Abstract
The packing of helices spanning lipid bilayers is crucial for the stability and function of α-helical membrane proteins. Using a modified Voronoi procedure, we calculated packing densities for helix-helix contacts in membrane spanning domains. Our results show that the transmembrane helices of protein channels and transporters are significantly more loosely packed compared with helices in globular proteins. The observed packing deficiencies of these membrane proteins are also reflected by a higher amount of cavities at functionally important sites. The cavities positioned along the gated pores of membrane channels and transporters are noticeably lined by polar amino acids that should be exposed to the aqueous medium when the protein is in the open state. In contrast, nonpolar amino acids surround the cavities in those protein regions where large rearrangements are supposed to take place, as near the hinge regions of transporters or at restriction sites of protein channels. We presume that the observed deficiencies of helix-helix packing are essential for the helical mobility that sustains the function of many membrane protein channels and transporters.
INTRODUCTION
The dense packing of secondary structures plays a central role in the folding and stability of proteins (Chothia et al., 1981; Popot and Engelman, 2000; Preissner et al., 1998). It has been recorded that the interactions of complementary surfaces (patches) contribute greatly to the stability of the tertiary structure in water-soluble and in membrane proteins (Bowie, 1997; Langosch and Heringa, 1998; Lemmon and Engelman, 1994). In these spatial interaction motifs, major side chains of one interface (knobs) often fit into the holes of the complementary interface. We have previously shown that the interfaces of secondary structures are conserved along evolution as documented for the proteasomal subunits (Gille et al., 2000) and that even patches from different types of secondary structures were found to resemble each other (Preissner et al., 1999). These findings imply that the spatial arrangement of secondary structures in proteins is stabilized by the geometry of each surface patch as well as by the chemical properties of the residues.
In membrane proteins, however, a somewhat different picture might be expected (MacKenzie and Engelman, 1998; White and Wimley, 1999). The distinctive surrounding milieu causes the weakening of the hydrophobic effect inside the protein within the lipid bilayer. In addition, the inclusion of prosthetic groups and the pervasion of ion channels and solute transporters with water-filled pores could result in deviations in molecular packing. Furthermore, it has been proposed that ion channels and solute transporters open throughout gating mechanisms that require broad molecular rearrangements of their transmembrane domains (Locher et al., 2003; Perozo et al., 2002; Swartz, 2004). Thus the question arises whether this flexibility is facilitated by a reduced packing density.
Nevertheless, the validation of atomic packing densities in proteins is a difficult problem (Fleming and Richards, 2000). Eilers and colleagues, for example, applied the occluded surface method (Pattabiraman et al., 1995) to assess the packing densities of transmembrane helices by investigating 11 helical membrane protein structures (Eilers et al., 2002). They presume, in conclusion, that helical membrane proteins are generally packed more densely than other proteins. This conclusion is surprising, because the hydrophobic effect as a driving force for helix-helix interaction is absent inside the lipid bilayer (MacKenzie and Engelman, 1998). In addition, polar- or hydrogen-bonded interactions generally occur less frequently than in water-soluble globular proteins (DeGrado et al., 2003). Leaving energetic considerations aside, the dense packing of transmembrane helices appears contradictory to the mobility that is thought to form the basis of the proper functioning of many membrane proteins (Jiang et al., 2002).
To solve this conundrum, we used a modified Voronoi procedure to calculate the packing values and to evaluate the number of cavities found in the different groups of proteins (Goede et al., 1997; Rother et al., 2003). Hereby we concentrated in those atoms buried in the helix-helix interfaces leaving surface atoms aside. The advantage of our method is that it also works for atoms that reside in those protein regions, where large packing deficiencies occur, as in protein voids or pockets (cavities). As pointed out in a mathematical appendix, the occluded surface method fails to give exact packing values in those cases. The analysis of an updated set of 20 helical membrane proteins finally leads us to a point of view that is contrary to the proposed opinion that helical membrane proteins are generally packed more densely than other helical proteins. We show clearly that packing deficiencies often occur at functionally important protein regions. To comprehend the molecular base of the observed packing differences between helices in membrane and globular proteins, detailed statistics about the amino acid and atomic composition of helix-helix patches and cavities were collected.
MATERIAL AND METHODS
We compared the packing of membrane protein channels and transporters to the packing of the remaining helical membrane proteins and to the packing of a reference group of helical domains in soluble globular proteins. For that purpose, two measures were used: atomic packing densities and the number (and environment) of atom-sized cavities.
Data sets
We have recently shown that, according to their different functions and architecture, membrane proteins can be subdivided into two groups (Hildebrand et al., 2004): membrane channels and transporters can be subsumed under the term “membrane gates”, because of the regulated opening or closing of the pore underlying their function. On the other hand, proton pumps, receptors, and photosystems can be classified as “membrane coils”, because the helix interaction motifs are very similar to those characteristic for soluble coiled-coils (Langosch and Heringa, 1998). Transmembrane helices in membrane coils predominantly cross at left-handed angles, those in membrane gates predominantly at right-handed angles. We further demonstrated that the members of these two groups differ markedly in the torsion angles and intrahelical hydrogen bonds (Hildebrand et al., 2004).
To establish whether the particular functionality is also reflected in different packing density values, the current data set of 20 high-resolution structures of nonrelated helical membrane proteins was specified into 10 membrane gates and 10 membrane coils (Table 1). All the calculations were carried out on monomers except for channels and transporters, where the functional multimers were used. We chose wild-type structures with highest resolution. Nonetheless, the structures of membrane coils are resolved at higher mean resolution than membrane gates. To qualify whether this could influence our analysis, the packing density values of 24 different structures of the photosynthetic reaction center was compared. As a result, we observed that the packing density (mean value = 0.81, standard deviation σ = 0.006) does not depend on the crystallographic resolution (2.1 Å–3.5 Å).
TABLE 1.
Atomic packing density values (PD) of the membrane-spanning regions of different nonhomologous helical membrane protein structures, subdivided into membrane gates and membrane coils
| Gates
| ||||
|---|---|---|---|---|
| Protein Data Bank code | Protein name | Resolution [Å] | PD | Cavtm [No.;%]* |
| 1eul | Calcium ATPase | 2.6 | 0.78 | 6;6 |
| 1iwg | AcrB multi-drug efflux transporter | 3.5 | 0.78 | 15;13 |
| 1j4n | AQP1 aquaporin water channel | 2.2 | 0.82 | 1;1 |
| 1jvm | KcsA potassium channel | 2.8 | 0.80 | 3;13 |
| 1kpl | ClC chloride channel | 3.0 | 0.79 | 5;5 |
| 1l7v | BtuCD vitamin B12 transporter | 3.2 | 0.80 | 7;6 |
| 1msl | MscL mechanosensitive channel | 3.5 | 0.80 | 0;0 |
| 1okc | Mitochondrial ADP/ATP carrier | 2.2 | 0.81 | 8;8 |
| 1pw4 | GlpT glycerol-3-phosphate transporter | 3.3 | 0.76 | 23;13 |
| 1rh5 | SecYEβ protein-conducting channel | 3.2 | 0.80 | 5;4 |
| all | 2.9 | 0.79 | 73;6 | |
| Coils
| ||||
| Protein Data Bank code | Protein name | Resolution [Å] | PD | Cavtm [No.;%] |
| 1aig | Photosynthetic reaction center | 2.2 | 0.82 | 0;0 |
| 1c3w | Bacteriorhodopsin | 1.6 | 0.83 | 4;13 |
| 1ezv | Cytochrome bc1 complex | 2.3 | 0.81 | 9;4 |
| 1f88 | Rhodopsin | 2.8 | 0.80 | 11;16 |
| 1jb0 | Photosystem I | 2.5 | 0.82 | 16;5 |
| 1kqf | Formate dehydrogenase-N | 1.6 | 0.83 | 1;1 |
| 1nek | Succinate dehydrogenase | 2.6 | 0.81 | 4;5 |
| 2occ | Cytochrome c oxidase | 2.3 | 0.79 | 22;6 |
| 1q16 | NarGHI nitrate reductase A | 1.9 | 0.82 | 1;4 |
| 1qla | Fumerate reductase complex | 2.2 | 0.80 | 1;6 |
| all | 2.2 | 0.81 | 69;5 | |
Number of cavities found within the membrane-spanning regions and fraction of buried atoms in contact with cavities Cavtm (number; percentage).
To exclude the influence of the polar milieu, solely those parts of the transmembrane helices were selected that are located within the hydrophobic part of the bilayer. For this purpose, we used the criteria described in Hildebrand et al. (2004) to draw two parallel planes to isolate the expansions of the hydrophobic part of the lipid bilayer. The packing interfaces of helices in water-soluble globular proteins were calculated by using the same set of 25 nonhomologous globular protein structures listed in the previous analysis.
Packing densities
The local packing density of an atom is defined as the fraction PDloc = VVdW/Vse, where VVdW is the space inside the atoms' van der Waals sphere and Vse is the van der Waals sphere expanded by 1.4 Å, the radius of a solvent molecule. When two atoms are bonded or close to each other, parts of their spheres are cut off by separating panes, with impact on the local packing values. The amount of space remaining in each sphere is referred to as the atomic volume.
To calculate atomic volumes, the Voronoi cell procedure using hyperboloid interfaces between atoms was applied (Goede et al., 1997). The interfaces between atoms are defined by all points that are equally distant from both van der Waals spheres of the two atoms. Atomic volumes were calculated numerically using a cubic lattice with a grid distance of 0.2 Å. We used the Stouten set of atom radii for volume calculations (Stouten et al., 1993). Ligands and prosthetic groups were kept in the structure, but all lipid molecules found at the protein surfaces were removed.
For each protein structure, the average packing density PD was calculated as the mean value of the local packing densities of all contributing atoms having the standard deviation σ and the variance σ2. Atoms at the protein surface (“surface”) are usually packed far less efficiently and were excluded from analysis. Surface atoms were defined as all atoms touched by a 1.4Å radius probe rolling along the VdW spheres at the whole protein surface. Thus, for the analysis of packing densities, solely “buried” atoms were considered.
Analyzing cavities
In protein structures, there are locations, where a 1.4 Å radius probe fits without intersecting any atoms' van der Waals sphere and without reaching the surface (Richmond, 1984). These locations are defined as cavities. All buried atoms touched by the 1.4 Å probe were grouped as cavity neighbors. For this part of our analysis, the water molecules were removed. The relative amount of neighboring atoms of the cavities was estimated. Finally, the frequency of polar and nonpolar amino acids that are in contact with cavities was calculated, using the hydrophobicity scale of Eisenberg et al. (1984). Cavities enclosed by a fraction of nonpolar amino acids above average are termed “polar cavities”; those below the average “nonpolar cavities”.
Molecular surface patches
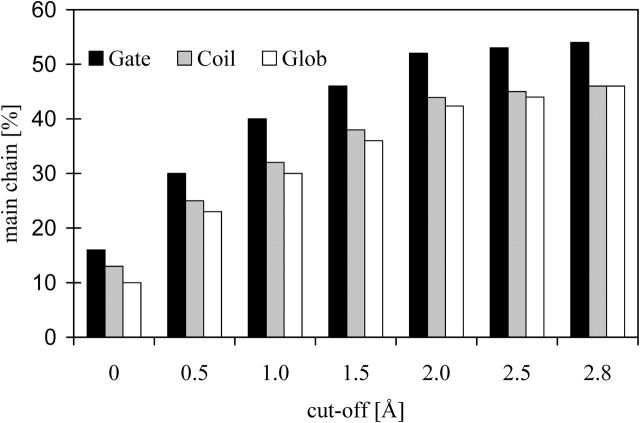
Helical interfaces are defined as pairs of molecular surface patches between neighboring helices that are in direct contact with each other (Preissner et al., 1998). Direct contact means that the atomic van der Waals surfaces are closer than a given cut-off distance. It was previously shown that molecular surface patches are generally flat atom assemblies with a length/width/depth ratio of ∼3:2:1. Nevertheless, the size and atomic composition of a molecular surface patch varies with the chosen cut-off (see Fig. 2). All packing values that will be mentioned were calculated at a medium cut-off value of 1.5 Å. At any rate, atoms being labeled as surface had been removed.
FIGURE 2.
Comparison of the average content of main-chain atoms (columns) of all buried atoms in different data sets of membrane (Gate, Coil) and globular proteins (Glob) in dependence of the proposed cut-off values.
RESULTS AND DISCUSSION
Packing densities
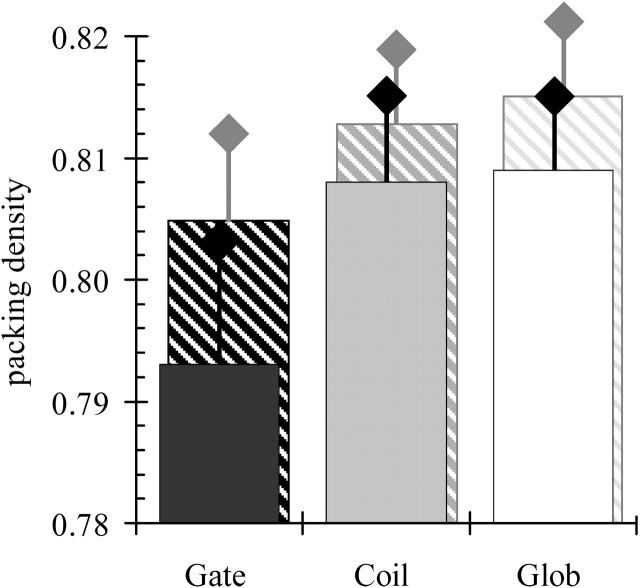
We used the Voronoi cell procedure to compare the packing densities of helical interfaces in nonredundant data sets of water-soluble globular proteins and of helical membrane proteins. According to their functions, we subdivided the data set of membrane protein structures into two classes: membrane gates comprising ion channels and solute transporters and membrane coils embracing metabolic-driven proton pumps, receptors, and photosystems (Table 1). Here we show that the helical interfaces of membrane coils are packed as tightly as in globular proteins (average packing density value 0.81). By contrast, transmembrane helices of membrane channels and transporters are significantly (t-test, p-value of 0.005) less well packed with an average packing value of 0.79 (Fig. 1).
FIGURE 1.
The average packing densities PD (filled columns) of all buried atoms in different data sets of membrane (Gate, Coil) and globular proteins (Glob) are depicted with the according variances σ2 (black lines) of the collected data. When the atoms with direct contact to cavities are removed, the PD values increase differently as indicated by the columns in dashed lines (variances σ2 now in shaded lines).
When we collated the composition and packing values of the buried amino acids, we found that virtually all types of residues promote for the lower packing values of membrane gates (Table 2). Only Ala, Gly, and Ile are packed as well as in the helical contacts of membrane coils and globular proteins. By sorting the amino acids according to their water solubility, it becomes apparent that the polar amino acids contribute most to the observed packing deficiency. These residues are again more abundant in the helix-helix interfaces of membrane gates, whereas the aromatic residues (Phe, Trp, and Tyr), which encompass the highest packing values in all data sets, are observed less frequently. These proportions approximately resemble those of the entire transmembrane helices (Hildebrand et al., 2004). That justifies our method of concentrating on the interface atoms. In addition, it seems that the lower packing density of membrane gates is already partly encoded in the primary structure.
TABLE 2.
Amino acid composition (%) and packing density values (PD) of helix-helix interfaces in different data sets of membrane (Gates, Coils) and globular proteins (Glob)
| Amino acid | Gates PD | Coils PD | Glob PD | Gates % | Coils % | Glob % |
|---|---|---|---|---|---|---|
| Ala | 0.79 | 0.79 | 0.79 | 11.8 | 8.0 | 8.6 |
| Arg | 0.74 | 0.78 | 0.79 | 1.5 | 0.9 | 5.1 |
| Asn | 0.74 | 0.77 | 0.78 | 2.0 | 1.0 | 2.4 |
| Asp | 0.72 | 0.81 | 0.79 | 0.9 | 0.8 | 2.8 |
| Cys | 0.78 | 0.79 | 0.80 | 1.4 | 1.0 | 0.7 |
| Gln | 0.77 | 0.80 | 0.79 | 1.4 | 0.9 | 3.3 |
| Glu | 0.74 | 0.75 | 0.79 | 0.9 | 0.4 | 5.6 |
| Gly | 0.77 | 0.78 | 0.77 | 7.9 | 5.6 | 1.8 |
| His | 0.76 | 0.82 | 0.81 | 0.5 | 4.8 | 2.6 |
| Ile | 0.79 | 0.81 | 0.80 | 10.4 | 8.5 | 7.0 |
| Leu | 0.78 | 0.80 | 0.80 | 15.7 | 18.2 | 15.2 |
| Lys | 0.74 | 0.80 | 0.80 | 0.8 | 0.5 | 3.4 |
| Met | 0.77 | 0.79 | 0.80 | 4.1 | 5.6 | 3.7 |
| Phe | 0.80 | 0.82 | 0.82 | 8.0 | 13.3 | 9.3 |
| Pro | 0.81 | 0.82 | 0.82 | 2.8 | 1.4 | 1.1 |
| Ser | 0.77 | 0.79 | 0.79 | 5.0 | 5.0 | 2.8 |
| Thr | 0.78 | 0.80 | 0.79 | 6.3 | 5.5 | 3.6 |
| Trp | 0.81 | 0.84 | 0.83 | 1.1 | 6.0 | 5.6 |
| Tyr | 0.79 | 0.82 | 0.83 | 4.9 | 5.2 | 8.5 |
| Val | 0.80 | 0.80 | 0.80 | 12.6 | 7.4 | 6.8 |
| Number | 5443 | 7330 | 26297 | 519 | 504 | 2527 |
| Hydrophobic* | 0.79 | 0.80 | 0.80 | 71.6 | 72.6 | 57.9 |
| Polar* | 0.74 | 0.79 | 0.79 | 7.5 | 4.5 | 22.7 |
| Indifferent* | 0.78 | 0.81 | 0.81 | 20.9 | 22.9 | 19.4 |
| Aromatic* | 0.80 | 0.83 | 0.83 | 14.0 | 24.5 | 23.4 |
PD was carried out on single atoms explicating the higher quantity of samples.
Hydrophobicity scale according to Eisenberg et al. (1984).
It has to be mentioned that, compared to the data set of globular proteins, the polar main-chain atoms of some amino acids (Ala, Cys, Gly, Leu, Phe, Tyr, Val) buried in the helix-helix interfaces of membrane gates are even more tightly packed (t-test, p-value of 0.05). We have recently shown that the helical geometry of membrane gates is characteristic for this class: The intrahelical hydrogen bonds between main-chain atoms are shorter and thus the Coulomb forces are stronger. These high packing values might be another feature of the characteristic architecture of transmembrane helices (Kim and Cross, 2002; Olivella et al., 2002). This shortening of hydrogen bonds and the optimized packing of main-chain atoms is supported by the low dielectric constant ɛ within the lipid bilayer (Chothia, 1975).
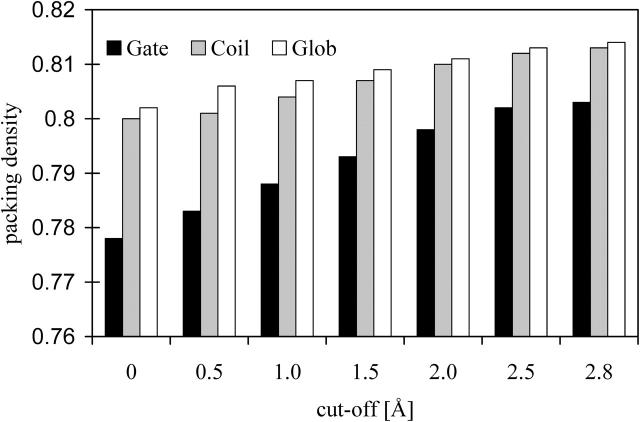
The comparison of the atomic packing density values reveals further details: The side-chain atoms buried between helices (PD = 0.77) are significantly (t-test, p-value of 0.005) less efficiently packed in the data set of membrane gates than the main-chain atoms (PD = 0.80). This is especially valid for the polar side-chain atoms (PD = 0.64). This difference is much smaller in the data sets of membrane coils and helices of globular proteins (Table 3). When a higher cut-off value was chosen, the helix patches expand in all three dimensions and the fraction of main-chain atoms at the interfaces increases (Fig. 2). Because the backbone atoms are packed more efficiently, the packing density of the investigated helix-helix interfaces accordingly rises (Fig. 3). This increase is plainest in the data set of membrane gates, because the side-chain atoms are packed most fluffily here.
TABLE 3.
Atomic packing density values (PD) of helix-helix interfaces in different data sets of membrane (Gates, Coils) and globular proteins (Glob)
| Types of atoms | Gates No. | Gates PD | Coils No. | Coils PD | Glob No. | Glob PD |
|---|---|---|---|---|---|---|
| Cmc* | 961 | 0.88 | 1056 | 0.89 | 3712 | 0.88 |
| Cα | 940 | 0.79 | 1109 | 0.80 | 3984 | 0.81 |
| Cβ | 710 | 0.76 | 964 | 0.80 | 3046 | 0.81 |
| Csc† | 1078 | 0.77 | 2071 | 0.81 | 8275 | 0.81 |
| Nmc | 785 | 0.81 | 828 | 0.82 | 2888 | 0.82 |
| Nsc | 46 | 0.67 | 144 | 0.74 | 636 | 0.75 |
| Omc | 798 | 0.70 | 860 | 0.72 | 2794 | 0.72 |
| Osc | 125 | 0.63 | 179 | 0.69 | 789 | 0.72 |
| S | 30 | 0.65 | 57 | 0.70 | 183 | 0.73 |
| Number | 5473 | 7268 | 26307 | |||
| mc* | 2544 | 0.80 | 2744 | 0.81 | 9394 | 0.81 |
| sc† | 2929 | 0.77 | 4524 | 0.80 | 16913 | 0.80 |
Main-chain atoms.
Side-chain atoms.
FIGURE 3.
Comparison of the average packing densities PD (columns) of all buried atoms in different data sets of membrane (Gate, Coil) and globular proteins (Glob) in dependence of the proposed cut-off values.
Our approach yields another characteristic trait of helix-helix packing of membrane gates that has not been described previously. Compared to helix-helix interfaces in membrane coils and globular proteins, the amount of main-chain atoms is higher in the data set of membrane gates (Fig. 2). At the cut-off value of 1.5 Å, nearly one-half of the atoms engaged in helix-helix packing are part of the main chain. In the former data sets, only one-third of those atoms are main chain (Table 3). This implies that helices in membrane gates tend to be closer to each other, which allows two adjacent helices to form direct contact between their backbones. In contrast, helices of membrane coils and globular proteins are farther apart and contact with nearby helices is formed instead by the side chains.
It has been noted that the tight backbone-to-backbone packing is important for the stabilization of the tertiary structure of membrane proteins. This structural signature was successfully applied to the prediction of their tertiary structure (Fleishman and Ben-Tal, 2002; Liu et al., 2004). Accordingly, the Cα-H⋯O hydrogen bond is a strong determinant of stability and specificity in transmembrane helix interactions (Senes et al., 2001). Interestingly, multiple hydrogen bonds of this type are predominantly found between parallel transmembrane helices that cross at right-handed angles. In this regard, membrane gates again differ markedly from the remaining helical membrane proteins: Two-thirds of the helix crossings are right-handed, whereas only one-third of the transmembrane helices in membrane coils cross right-handed (Hildebrand et al., 2004). Our approach thus supports the proposition that helices in membrane channels and transporters are stabilized throughout close approximations of their backbones, which again facilitates the formation of multiple interhelical hydrogen bonds, stabilizing the topology of the helices. The helical mobility that is necessary for the proper functioning of membrane channels and transporters (Swartz, 2004) could again be realized by the loose packing of their side-chain atoms.
Finally, the question has to be addressed as to whether the observed packing deficiencies found in the three data sets are equally dispersed over the entire protein structure, or whether they cluster in certain protein regions (cavities). In the first case, the observed packing deficiencies could be traced back to intrinsic factors, such as the different values for the dielectric constant, the specific amino acid composition, or the different quality of the protein structures (Fleming and Richards, 2000; Vaguine et al., 1999). In the second case, the packing defects probably point toward functionally important sites of the protein. Subsequently, all atoms that are in direct contact with cavities were removed. As a result, the packing densities of the different data sets converge articulately (Fig. 1). This strongly favors the hypothesis that the packing deficiencies detected in membrane gates might be functionally important.
Cavities
Cavities are empty or solvent-filled spaces within a protein structure that are large enough to accommodate at least one water molecule. When the atoms exposed to cavities were excluded, the actual differences in packing densities are rather small (Fig. 1). Thus the cavities account for the reduced packing of membrane gates. The density distribution of membrane gates is therefore reflected by the localization and characterization of internal cavities.
Transmembrane domains of membrane coils resemble nearly the same content of internal cavities as helices of globular proteins (5.2% and 5.5% of the buried atoms are in direct contact with a cavity). By contrast, slightly more cavities were found in the data set of membrane gates (6.4%, Table 1). Most cavities are neighbored by 15–25 atoms (∼31%) or <5 (57%) atoms. The results therefore indicate that the size of cavities found in proteins depends neither on the surrounding milieu, nor on the fold of the proteins (Rother et al., 2003).
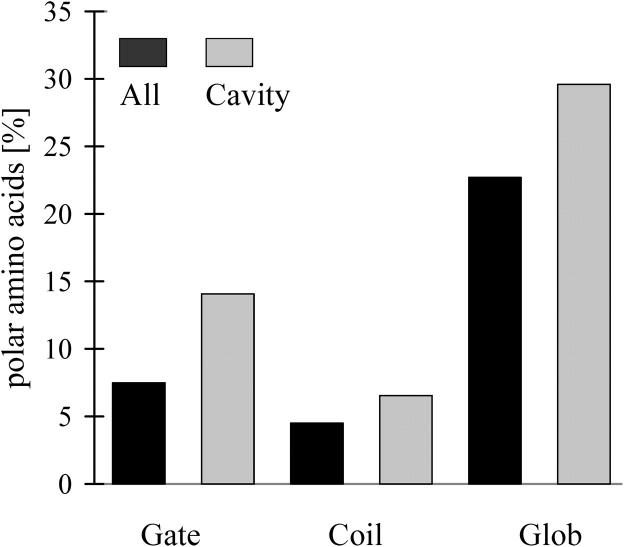
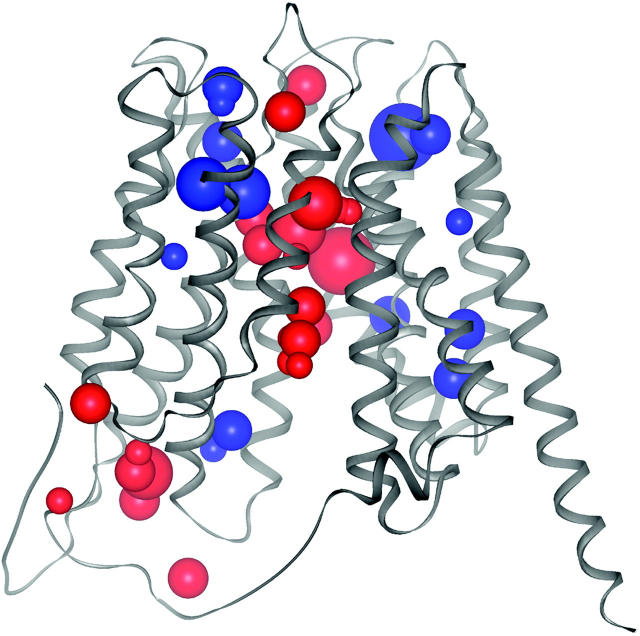
Cavities in membrane gates are highly polar in contrast to cavities in membrane coils (Fig. 4). In globular proteins, most polar cavities are found close to the protein surface (Rother et al., 2003). Unlike the helices in water-soluble globular proteins, transmembrane helices are encircled by hydrophobic lipid tails (see Material and Methods). Thus polar cavities within membrane proteins are positioned close to those protein regions that communicate with the extra- and intracellular environments, as the pores that pervade these proteins (Fig. 5). Therefore it seems feasible that the cavities of membrane gates are encircled by more polar amino acids than the cavities of membrane coils, and that many of them might be filled with water molecules.
FIGURE 4.
Portions of polar amino acids at interfaces between helices or at the walls of cavities of different data sets of membrane (Gate, Coil) and globular proteins (Glob).
FIGURE 5.
Polar (red) cavities are predominantly positioned in helix cap regions that are exposed to the polar milieu or within the gated pore of the glycerol-3-phosphate transporter (Protein Data Bank code 1pw4). Nonpolar (blue) cavities are placed in the proposed hinge regions that facilitate the rocker-switch type movement of the helices that occur upon substrate binding (Huang et al., 2003). The cavities are depicted as balls that are sized according to the number of atomic neighbors. The centers of the cavities were calculated from the atom coordinates of the cavities' neighbor atoms.
On the other hand, it is likely that many of the polar cavities that are placed within the structures of membrane gates will become surface when, via molecular rearrangements, the pores are opened and the residues previously surrounding the cavities become solvent-accessible. Therefore it is not surprising that more cavities were found in those membrane gates that had been crystallized in a closed or partly closed conformation and that these structures (1iwg, 1pw4) are among those having the lowest packing density values (Table 1). However, the packing density is influenced by the presence or absence of water in cavities and grooves at the protein surface (Tsai et al., 1999). This is a critical point, since water molecules are not always recorded in protein structure files.
However, it is assumed that deeply buried cavities contribute only little to this potential error, because they are hydrophobic, indicating that they are devoid of water (Rother et al., 2003). This is supported by molecular dynamic simulations revealing that nonpolar cavities in the protein interior are even more compressible than other parts of the protein (Kocher et al., 1996). Ongoing, we detected nonpolar cavities in protein regions that are supposed to be functionally important, too: around the restriction region and the so called “plug” of the translocon (1rh5, Fig. 6), and around the hinge regions of the structure of glycerol-3-phosphate transporter (1pw4, Fig. 5). We therefore conclude that focal packing defects allow for the structural flexibility that is required for the proper functioning of membrane channels and transporters.
FIGURE 6.
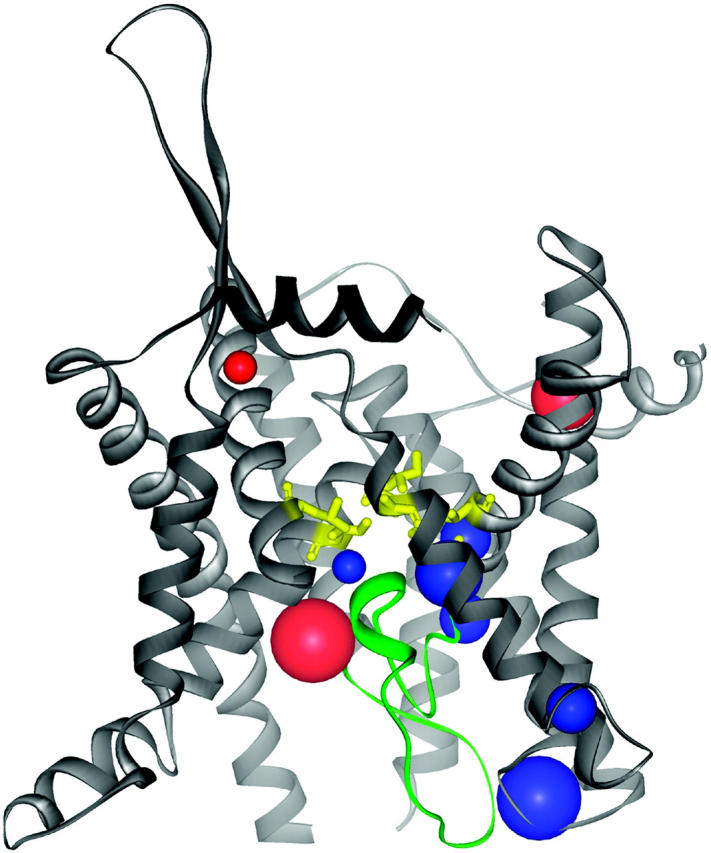
Polar (red) and apolar (blue) cavities in the translocon channel (Protein Data Bank code 1rhz) are positioned near the plug (green) and close to the restriction site (yellow, with transmembrane helix 8 and 9 in the foreground removed for a better view), where large rearrangements are supposed to occur (Van den Berg et al., 2004). The cavities are depicted as described in Fig. 5.
CONCLUSION
With the help of the Voronoi cell procedure, we discovered differences in helix-helix packing densities in membrane proteins of different functions: membrane channels and transporters are packed less efficiently than other membrane proteins where molecular rearrangements are supposed to occur only on a small scale. These membrane coils have the same average atomic packing densities as helices of globular proteins. Our method thus leads to another conclusion than a previous approach, where the packing densities of entire amino acids were calculated with the occluded surface method (Eilers et al., 2002). As pointed out in a mathematical appendix, using their method will lead to the packing density of atoms in direct neighborhood to protein cavities being overrated.
The loose packing of membrane channels and transporters is in turn mainly caused by the frequent placement of polar side chains close to voids or pockets that are lined along the pores that pervade these proteins (Fig. 1). It is inferred by focal packing defects, i.e., cavities, rather than by steadily increased distances between midpoints of atoms. Nevertheless, if the proper functioning of membrane channels and transporters is based on a relatively loose packing of helical interfaces, how are these proteins stabilized compared to other membrane proteins? The comparatively close packing of the transmembrane backbones indicates that main-chain interactions probably compensate for the loose packing of side chains. The investigation of the packing density of transmembrane helices finally leads to the detection of structural details that promote a better understanding of the relation between stabilization and function of membrane proteins.
Acknowledgments
We thank Dr. Christoph Gille and Dr. Simon Ward for carefully reading the manuscript.
This work was supported with grants from the Federal Ministry of Education and Research of Germany (BMBF), Berlin Center for Genome Based Bioinformatics (BCB), and the Charité.
APPENDIX: ESTIMATION OF THE MOLECULAR PACKING IN DEPENDENCE ON THE METHOD EMPLOYED AND THE DISTANCE TO THE CENTER OF A CAVITY
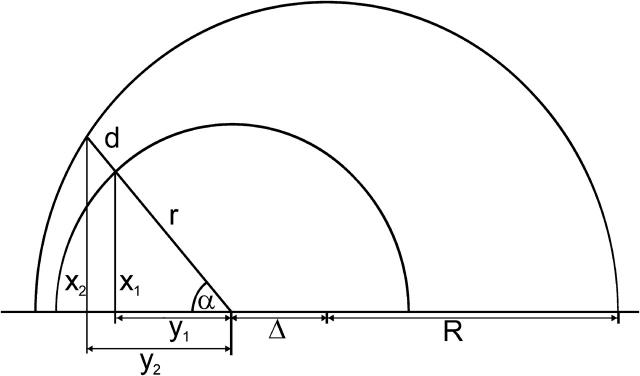
An atom is considered to have the radius r, lying in a cavity of radius R, and in a distance of Δ from the center of the cavity (Fig. 7). If the atomic packing density (PD) is estimated by the Voronoi cell method, the situation will be the same regardless of whether the atom lies in the middle or more on the periphery of a cavity. If the occluded surface method (PV) is used, different values will be obtained. When calculating the packing density the following integral has to be solved:
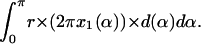 |
FIGURE 7.
Two-dimensional sketch illustrating the position dependence of the packing values calculated by the occluded surface method. An atom is considered with radius r, lying in a cavity of radius R, and in a distance of Δ from the center of the cavity. The variables (x1, x2, y1, y2) are used to calculate d in dependence on the angle α (see Eqs. 1–5).
The factor r is the integral on the circle with radius r; the factor (2πx1(α)) is the integral around the circle with radius x1, orthogonal to the x axis, and d(α) is the distance between the two spheres, measured from the center of the atom. The following equations must be combined, when the function is integrated:
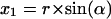 |
(1) |
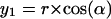 |
(2) |
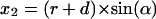 |
(3) |
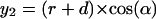 |
(4) |
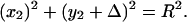 |
(5) |
Inserting Eqs. 3 and 4 into Eq. 5 results in
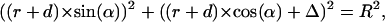 |
and thus
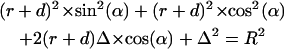 |
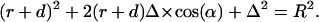 |
This last quadratic equation can be solved by:
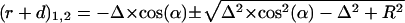 |
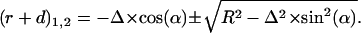 |
Because 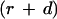 should not be negative, we get
should not be negative, we get
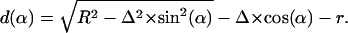 |
The integral becomes
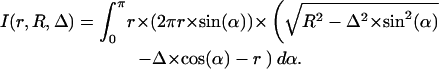 |
For the packing value, we have to divide through the surface area of the atom 4πr2 and the maximal length 2.8 Å, getting
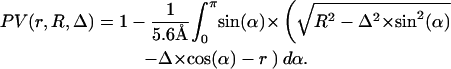 |
Although the packing value PV (occluded surface method) estimates the mean distance to the next atoms, the packing density PK (Voronoi cell) rates the nonoccupied space around the atoms. This space increases with the power of 3 in relation to the distance to the center of the atom, whereas this distance has only linear influence on the packing value PV. Therefore, especially near cavities, the packing value PV will be larger compared to other cases with the same nonoccupied space around the atom and therefore the same packing density (Table 4). Because cavities are particularly frequent in protein channels and transporters, the molecular packing density of these proteins will be overestimated when calculated with the occluded surface method (PV).
TABLE 4.
Some examples for different packing values (PV) in dependence on the distance Δ of the center of the cavity to the center of the atom
| r | R | Δ | PV(Δ) | PV(0) | Difference |
|---|---|---|---|---|---|
| Å | Å | Å | % | % | % |
| 1.4 | 2.0 | 0.6 | 80.75 | 78.57 | 2.18 |
| 1.4 | 2.5 | 0.6 | 62.45 | 60.71 | 1.73 |
| 1.4 | 2.5 | 0.9 | 64.68 | 60.71 | 3.96 |
| 1.4 | 2.5 | 1.1 | 66.72 | 60.71 | 6.01 |
| 1.4 | 3.0 | 0.6 | 44.30 | 42.86 | 1.44 |
| 1.4 | 3.0 | 0.9 | 46.13 | 42.86 | 3.27 |
| 1.4 | 3.0 | 1.2 | 48.77 | 42.86 | 5.91 |
Packing values (occluded surface) differ by up to 6%. Increased packing values occur for atoms near the wall of a cavity, although the nonoccupied space near the atom and therefore the packing density would be the same in both cases (PV (Δ), PV (0)). Note that in the listed examples, the distance to the wall is not larger than 2.8 Å.
References
- Bowie, J. U. 1997. Helix packing in membrane proteins. J. Mol. Biol. 272:780–789. [DOI] [PubMed] [Google Scholar]
- Chothia, C. 1975. Structural invariants in protein folding. Nature. 254:304–308. [DOI] [PubMed] [Google Scholar]
- Chothia, C., M. Levitt, and D. Richardson. 1981. Helix to helix packing in proteins. J. Mol. Biol. 145:215–250. [DOI] [PubMed] [Google Scholar]
- DeGrado, W. F., H. Gratkowski, and J. D. Lear. 2003. How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 12:647–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers, M., A. B. Patel, W. Liu, and S. O. Smith. 2002. Comparison of helix interactions in membrane and soluble α-bundle proteins. Biophys. J. 82:2720–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1984. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA. 81:140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman, S. J., and N. Ben-Tal. 2002. A novel scoring function for predicting the conformations of tightly packed pairs of transmembrane α-helices. J. Mol. Biol. 321:363–378. [DOI] [PubMed] [Google Scholar]
- Fleming, P. J., and F. M. Richards. 2000. Protein packing: dependence on protein size, secondary structure and amino acid composition. J. Mol. Biol. 299:487–498. [DOI] [PubMed] [Google Scholar]
- Gille, C., A. Goede, R. Preissner, K. Rother, and C. Frömmel. 2000. Conservation of substructures in proteins: interfaces of secondary structural elements in proteasomal subunits. J. Mol. Biol. 299:1147–1154. [DOI] [PubMed] [Google Scholar]
- Goede, A., R. Preissner, and C. Frömmel. 1997. Voronoi cell: new method for allocation of space among atoms: elimination of avoidable errors in calculation of atomic volume and density. J. Comput. Chem. 18:1113–1123. [Google Scholar]
- Hildebrand, P. W., R. Preissner, and C. Frömmel. 2004. Structural features of transmembrane helices. FEBS Lett. 559:145–151. [DOI] [PubMed] [Google Scholar]
- Huang, Y., M. J. Lemieux, J. Song, M. Auer, and D. N. Wang. 2003. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 301:616–620. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- Kim, S., and T. A. Cross. 2002. Uniformity, ideality, and hydrogen bonds in transmembrane α-helices. Biophys. J. 83:2084–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher, J. P., M. Prevost, S. J. Wodak, and B. Lee. 1996. Properties of the protein matrix revealed by the free energy of cavity formation. Structure. 4:1517–1529. [DOI] [PubMed] [Google Scholar]
- Langosch, D., and J. Heringa. 1998. Interaction of transmembrane helices by a knobs-into-holes packing characteristic of soluble coiled coils. Proteins. 31:150–159. [DOI] [PubMed] [Google Scholar]
- Lemmon, M. A., and D. M. Engelman. 1994. Specificity and promiscuity in membrane helix interactions. FEBS Lett. 346:17–20. [DOI] [PubMed] [Google Scholar]
- Liu, W., M. Eilers, A. B. Patel, and S. O. Smith. 2004. Helix packing moments reveal diversity and conservation in membrane protein structure. J. Mol. Biol. 337:713–729. [DOI] [PubMed] [Google Scholar]
- Locher, K. P., R. B. Bass, and D. C. Rees. 2003. Structural biology. Breaching the barrier. Science. 301:603–604. [DOI] [PubMed] [Google Scholar]
- MacKenzie, K. R., and D. M. Engelman. 1998. Structure-based prediction of the stability of transmembrane helix-helix interactions: the sequence dependence of glycophorin A dimerization. Proc. Natl. Acad. Sci. USA. 95:3583–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivella, M., X. Deupi, C. Govaerts, and L. Pardo. 2002. Influence of the environment in the conformation of α-helices studied by protein database search and molecular dynamics simulations. Biophys. J. 82:3207–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman, N., K. B. Ward, and P. J. Fleming. 1995. Occluded molecular surface: analysis of protein packing. J. Mol. Recognit. 8:334–344. [DOI] [PubMed] [Google Scholar]
- Perozo, E., D. M. Cortes, P. Sompornpisut, A. Kloda, and B. Martinac. 2002. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 418:942–948. [DOI] [PubMed] [Google Scholar]
- Popot, J. L., and D. M. Engelman. 2000. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69:881–922. [DOI] [PubMed] [Google Scholar]
- Preissner, R., A. Goede, and C. Frommel. 1999. Spare parts for helix-helix interaction. Protein Eng. 12:825–832. [DOI] [PubMed] [Google Scholar]
- Preissner, R., A. Goede, and C. Frömmel. 1998. Dictionary of interfaces in proteins (DIP). Data bank of complementary molecular surface patches. J. Mol. Biol. 280:535–550. [DOI] [PubMed] [Google Scholar]
- Richmond, T. J. 1984. Solvent accessible surface area and excluded volume in proteins. Analytical equations for overlapping spheres and implications for the hydrophobic effect. J. Mol. Biol. 178:63–89. [DOI] [PubMed] [Google Scholar]
- Rother, K., R. Preissner, A. Goede, and C. Frömmel. 2003. Inhomogeneous molecular density: reference packing densities and distribution of cavities within proteins. Bioinformatics. 19:2112–2121. [DOI] [PubMed] [Google Scholar]
- Senes, A., I. Ubarretxena-Belandia, and D. M. Engelman. 2001. The Cα-H⋯O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. USA. 98:9056–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouten, P. F. W., C. Frömmel, H. Nakamura, and C. Sander. 1993. An effective solvation term based on atomic occupancies for use in protein simulations. Mol. Simulat. 10:97–120. [Google Scholar]
- Swartz, K. J. 2004. Opening the gate in potassium channels. Nat. Struct. Mol. Biol. 11:499–501. [DOI] [PubMed] [Google Scholar]
- Tsai, J., R. Taylor, C. Chothia, and M. Gerstein. 1999. The packing density in proteins: standard radii and volumes. J. Mol. Biol. 290:253–266. [DOI] [PubMed] [Google Scholar]
- Vaguine, A. A., J. Richelle, and S. J. Wodak. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55:191–205. [DOI] [PubMed] [Google Scholar]
- Van den Berg, B., W. M. Clemons Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature. 427:36–44. [DOI] [PubMed] [Google Scholar]
- White, S. H., and W. C. Wimley. 1999. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28:319–365. [DOI] [PubMed] [Google Scholar]