Abstract
The mouse hypervariable minisatellite (MN) Pc-1 consists of tandem repeats of d(GGCAG) and flanked sequences. We have previously demonstrated that single-stranded d(GGCAG)n folds into the intramolecular folded-back quadruplex structure under physiological conditions. Because DNA polymerase progression in vitro is blocked at the repeat, the characteristic intramolecular quadruplex structure of the repeat, at least in part, could be responsible for the hypermutable feature of Pc-1 and other MNs with similar repetitive units. On the other hand, we have isolated six MN Pc-1 binding proteins (MNBPs) from nuclear extracts of NIH 3T3 cells. Here, we describe one of those MNBPs, MNBP-B, that binds to the single-stranded d(GGCAG)n. Amino acid sequences of seven proteolytic peptide fragments of MNBP-B were determined, and the cDNA clones were isolated. MNBP-B was proven identical to the single-stranded DNA-binding protein, UP1. Recombinant UP1 bound to single-stranded d(GGCAG)n and other G-rich repetitive sequences, such as d(GTCAGG)n and d(GTTAGG)n. In addition, UP1 was demonstrated by CD spectrum analysis to unfold the intramolecular quadruplex structure of d(GGCAG)5 and d(TTAGGG)4 and to abrogate the arrest of DNA synthesis at the d(GGG)n site. This ability of UP1 suggests that unfolding of quadruplex DNA is required for DNA synthesis processes.
Minisatellites (MNs), also called variable number of tandem repeats, are arrays of 5-to 100-bp repeats widely dispersed in eukaryotic genomes (1). Some have been suggested to be implicated in genetic predisposition to various human diseases. Although MNs are known to be hot spots for meiotic recombination in germ cells (1, 2), they are considered to be genetically stable in somatic cells (3). However, they also have been demonstrated to be altered in somatic cells with exposure to chemical carcinogens, γ irradiation, or UV irradiation (4–7). They also exhibit alterations in a range of tumors of humans and experimental animals (8–10).
The MN Pc-1, located on mouse chromosome 4, consists of tandem repeats of d(GGCAG) (11, 12). The germ-line mutation rate for Pc-1 is 15% per gamete, whereas the mutation rate in somatic cells is 2–3% per 22–24 population doublings (12–14). Many hypervariable MNs, consisting of G-rich repeat units similar to Pc-1, have been found in the mouse and other mammalian genomes, and mouse MNs containing d(GGCAG)n or d(GGCAGG)n motifs are very unstable in germ-line cells (11). Pc-1 and Pc-1-like MNs were demonstrated to be strikingly unstable by DNA fingerprint analysis, both in fibroblasts deficient in DNA-dependent protein kinase activity and in NIH 3T3 cells treated with okadaic acid, an inhibitor of protein phosphatases (13, 14).
Although the molecular mechanisms underlying the induction of MN mutations are largely unknown, the G-rich strand of Pc-1 is known to form an intramolecular folded-back quadruplex structure under physiological conditions and cause arrest of DNA synthesis (15). We have isolated six MN Pc-1 binding proteins (MNBPs) from NIH 3T3 cells (16). Two of them, MNBP-A and MNBP–B, bind to the G-rich strand of Pc-1, and the other four, MNBP-D, MNBP-E, MNBP-F, and MNBP-G, to the complementary C-rich strand.
In this article, we document isolation of cDNA clones encoding MNBP-B and characterization of a recombinant MNBP-B. Sequences of seven proteolytic peptides of purified MNBP-B were determined, and cDNA clones were subsequently isolated. MNBP-B was revealed to be identical to the single-stranded DNA binding protein, UP1 (17), which is a proteolytic product corresponding to the N-terminal 195 aa of the 34-kDa heterogeneous nuclear ribonucleoprotein (hnRNP) A1 (18, 19). DNA binding specificity and in vitro effect on quadruplex structures of UP1 were analyzed to cast light on its biological roles.
Materials and Methods
Oligonucleotides and Plasmids.
All synthetic oligonucleotides were purified by reverse-phase cartridge or HPLC chromatography [Sawady Technology (Tokyo), Takara Biomedicals (Kyoto), and Grainer (Tokyo)]. The names and nucleotide sequences of oligonucleotides for electrophoretic mobility-shift assay (EMSA) are as follows: pG8, d(GGCAG)8; pG5, d(GGCAG)5; pG4, d(GGCAG)4; pG3, d(GGCAG)3; pG4R, d(CAGGG)4; pG3R, d(CAGGG)3; pG5+G, d(GGCAGG)5; pG5+A, d(GACAGG)5; pG5+C, d(GCCAGG)5; pG5+T, d(GTCAGG)5; pG5TEL, d(GTTAGG)5; poly(dA), d(A)20; poly(dT), d(T)20; poly(dC), d(C)25; poly(dG), d(G)25; pC8, d(CTGCC)8; rG5, r(GGCAG)5; and rG5+U, r(GUCAGG)5. Poly(dI-dC)/poly(dI-dC) and poly(dT)16 were purchased from Amersham Pharmacia. pG8 and pRandom40 [d(GATCCTAGAGCTTCGATAGCCGATCAGTTGACACTGGTCA)] were used for analysis performed by the surface plasmon resonance (SPR) system (Biacore, Uppsala, Sweden). pG5, pTRM4 [d(TTAGGG)4], and pTPs [d(TAGCAGCTATGGGGGAGCTGGGGAAGGTGGGAATGTGA)] were used for CD analysis. pSub15 [d(CAGGG)15TCTAGTACTGGCCGTCGTTTTACAACGTCG] and its 3′ complementary 24-mer sequence pM13–20 were used for primer extension experiments. The plasmid pYA-3 was a derivative of pUC119 (20) carrying 12 repeats of d(GGCAG) (15). Single-stranded DNA of pYA-3 was generated as described (20). Double-stranded oligonucleotide DS12 [d(GGCAG)12]/[d(CTGCC)12] was prepared as described (16).
EMSA.
EMSA and analyses of oligonucleotide competition using EMSA were performed as detailed (16) with 32P-labeled pG8 (final concentration 2 nM) applied as a probe. IC50 values, the concentrations of competitors at which the complex yield is halved, were estimated from the inhibition curves when the relative amount of complex was plotted against concentration of competitor oligonucleotides. KD values were estimated as the concentrations of the proteins at which free pG8 is halved, when a relative amount of free DNA was plotted against protein concentration.
cDNA Cloning for MNBP-B.
MNBPs were purified from NIH 3T3 cells as described (16), and partial polypeptide sequences were determined according to the methods described by Shinohara et al. (21). Seven polypeptide sequences of MNBP-B were determined by a method of Edman degradation under a contract with APRO Science (Naruto, Japan). cDNA fragments for the MNBP-B were amplified with a set of degenerated primers, 5′-GAYGGNMGNGTNGTNGARCC-3′ and 5′-ACNSWRTCRTGRTCRTCRAA-3′, using cDNAs from NIH 3T3 cells as templates. To isolate a full-length MNBP-B cDNA, an NIH 3T3 cDNA library in Lambda ZAP II (Stratagene) was screened by plaque hybridization (22) with the partial cDNA fragment described above as a probe. Finally, one clone, MNBP-B Cl-24, containing the full-length cDNA was identified. Detailed procedures are provided in additional Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org.
Expression and Purification of Recombinant UP1.
UP1 was expressed in Escherichia coli as a GST-fusion protein, affinity-purified on glutathione Sepharose (Amersham Pharmacia), and further purified on ion exchange and gel filtration columns. Release of the UP1 portion from the GST-fusion protein was performed by PreScission Protease (Amersham Pharmacia), and further purifications were performed by several column chromatographies. Purifications of GST-hnRNP A1 protein and release of hnRNP A1 from the GST-fusion protein were performed following the procedure similar to that for GST-UP1. Detailed procedures are provided in Supporting Materials and Methods.
Protein–DNA Interaction Analysis by SPR-Based Biosensor.
Binding experiments were performed by monitoring the association and dissociation reactions in real time by using the SPR Biosensor, BIACORE3000 system (Biacore). UP-1 was diluted to various concentrations with HBS-EP buffer (10 mM Hepes, pH 7.4/150 mM NaCl/3 mM EDTA/0.005% Tween 20, pH 7.4) and injected to the oligonucleotide-immobilized sensor chip at a constant flow rate of 20 μl/ml. The kinetic parameters of the interactions were calculated by nonlinear curve fitting analysis of the association and dissociation curves by using BIAEVALUATION 3.0 software (Biacore). Detailed procedures are provided in Supporting Materials and Methods.
Formation and Purification of Intramolecular Quadruplex DNA.
Oligonucleotide pG8 labeled at its 5′ end with 32P in Tris-EDTA (TE) buffer (10 mM Tris·HCl/1 mM EDTA, pH 8.0) containing 100 mM KCl (final 20 nM) was heated at 95°C for 3 min, followed by gradual cooling to room temperature, and stored at 4°C for more than 2 days to allow sufficient formation of intramolecular quadruplex structures. Sample solutions containing both intramolecular quadruplex and single-stranded forms were separated by 7.5% PAGE in 0.5 × TBE buffer containing 10 mM KCl. Single-stranded forms were purified from a gel as described (23) and dissolved in TE. Intramolecular quadruplex forms were purified as described above with the following modification: KCl was added to all solutions used for the elution and purification procedures to give a final concentration of 100 mM.
CD Analysis.
For CD measurement, lyophilized DNA, pG5, pTRM4, and pTPs were dissolved in 20 mM sodium phosphate buffer (pH 7.0) containing 150 mM KCl. The strand concentration was 10 μM. Each sample was heated at 90°C for 5 min, followed by gradual cooling to room temperature, and stored at 4°C for at least 2 days. Either GST-UP1 or GST, dissolved in 20 mM sodium phosphate buffer (pH 7.0) containing 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM benzamidine and 0.2% acetone, was added step by step to each DNA sample with DNA to protein ratios of 1:0.5, 1:1, and 1:2. CD spectra of the DNA and DNA–protein complexes were recorded with a Jasco J-720 spectropolarimeter and a 1-mm cell. CD spectra of GST-UP1 and GST were also recorded. From the CD spectrum of the DNA–protein complex, that for the corresponding protein was subtracted.
In Vitro DNA Synthesis Assay.
Primer extension reaction was performed as described (15) with some modifications. Single-stranded pYA-3 (final 100 nM) in TE buffer containing 100 mM KCl was stored at 7°C for a few days to allow the sufficient formation of the quadruplex structure, after heat denaturing at 95°C for 3 min. Five microliters of this sample was mixed with 7.5 μl of Milli-Q water (Millipore) and 5 μl of the pUC/M13 forward (−40) primer (Promega) labeled with 32P at the 5′ end (0.5 μM) in TE buffer. The mixture was heated at 60°C for 7 min and incubated at 37°C for more than 30 min to allow annealing and sufficient quadruplex formation. An aliquot of 1.75 μl of this primer-annealed template (final 23 nM) was mixed with 0.75 μl of 10 × BcaBEST buffer (200 mM Tris⋅HCl, pH 8.5/100 mM MgCl2) and 0.5 μl of dNTPs mixture (50 μM each). After adding 4 μl of GST-UP1 suspended in buffer P (20 mM sodium phosphate, pH 7.0/0.5 mM DTT), the mixture was incubated at 37°C for 5 min, and then the primer extension reaction was carried out at 37°C for 8 min in the presence of BcaBEST DNA polymerase (final 66 units/ml, Takara Biomedicals).
A primer extension reaction using a synthetic oligonucleotide pSub15 as a template was also performed in the presence and absence of GST-UP1 as described above with some modifications. A mixture of the pSub15 and the pM13–20 primer, which was labeled with 32P at the 5′ end (final 100 nM each), in TE buffer containing 150 mM KCl was heated at 95°C for 5 min and then at 72°C for 5 min, followed by gradual cooling to room temperature, and stored at 7°C for more than 24 h. The concentrations of BcaBEST DNA polymerase, the primer-annealed template, and dNTPs were 18 units/ml, 10 nM, and 1.7 μM, respectively. The reaction was stopped by adding 1.5 μl of a solution (160 mM EDTA, 0.7% SDS, 6 mg/ml proteinase K), and then the samples were incubated at 37°C for 30 min.
Results
Cloning of cDNA for MNBP-B.
A 24-kDa MNBP–B is known to bind to the pG8 [d(GGCAG)8], representing the G-rich strand of Pc-1 (16). MNBP-B was purified from the NIH 3T3 cells as described (16) and digested with lysyl endopepetidase in a gel. Amino acid sequences of five peak fractions of the proteolytic peptide fragments separated by reverse-phase HPLC were determined (Table 1). Two of the five fractions also contained minor peptide-fragments (2c and 3c). The N-terminal sequence of the purified MNBP-B could not be determined, suggesting protection from the Edman modification. Partial cDNA sequences for MNBP-B were amplified with a set of degenerated primers designed from the peptide sequences of the fragments 2 and 3, subcloned into a pCRII vector, and sequenced. Using this cDNA fragment as a probe, a cDNA clone, MNBP-B Cl-24, containing the full-length cDNA sequence was isolated from the NIH 3T3 cDNA library.
Table 1.
Amino acid sequence of proteolytic peptides of MNBP-B
| Fragment | Amino acid sequence |
|---|---|
| 1 | IFVGGIK |
| 2 | RGFAFVTFDDHDSV |
| 2c* | IEVIEIMTDRGSGK |
| 3 | DGRVVEPK |
| 3c* | VIQK |
| 4 | RAVSREDSQRPGAHLTVK |
| 5 | EDTEEHHLRDYFEQYGK |
Fragments 2c and 3c were minor peptides coexisting with fragments 2 and 3, respectively.
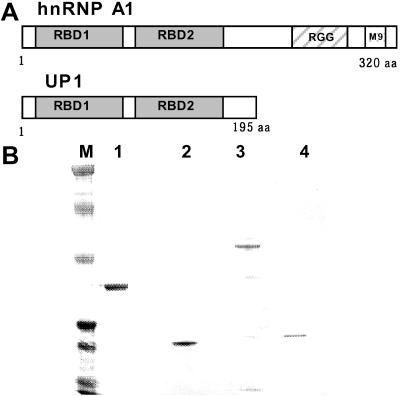
The nucleotide sequence of cDNA for MNBP-B was identical to that reported for hnRNP A1 (see Fig. 6, which is published as supporting information on the PNAS web site). Although hnRNP A1 is a 34-kDa protein and differs from MNBP-B in size, the size of UP1, a proteolytic product of hnRNP A1, was equivalent to that of MNBP-B, 24 kDa (18, 19, 24). N-terminal and C-terminal sequences of MNBP-B were not determined. However, all of the squences of proteolytic peptide fragments of MNBP-B were coincident with the UP1 sequence (see Fig. 6). UP1 is composed mainly of two copies of single-stranded nucleotide binding domains (25), along with short amino acid sequences at both terminal and internal regions (Fig. 1A). Taking these results together, it was concluded that MNBP-B is identical to UP1. We hereafter refer MNBP-B as UP1.
Figure 1.
Structural features of hnRNP A1/UP1 (A) and purification of the recombinant proteins (B). (A) The RBD segments are RNA binding domain (25). (B) An SDS 4–12% polyacrylamide gel showing purified GST-UP1 (lane 1), UP-1 (lane 2), GST-hnRNP A1 (lane 3), and hnRNP A1 (lane 4). Lane M, Kaleidoscope prestained standards (244, 139, 80, 43, 32, 19, 7.7 kDa, top to bottom). Note that hnRNP A1 shows faster electrophoretic mobility than that expected from its molecular mass, probably because of the characteristic amino acid composition in its C-terminal region.
Kinetic Analysis of the Interaction between UP1 and pG8.
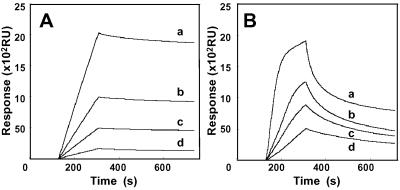
A recombinant protein of UP1 expressed in E. coli as a GST fusion protein was purified (Fig. 1B, lane 1). After releasing GST tag, recombinant UP1 was purified to homogeneity through affinity and gel filtration chromatographies (Fig. 1B, lane 2). In a similar way, recombinant hnRNP A1 was purified to homogeneity (Fig. 1B, lane 4). From EMSA (data not shown), the value of KD (equilibrium dissociation constant) for UP1 to pG8 was estimated to be 7 × 10−8 M. The affinity of UP1 for pG8 was almost equivalent to that of GST-UP1 and slightly lower than that of hnRNP A1 (data not shown). The UP1 portion appears essential for the specific binding of hnRNP A1 to pG8, whereas the C-terminal portion of hnRNP A1, which is deleted in UP1, may have a stabilizing effect on its interaction with pG8. Both associating and dissociating processes were monitored in real time with an SPR system. pG8 was immobilized on the sensor chip surface and its binding reaction with GST-UP1 was studied after injecting the latter at various concentrations (Fig. 2A). The values for ka (association rate constant), kd (dissociation rate constant), and KD were 2 × 104 M−1⋅s−1, 1 × 10−4 s−1, and 5 × 10−9 M, respectively. Although this KD value is several times lower than that evaluated by EMSA, the difference could be due to the assay systems. The kd value and the sensorgrams in the dissociation phase indicated that the complex between UP1 and pG8 is stable. The association of GST-UP1 with pG8 is four times faster than that with pRandom40, a random 40-mer oligonucleotide sequence, and the dissociation from pG8 is 20 times slower than that from pRandom40 (Fig. 2B).
Figure 2.
Interaction between UP1 and oligonucleotides monitored by the SPR-based biosensor system. (A) Sensorgrams showing the interaction between pG8 and GST-UP1 at concentrations of 140 pM (a), 70 pM (b), 35 pM (c), and 17 pM (d). (B) Sensorgrams showing the interaction between GST-UP1 and pRandom40. Concentrations of GST-UP1 were 560 pM (a), 280 pM (b), 140 pM (c), and 70 pM (d).
Sequence Specificity for DNA Binding of UP1.
Binding of UP-1 and hnRNP A1 to the double-stranded oligonucleotide DS12, single-stranded oligonucleotide pC8, poly(dC), poly(dA), or poly(dT) was not detected by EMSA (data not shown). Oligonucleotide competition assays using EMSA revealed that UP1 bound to pG5+T and pG5TEL with a higher affinity than to pG5, but not to pG5+C (Table 2). The affinity of UP1 for pG5 was less than for pG8 (Table 2). These data are in good agreement with our previous results regarding binding properties of UP1 purified from NIH 3T3 cells (16). The affinity of UP1 for pG3R [d(CAGGG)3] was 10 times higher than for pG3 [d(GGCAG)3] and the increase of the repeat number of d(CAGGG) enhanced the binding (Table 2). The affinities of UP1 for oligo RNAs, rG5 and rG5+U, were four times stronger than for corresponding oligo DNAs, pG5 and pG5+T, respectively (Table 2), suggesting that UP1 binds to not only single-stranded DNA but RNA with similar sequence specificity. Sequence specificity of hnRNP A1 was similar to that of UP1 (data not shown).
Table 2.
IC50 values for formation of pG8-UP1 complexes
| Competitor | IC50 | Competitor | IC50 | Competitor | IC50 |
|---|---|---|---|---|---|
| pG5TEL | 3 nM | pG5+C | >1 μM | pG3 | 1 μM |
| pG5+T | 4 nM | pG8 | 4 nM | poly(dG) | 100 nM |
| pG5 | 20 nM | pG4R | 40 nM | rG5 | 5 nM |
| pG5+A | 30 nM | pG4 | 50 nM | rG5+U | 1 nM |
| pG5+G | 40 nM | pG3R | 100 nM |
IC50 values, the concentrations of unlabeled competitor oligonucleotide at which the labeled pG8-UP1 complex yield is halved, were estimated from the inhibition curves when the relative amount of complex was plotted against the concentration of a competitor.
UP1 Binds to Intramolecular Quadruplex DNA.
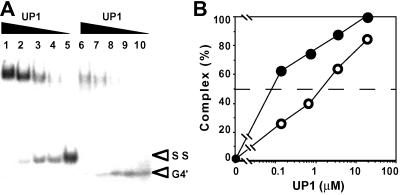
NMR and CD analyses of pG8 and pG5 indicated that the G-rich strand of Pc-1 with five or more d(GGCAG) repeats folds into an intramolecular folded-back quadruplex structure under physiological conditions (15). The intramolecular quadruplex form of pG8 oligonucleotide shows faster mobility than the single-stranded form on PAGE, and each could be separated as described in Materials and Methods. UP1 bound to both the intramolecular quadruplex and single-stranded form of pG8 (Fig. 3A). This was also the case for GST-hnRNP A1 (data not shown). The affinity of UP1 for the single-stranded form was about 20 times as high as for the intramolecular quadruplex form by evaluating KD values of these two forms of DNA (Fig. 3B).
Figure 3.
Binding of UP1 to intramolecular quadruplex DNA. (A) An EMSA showing the binding of rMNBP-B to single-stranded (SS) and intramolecular quadruplex (G4′) form of oligonucleotide pG8. Concentrations of SS and G4′ forms of pG8 were 20 nM and 10 nM, respectively. Concentrations of UP1 were 18 μM (lanes 1 and 6), 3.6 μM (lanes 2 and 7), 0.72 μM (lanes 3 and 8), 0.14 μM (lanes 4 and 9), and 0 μM (lanes 5 and 10). (B) Graphs showing the result of quantitative analysis of a similar experiment to A. UP1-pG8 complex formations with single-stranded and intramolecular quadruplex forms are indicated by ● and ○, respectively. Concentrations of both SS and G4′ forms of pG8 were 10 nM. Each point represents the mean of two separate experiments.
UP1 Unfolds Intramolecular Quadruplex DNA.
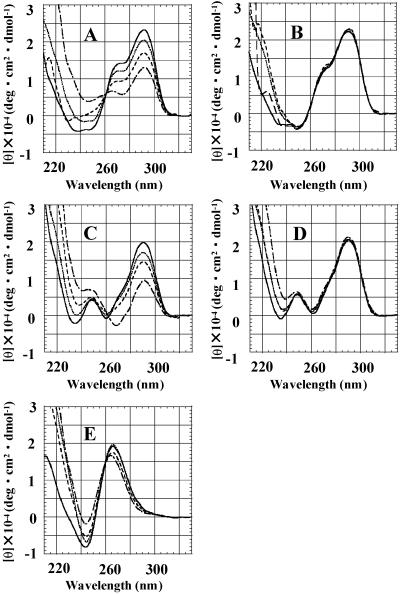
To investigate the effect of UP1 on the intramolecular folded-back quadruplex structure of the pG5 oligonucleotide, d(GGCAG)5, CD analysis of the pG5 in the presence or absence of GST-UP1 was performed. The pG5 oligonucleotide gave the positive CD band at 290–295 nm after incubating with 150 mM KCl, which is characteristic for the quadruplex with the folded-back structure (Fig. 4A). The CD band at 290–295 nm of the pG5 was decreased by addition of GST-UP1 in a concentration-dependent manner, and twice the molar excess of GST-UP1 protein could halve the peak (Fig. 4A). No change in the CD band was observed on addition of GST protein (Fig. 4B). It is therefore concluded that UP1 has the capacity to unfold the intramolecular folded-back quadruplex form of the d(GGCAG) repeat.
Figure 4.
CD spectra showing unfolding of DNA quadruplex by UP1. (A) pG5 titrated with GST-UP1. (B) pG5 with GST. (C) pTRM4 with GST-UP1. (D) pTRM4 with GST. (E) pTPs with GST-UP1. From each spectrum, the spectrum of the corresponding protein is subtracted. Solid line, a DNA/protein ratio of 1:0; dotted line, 1:0.5; dashed line, 1:1; centered, 1:2.
UP1 also binds to pG5TEL, which contains four telomeric repeats, d(TTAGGG)4. Because the d(TTAGGG)n oligonucleotides, representing the G-rich strand of telomere repeats, were demonstrated to form quadruplex structures in vitro (26), we investigated the effect of UP1 on this ability. Oligonucleotide pTRM4 [d(TTAGGG)4] gave a positive CD band at 290–295 nm, like pG5, after incubating with 150 mM KCl, and the band was also decreased by GST-UP1 dose-dependently (Fig. 4C). No change was again observed with GST protein (Fig. 4D). These results indicate the intramolecular quadruplex structure of the pTRM4 oligonucleotide to be unfolded by GST-UP1 and suggest a possibility that UP1 plays a role in telomere maintenance by destroying the quadruplex structure of telomere ends in vivo. Four-stranded parallel quadruplex DNA constructed by oligonucleotide TPs was not appreciably unwound by GST-UP1 (Fig. 4E).
Abrogation of DNA Synthesis Arrest on Single-Stranded d(GGCAG)n Template by UP1.
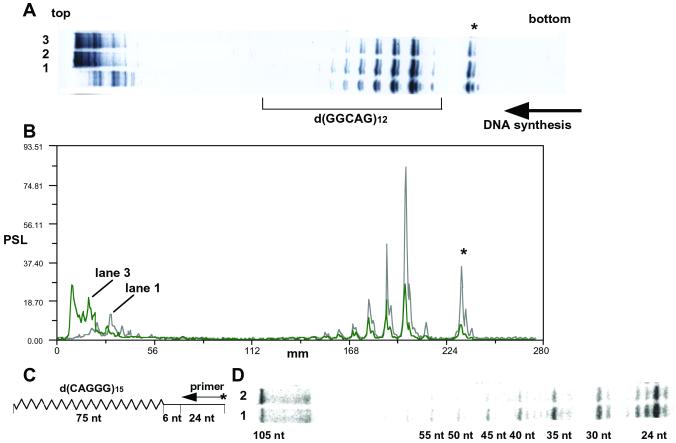
We have reported arrest of DNA synthesis in vitro on a d(GGCAG)n template (15). Because UP1 unfolded the intramolecular quadruplex structure of d(GGCAG)5, we examined the effect of UP1 on the DNA synthesis arrest in vitro. Single-stranded phagemid pYA-3 carrying d(GGCAG)12 was used as a template. Primer extension reaction with BcaBest DNA polymerase was obstructed mainly at the first d(GGG) present in d(GGCAG)12, with additional weaker stops at the second, third, fourth, fifth, and sixth d(GGG) sequences (Fig. 5A, lane 1). Other bands showed pausing of the primer exension reaction at d(GGG) present in multiple cloning sites of the vector plasmid (indicated by an asterisk in Fig. 5A). Because pausing of DNA synthesis was not observed at the same site on the vector plasmid without the insert (data not shown), this d(GGG) site in the multiple cloning site could have made some interaction with the d(GGG) sequences of d(GGCAG)12 repeat. This pausing was weakened dose-dependently by addition of UP1 to the reaction (Fig. 5A, lanes 2 and 3). Abrogation of DNA synthesis arrest was demonstrated quantitatively by scanning radioactivity in the gel (Fig. 5B). UP1 reduced the arrest of DNA synthesis in vitro at d(GGCAG)n. In Fig. 5B, lane 2, where the amount of UP1 was 1/8 as much as lane 3, showed bands of intensity between the profiles of lanes 1 and 3 (data not shown).
Figure 5.
Effect of UP1 on the arrest of DNA synthesis at d(GGCAG) repeats. (A) Results of PAGE indicating the primer extension reaction with or without UP1, using the single-stranded phagemid pYA-3 carrying d(GGCAG)12. The concentrations of UP1 were 0 M (lane 1), 6.0 μM (lane 2), and 48 μM (lane 3). Horizontal arrow indicate the direction of the DNA synthesis. (B) Scanning profiles of lanes 1 and 3 of the gel shown in A. (C) Schematic representation of oligonucleotides used for the primer extension depicted in D. The synthetic oligonucleotides, pM13–20 of 24 mer and pSub15 of 105 mer carrying d(CAGGG)15, were used as primer and template, respectively. The zigzag line indicates d(CAGGG)15 on pSub15. (D) Results of the primer extension reaction with or without GST-UP1, using the pSub15 (final 10 nM) as a template. The concentrations of GST-UP1 were 0 M (lane 1) and 750 nM (lane 2). The sizes of the fragments are indicated in nt.
To confirm the above results, we prepared a synthetic 105-mer oligonucleotide containing d(CAGGG)15 repeat and used it as a template for the primer extension reaction (Fig. 5C). Progression of a DNA polymerase was obstructed mainly at the first d(GGG) site with additional weaker stops at the second, third, fourth, fifth, and sixth d(GGG) sequences, which corresponded to 30-, 35-, 40-, 45-, 50-, and 55-nt fragments, respectively (Fig. 5D, lane 1). Addition of a 75-fold molar excess of GST-UP1 over the template reduced the arrest of DNA synthesis in vitro by 24–60% at the first, second, third, fourth, fifth, and sixth d(GGG) sites in d(CAGGG)15, and enhanced the synthesis of complete length DNA (105 nt) by 100% (Fig. 5D, lane 2 and Fig. 7, which is published as supporting information on the PNAS web site). GST-UP1 reduced the arrest of DNA synthesis in vitro at d(TTAGGG)n when a synthetic oligonucleotide containing d(TTAGGG)12 was used as a template (data not shown). Similar results were obtained in the case of other bacterial DNA polymerases, Taq and Klenow fragment, as well as BcaBest DNA polymerase.
Discussion
Sequence Preference of DNA Binding Activity of UP1.
MNBP-B, first detected as an MNBP, was here demonstrated to be identical to UP-1 originally isolated from calf thymus as a single-stranded DNA binding protein with a low sequence preference (27). However, recombinant UP1 bound preferentially to single-stranded d(GGCAG)n and similar sequences, such as d(GTTAGG)n and d(GTCAGG)n, in agreement with our previous results obtained with UP1 purified from NIH 3T3 cells (16). Although the function of UP1 in vivo is unknown, UP1 was demonstrated to be a proteolytic product of 34-kDa hnRNP A1, and it corresponds to the N-terminal 195 aa of hnRNP A1 (18, 19, 24). The latter was first isolated as a major constituent protein of a hnRNP complex and is suspected to be involved in pre-mRNA transport, protection, and splicing (for reviews, see ref. 28). It has also been reported that hnRNP A1 binds to the telomeric repeat sequence and promotes telomere elongation (29–32). Several members of the hnRNP family are thought to play roles in transcriptional regulation, recombination, and telomere maintenance as a DNA binding protein, in addition to their action as RNA binding proteins (for review, see refs. 33 and 34). The results of our current study strongly suggest further significance of hnRNP A1 and UP1 as DNA binding proteins. hnRNP A1 is very abundant, with an estimated 7 × 107 molecules in a HeLa cell (35). Although the affinity of UP1 for DNA was estimated to be only about one-fourth of that for RNA, it is reasonable, by taking its apparent abundance into account, that it could have through DNA binding.
Binding and Unfolding Activities of UP1 for Quadruplex Oligonucleotides.
Many proteins have been reported to bind to quadruplex DNA and some of them, including the Ku antigen, demonstrated an ability to stabilize the quadruplex structure (36). In addition, only some proteins, such as BLM helicase, WRN helicase, and murine telomeric DNA binding proteins, qTBP42 and uqTBP25, were reported to unwind tetra- or bi-molecular quadruplex DNA (37–39). UP1 efficiently unfolds unimolecular folded-back quadruplex DNA and does not appreciably unwind tetramolecular parallel quadruplex DNA. NTP hydrolysis energy is not required for unfolding the quadruplex by UP1, and a mechanism for the unfolding is unknown. However, the mechanism may be provided by the result that UP1 bound with higher affinity to single-stranded d(GGCAG)8 than the intramolecular quadruplex form of the same oligonucleotide. It is possible that binding of UP1 changes the equilibrium constant between the intramolecular quadruplex and single-stranded DNA, and the UP1-quadruplex DNA complex is converted to a UP1–single-stranded DNA complex.
hnRNP A1 is suspected to be involved in pre-mRNA processing. We indicate that UP1 and hnRNP A1 binds to both single-stranded DNA and RNA with similar sequence specificity. These results strongly suggest that hnRNP A1 and UP1 have an unfolding activity of intramolecular quadruplex RNA and could be involved in pre-mRNA processing through this activity. Further study about the effects of hnRNP A1/UP1 on quadruplex RNA is necessary to verify whether this hypothesis is true or not.
Possible Involvement of UP1 in Stable Maintenance of GC-Rich Repetitive Sequences and Telomere.
MNs are genetically unstable even in somatic cells, although much more unstable in germ cells (1–3). They are altered in various tumors and somatic cells exposed to various carcinogens (4–10). Many MNs, consisting of G-rich repeat units similar to Pc-1, have been found in the mouse and other mammalian genomes (11). Pc-1 and Pc-1-like MNs have also been demonstrated to be strikingly unstable both in fibroblasts deficient in DNA-dependent protein kinase, and in NIH 3T3 cells treated with okadaic acid, an inhibitor of serine/threonine protein phosphatases (13, 14). These findings suggest that there is a certain molecular mechanism involved in stable maintenance of MNs in somatic cells, and this mechanism could be modulated by the phospholylation status of certain cellular proteins. Dysfunction or misfunction of this mechanism or large amounts of DNA damage surpassing repair capacity may cause MN mutations. It would be interesting to determine whether particular tumors contain mutations in MNBPs, including UP1, thus accounting for the high frequency of mutations observed in these sequences.
Based on our knowledge that the G-rich strand of MN Pc-1 and other short tandem repeats such as d(CGG)n triplet repeat and insulin-variable number of tandem repeat fold into a quadruplex structure and causes the arrest of DNA synthesis in vitro, the characteristic structure of the repeats could be responsible for the hypermutable feature of Pc-1 and other tandem repeats with similar repetitive units (15, 40, 41). In the case of d(CGG)n triplet repeat, WRN helicase was demonstrated to facilitate copying of the quadruplex structure of the triplet repeat by DNA polymerase δ in vitro (42). Our finding that UP1 has the activity to unfold the intramolecular quadruplex form of d(GGCAG)5 in vitro and to abrogate the pausings of DNA polymerase progression at the repeat strongly suggests that UP1 may prevent MN mutations by unfolding the quadruplex structure cooperating with or making up for WRN helicase in vivo. Although we did not use eukaryotic DNA polymerases, we assumed that these bacterial DNA polymerases could be representatives of polymerases that occur in cells.
Loss of hnRNP A1 in mouse cells correlated with short telomeres and expression of hnRNP A1 or UP1 restored the normal length of telomeric repeats (30). In the present study, UP1 was demonstrated to bind to oligonucleotide sequences harboring the mammalian telomeric repeat d(TTAGGG)n and unfold the intramolecular folded-back quadruplex structure. It is possible that UP1 could play a role as a modulator of a tertial structure of telomeric repeats, as revealed in this study, by converting the intramolecular quadruplex form of telomere end into the single-stranded form.
As noted above, hnRNP A1/UP1 is an abundant protein in cells, and considered to have multiple physiological functions, including pre-mRNA processing and telomere maintenance. The present study revealed that MNBP-B/UP1 also has two important activities: (i) unfolding the intramolecular quadruplex structure of d(GGCAG) repeat and abrogating the arrest of DNA synthesis in vitro at this repeat and (ii) unfolding the quadruplex structure of d(TTAGGG) repeats, that could facilitate telomere replication or elongation.
Supplementary Material
Acknowledgments
We thank Professor S. Uesugi (Yokohama National Univ.) for helpful discussions. This work was supported by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor, and Welfare of Japan and by the Public Trust Haraguchi Memorial Cancer Research Fund. M.K. and H.F. were supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grants 10179102, 14035219, and 12470487 to M.K. and 12780528 to H.F.).
Abbreviations
- MN
minisatellite
- MNBP
MN Pc-1 binding protein
- hnRNP
heterogeneous nuclear ribonucleoprotein
- EMSA
electrophoretic mobility-shift assay
- SPR
surface plasmon resonance
- TE
Tris-EDTA
References
- 1.Jeffreys A J, Wilson V, Thein S L. Nature (London) 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 2.Wahls W P, Wallace L J, Moore P D. Cell. 1990;60:95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- 3.Jeffreys A J, Tamaki K, MacLeod A, Monckton D G, Neil D L, Armour J A L. Nat Genet. 1994;6:136–145. doi: 10.1038/ng0294-136. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki S, Takada T, Sugawara Y, Muto T, Kominami R. Jpn J Cancer Res. 1991;82:1061–1064. doi: 10.1111/j.1349-7006.1991.tb01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paquette B, Little J B. Cancer Res. 1992;52:5788–5793. [PubMed] [Google Scholar]
- 6.Honma M, Kataoka E, Ohnishi K, Kikuno T, Hayashi M, Sofuni T, Mizusawa H. Mutat Res. 1993;286:165–172. doi: 10.1016/0027-5107(93)90180-n. [DOI] [PubMed] [Google Scholar]
- 7.Kitazawa T, Kominami R, Tanaka R, Wakabayashi K, Nagao M. Mol Carcinog. 1994;9:67–70. doi: 10.1002/mc.2940090203. [DOI] [PubMed] [Google Scholar]
- 8.Thein S L, Jeffreys A J, Gooi H C, Cotter F, Flint J, O'Connor N T, Weatherall D J, Wainscoat J S. Br J Cancer. 1987;55:353–356. doi: 10.1038/bjc.1987.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumura Y, Tarin D. Cancer Res. 1992;52:2174–2179. [PubMed] [Google Scholar]
- 10.Ledwith B J, Joslyn D J, Troilo P, Leander K R, Clair J H, Soper K A, Manam S, Prahalada S, van Zwieten M J, Nichols W W. Carcinogenesis. 1995;16:1167–1172. doi: 10.1093/carcin/16.5.1167. [DOI] [PubMed] [Google Scholar]
- 11.Mitani K, Takahashi Y, Kominami R. J Biol Chem. 1990;265:15203–15210. [PubMed] [Google Scholar]
- 12.Suzuki S, Mitani K, Kuwabara K, Takahashi Y, Niwa O, Kominami R. J Biochem (Tokyo) 1993;114:292–296. doi: 10.1093/oxfordjournals.jbchem.a124169. [DOI] [PubMed] [Google Scholar]
- 13.Nakagama H, Kaneko S, Shima H, Inamori H, Fukuda H, Kominami R, Sugimura T, Nagao M. Proc Natl Acad Sci USA. 1997;94:10813–10816. doi: 10.1073/pnas.94.20.10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai H, Nakagama H, Komatsu K, Shiraishi T, Fukuda H, Sugimura T, Nagao M. Proc Natl Acad Sci USA. 1997;94:10817–10820. doi: 10.1073/pnas.94.20.10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katahira M, Fukuda H, Kawasumi H, Sugimura T, Nakagama H, Nagao M. Biochem Biophys Res Commun. 1999;264:327–333. doi: 10.1006/bbrc.1999.1521. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda H, Sugimura T, Nagao M, Nakagama H. Biochim Biophys Acta. 2001;1528:152–158. doi: 10.1016/s0304-4165(01)00186-6. [DOI] [PubMed] [Google Scholar]
- 17.Herrick G, Alberts B. J Biol Chem. 1976;251:2124–2132. [PubMed] [Google Scholar]
- 18.Valentini O, Biamonti G, Pandolfo M, Morandi C, Riva S. Nucleic Acids Res. 1985;13:337–346. doi: 10.1093/nar/13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merril B M, LoPresti M B, Stone K L, Williams K R. J Biol Chem. 1986;261:878–883. [PubMed] [Google Scholar]
- 20.Vieira J, Messing J. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 21.Shinohara Y, Sagawa I, Ichihara J, Yamamoto K, Terao K, Terada H. Biochim Biophys Acta. 1997;1319:319–330. doi: 10.1016/s0005-2728(97)00002-9. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Maxam A M, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 24.Pandolfo M, Valentini O, Biamonti G, Morandi C, Riva S. Nucleic Acids Res. 1985;13:6577–6590. doi: 10.1093/nar/13.18.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burd C G, Dreyfuss G. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blackburn E H, Greider C W, editors. Telomeres. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 27.Herrick G, Alberts B. J Biol Chem. 1976;251:2133–2141. [PubMed] [Google Scholar]
- 28.Dreyfuss G, Matunis M J, PinolRoma S, Burd C G. Annu Rev Biochem. 1993;269:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa F, Matunis M J, Dreyfuss G, Cech T R. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaBranche H, Dupuis S, Ben-David Y, Bani M R, Wellinger R J, Chabot B. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 31.Dallaire F, Dupuis S, Fiset S, Chabot B. J Biol Chem. 2000;275:14509–14516. doi: 10.1074/jbc.275.19.14509. [DOI] [PubMed] [Google Scholar]
- 32.Fiset S, Chabot B. Nucleic Acids Res. 2001;29:2268–2275. doi: 10.1093/nar/29.11.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krecic A M, Swanson M S. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 34.Ladomery M. BioEssays. 1997;19:903–909. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- 35.Kiledjian M, Burd C G, Gorlach M, Portman D S, Dreyfuss G. In: Protein-RNA Interactions-Frontiers in Molecular Biology. Mattaj I, Nagai K, editors. Oxford: Oxford Univ. Press; 1994. pp. 127–149. [Google Scholar]
- 36.Uliel L, Weisman-Shomer P, Oren-Jazan H, Newcomb T, Loeb L A, Fry M. J Biol Chem. 2000;275:33134–33141. doi: 10.1074/jbc.M005542200. [DOI] [PubMed] [Google Scholar]
- 37.Sun H, Karow J K, Hickson I D, Maizels N. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 38.Fry M, Loeb L A. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 39.Weisman-Shomer P, Naot Y, Fry M. J Biol Chem. 2000;275:2231–2238. doi: 10.1074/jbc.275.3.2231. [DOI] [PubMed] [Google Scholar]
- 40.Fry M, Loeb L A. Proc Natl Acad Sci USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Catasti P, Chen X, Moyzis R K, Bradbury E M, Gupta G. J Mol Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 42.Kamath-Loeb A S, Loeb L A, Johansson E, Burgers P M J, Fry M. J Biol Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.