linked to crosspeak disappearance? For the FlgM-σ28 complex, it has been suggested that the crosspeaks disappear because of conformational exchange between the bound, structured form of FlgM and the free, unstructured form of FlgM, and that crosspeaks from the N-terminal half persist because this half of the protein remains unstructured (8). The observation that high solute concentrations can cause the loss of crosspeaks from the C-terminal half of FlgM in the absence of σ28 proves that complex driven exchange is not the only mechanism for the disappearance of crosspeaks. The structured form of FlgM may be a molten globule, but we cannot rule out the idea that the structured species is globular but exists in intermediate exchange on the NMR timescale. The disappearance of crosspeaks from the spectra of molten globules is well known and has been described in detail for the molten globule state of α-lactalbumin (5, 6) and β2-microglobulin (7). How such a molten globule would form a specific interaction with σ28 remains unclear, but multiple binding modes may be involved. In summary, FlgM forms an α-helix-rich molten globule on binding σ28 in dilute solution and on addition of high sugar concentrations in the absence of σ28.
The sugar data are important for providing information about the type of structure induced in FlgM, but small molecule solutes are of minor biological importance in cells compared with larger solutes such as proteins. To show that high protein concentrations can drive structure formation, we examined the HSQC spectrum of 15N-enriched FlgM in the presence of 400 g/liter BSA (68 kDa). The spectrum is shown in Fig. 4. BSA also induces structure in FlgM, as seen by comparing this spectrum to spectra acquired in 450 g/liter glucose (Fig. 2) and in dilute solution on σ28 binding (8). This observation is not limited to BSA. Identical results were obtained by using 450 g/liter ovalbumin (45 kDa; see Fig. 6, which is published as supporting information on the PNAS web site). Specific BSA-FlgM interactions can be ruled out as the driving force for structure formation because crosspeaks only begin to disappear at a BSA concentration of 330 g/liter. Two observations show that the presence or absence of crosspeaks is not simply a matter of viscosity. First, crosspeaks from the N-terminal half of FlgM are present under all conditions tested, even though the relative viscosities of the solutions differ dramatically, from a relative viscosity of 1 for dilute buffer to ≈24 for 400 g/liter BSA (19). Second, the absence of crosspeaks does not correlate with increased viscosity; the C-terminal crosspeaks are absent in 2.5 M glucose, which has a relative viscosity of ≈5, but are present in 330 g/liter BSA, which has a relative viscosity of ≈12 (19, 20). Proteins are macro ions, so we also considered the possibility that the small ions present in the sample caused structure formation. CD studies show that neither 1 M NaCl nor 0.5 M Na2SO4 induce structure (data not shown). These results are consistent with NMR data showing that an ionic strength of 0.5 M does not alter the FlgM spectrum in dilute solution (4). Additionally, the protein-induced structure in FlgM cannot be explained by favorable electrostatic interactions between BSA (or ovalbumin) and the C-terminal half of FlgM because the C-terminal half of FlgM has a calculated pI of 4.5, whereas the pI of BSA is 4.7 and the pI of ovalbumin is 4.6. In summary, these data show FlgM is structured inside living cells and solute concentrations that mimic intracellular solute concentrations can induce that structure.
Figure 4.
BSA induces structure in FlgM. HSQC spectrum of FlgM in 400 g/liter BSA. Conditions: 400 μM FlgM/10 mM sodium acetate, pH 5.0/10 mM NaCl/0.02% NaN3/10% (vol/vol) D2O at 298 K.
Discussion
Our observation that the intrinsically disordered protein FlgM gains structure when exposed to physiologically relevant environments is crucial to the study of disordered proteins. However, Anfinsen (21) stressed the importance of solution conditions 29 years ago in his statement, “conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence in a given environment.” The fact that the C-terminal half of uncomplexed FlgM is structured only at high solute concentrations suggests that the environment is highly biologically significant for intrinsically disordered proteins. The structures of globular proteins are maintained in dilute solution because the equilibrium between folded and unfolded states favors structure formation (22). Proteins such as the C-terminal half of FlgM are unstructured in dilute solution because the equilibrium favors the unstructured forms; however, the equilibrium is only slightly unfavorable. Under these conditions, solution properties can alter dramatically the population of the structured state. This stabilization involves two phenomena: excluded volume effects and binding interactions between solution components (13, 15, 23, 24). Identifying the dominant effect requires additional experiments where the extent of structure formation is assessed as a function of the size and charge of the solutes. Regardless of the mechanism, our observations show that the structure in the C-terminal half of FlgM is thermodynamically unstable compared with most proteins, because most proteins maintain their structure in dilute solution. This diminished stability may be important for the function of intrinsically disordered proteins. For instance, the low stability of FlgM might facilitate its exit into the extracellular medium (8). In other instances, low stability may promote protein turnover.
Our results support the hypothesis that there are two classes of intrinsically disordered proteins, with FlgM providing an example of each class. One class, exemplified by the C-terminal half of FlgM, is structured in cells. The driving force for solute-induced structure is likely the formation of a hydrophobic core, which is the most common characteristic of folded proteins (25). The other class, exemplified by the N-terminal half of FlgM, does not become structured at physiologically relevant solute concentrations. Some of these proteins may require another protein to provide a framework for structure formation. Support for this notion comes from the observation that certain transcription factors remain disordered even at physiologically relevant solute concentrations, yet they gain structure in the presence of other components of the complex (26). Along these lines, we predict that many of the intrinsically disordered ribosomal proteins will remain disordered even under physiologically relevant conditions because their protein and nucleic acid-binding partners provide a structural template. Some proteins may be a combination of these two classes. For instance, the components of the signal transduction heterodimer p160-CBP/p300 are disordered in dilute solution, but they gain structure on complex formation (27). It is likely that the individual components are disordered because they cannot form a stable core on their own.
In summary, our results show that a so-called intrinsically disordered protein possesses structure under biologically relevant conditions. The results also suggest a reason for the observation that some proteins are only folded under physiologically relevant conditions and prove the biological relevance of studying proteins in vivo and at physiologically relevant solute concentrations. Our results also suggest that the search for drugs that target intrinsically disordered proteins should be conducted at physiologically relevant solute concentrations.
Supplementary Material
Acknowledgments
We thank Frederick Dahlquist for providing the expression system, Matthew Redinbo, Edward Samulski, Dorothy Erie, Terrence Oas, Elizabeth Pielak, and the Pielak group for helpful discussions. This work was supported by the National Science Foundation, the grantors of the Petroleum Research Fund administered by the American Chemical Society, and the Smallwood Foundation.
Abbreviation
- HSQC
heteronuclear single quantum correlation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dunker A K, Brown C J, Lawson J D, Iakoucheva L M, Obradovic Z. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 2.Uversky V N, Gillespie J R, Fink A L. Proteins Struct Funct Genet. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 4.Daughdrill G W, Hanely L J, Dahlquist F W. Biochemistry. 1998;37:1076–1082. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- 5.Schulman B A, Kim P S, Dobson C M, Redfield C. Nat Struct Biol. 1997;4:630–634. doi: 10.1038/nsb0897-630. [DOI] [PubMed] [Google Scholar]
- 6.Redfield C, Schulman B A, Millhollen M A, Kim P S, Dobson C M. Nat Struct Biol. 1999;6:948–952. doi: 10.1038/13318. [DOI] [PubMed] [Google Scholar]
- 7.McParland V J, Kalverda A P, Homans S W, Radford S E. Nat Struct Biol. 2002;9:326–331. doi: 10.1038/nsb791. [DOI] [PubMed] [Google Scholar]
- 8.Daughdrill G W, Chadsey M S, Karlinsey J E, Hughes K T, Dahlquist F W. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 9.Serber Z, Ledwidge R, Miller S M, Dötsch V. J Am Chem Soc. 2001;123:8895–8901. doi: 10.1021/ja0112846. [DOI] [PubMed] [Google Scholar]
- 10.Kay L E, Keifer P, Saarinen T. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 11.Bodenhausen G, Ruben D J. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 12.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 13.Minton A P. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 14.Ellis R J. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 15.Davis-Searles P R, Saunders A J, Erie D A, Winzor D J, Pielak G J. Annu Rev Biophys Biomol Struct. 2001;30:271–306. doi: 10.1146/annurev.biophys.30.1.271. [DOI] [PubMed] [Google Scholar]
- 16.Davis-Searles P R, Morar A S, Saunders A J, Erie D A, Pielak G J. Biochemistry. 1998;37:17048–17053. doi: 10.1021/bi981364v. [DOI] [PubMed] [Google Scholar]
- 17.Qu Y, Bolen C L, Bolen D W. Proc Natl Acad Sci USA. 1998;95:9268–9273. doi: 10.1073/pnas.95.16.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baskakov I V, Kumar R, Srinivasan G, Ji Y S, Bolen D W, Thompson E B. J Biol Chem. 1999;274:10693–10696. doi: 10.1074/jbc.274.16.10693. [DOI] [PubMed] [Google Scholar]
- 19.Wetzel R, Becker M, Behlke J, Billwitz H, Böhm S, Ebert B, Hamann H, Krumbiegel J, Lassman G. Eur J Biochem. 1980;104:469–478. doi: 10.1111/j.1432-1033.1980.tb04449.x. [DOI] [PubMed] [Google Scholar]
- 20.Wolf A V, Brown M G, Prentiss P G. CRC Handbook of Chemistry and Physics. Boca Raton, FL: CRC; 1980. [Google Scholar]
- 21.Anfinsen C B. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 22.Lattman E E, Rose G D. Proc Natl Acad Sci USA. 1993;90:439–441. doi: 10.1073/pnas.90.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders A J, Davis-Searles P R, Allen D L, Pielak G J, Erie D A. Biopolymers. 2000;53:293–307. doi: 10.1002/(SICI)1097-0282(20000405)53:4<293::AID-BIP2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 24.Record M T, Jr, Zhang W, Anderson C F. Adv Protein Chem. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 25.Dill K A. Biochemistry. 1990;31:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 26.Flaugh S L, Lumb K J. Biomacromolecules. 2001;2:538–540. doi: 10.1021/bm015502z. [DOI] [PubMed] [Google Scholar]
- 27.Demarest S J, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson H J, Evans R M, Wright P E. Nature (London) 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.