Abstract
Intrinsically unstructured/disordered proteins (IUPs) exist in a disordered and largely solvent-exposed, still functional, structural state under physiological conditions. As their function is often directly linked with structural disorder, understanding their structure-function relationship in detail is a great challenge to structural biology. In particular, their hydration and residual structure, both closely linked with their mechanism of action, require close attention. Here we demonstrate that the hydration of IUPs can be adequately approached by a technique so far unexplored with respect to IUPs, solid-state NMR relaxation measurements. This technique provides quantitative information on various features of hydrate water bound to these proteins. By freezing nonhydrate (bulk) water out, we have been able to measure free induction decays pertaining to protons of bound water from which the amount of hydrate water, its activation energy, and correlation times could be calculated. Thus, for three IUPs, the first inhibitory domain of calpastatin, microtubule-associated protein 2c, and plant dehydrin early responsive to dehydration 10, we demonstrate that they bind a significantly larger amount of water than globular proteins, whereas their suboptimal hydration and relaxation parameters are correlated with their differing modes of function. The theoretical treatment and experimental approach presented in this article may have general utility in characterizing proteins that belong to this novel structural class.
INTRODUCTION
Intrinsically unstructured/disordered or natively unfolded proteins (IUPs), common in living organisms, exist in a highly flexible conformational state mostly devoid of well-defined secondary and tertiary structure (Dunker et al., 2002; Tompa, 2002; Uversky, 2002a; Wright and Dyson, 1999). These proteins fulfill essential functions (Dunker et al., 2002; Tompa, 2002, 2003; Wright and Dyson, 1999), intimately linked with the lack of a well-defined structure. In terms of their modes of action, IUPs can be classified into six broad functional categories (Dunker et al., 2002; Tompa, 2002, 2003; Tompa and Csermely, 2004), in five of which they act via permanent or transient binding of a physiological partner, i.e., another protein, DNA, RNA, or some other ligand. In these cases, termed effectors, scavengers, assemblers, display sites, and chaperones, the disordered protein recognizes a structured partner and undergoes induced folding (Demchenko, 2001; Dunker et al., 2002; Dyson and Wright, 2002; Leulliot and Varani, 2001; Tompa, 2002). Such recognition by an initially disordered protein confers exceptional specificity and versatility to the interaction process, which explains the prevalence of structural disorder in signaling and regulatory proteins (Iakoucheva et al., 2002; Ward et al., 2004).
To understand these binding processes in full mechanistic and thermodynamic detail, the quantitative elucidation of the hydration properties of IUPs is needed. For example, their largely open and solvent-exposed structure has to undergo rapid dehydration-hydration cycles to fulfill functions that rely on transient partner recognition, such as of display sites and chaperones (Dunker et al., 2002; Tompa, 2002; Tompa and Csermely, 2004). In concordance, their flexible and adaptable structure abounds in an extended and fully hydrated motif, the polyproline II helix (Syme et al., 2002; Tompa, 2003; Uversky et al., 1998), often implicated in intermolecular recognition processes (Bochicchio and Tamburro, 2002; Williamson, 1994). In addition, hydration and water retention is the very function of certain IUPs involved in dehydration or other types of osmotic stresses (Goyal et al., 2003; Kiyosue et al., 1994; Lisse et al., 1996). Last, but not least, IUPs often are not fully disordered but have permanent or transient global or local structural organization (Tompa, 2002, 2003; Uversky, 2002b). Such a residual structure closely associated with binding functions (Fuxreiter et al., 2004) must manifest itself in suboptimal hydration of the polypeptide chain (Konno et al., 1997).
Here we demonstrate that a technique so far unexplored with respect to IUPs, solid-state NMR relaxation measurements, yields information on the amplitude and dynamics of their hydration processes. The information provided by this NMR technique is complementary to that obtained by other, more often used methods (homo- and heteronuclear nuclear Overhauser effect, relaxation dispersion (nuclear magnetic relaxation dispersion), and spin-spin-lattice relaxation (for reviews, see Antzutkin, 2002; Dyson and Wright, 2004; Otting, 1997; Wider, 1998). We have selected three IUPs for these studies. Calpastatin, the specific inhibitor of the Ca2+-activated intracellular cysteine protease, calpain (Emori et al., 1988; Maki et al., 1989), is capable of very rapid and specific interaction with the enzyme. Microtubule-associated protein 2c (MAP2c) binds microtubules (Matus, 1994; Sanchez et al., 2000), and its major function is to ensure proper spacing in the cytoskeleton by long-range entropic repulsion (Chen et al., 1992; Mukhopadhyay and Hoh, 2001). Early responsive to dehydration 10 is a dehydration-stress protein, which shifts the osmotic balance of plant cells due to its putative large hydration capacity (Alsheikh et al., 2003; Kiyosue et al., 1994). To study the hydration of these proteins, their solutions are gradually frozen down, to separate free induction decays (FIDs) of the hydrate layer from those of the overwhelming background of bulk water. The full methodological and theoretical background of how to carry out and interpret these measurements in four different temperature ranges above and below the freezing point has been outlined (Noack, 1971; Racz et al., 1983; Slichter, 1990; Tompa et al., 2001, 2003). As a result, we report the amplitude of hydrate water and dynamic parameters, such as activation energy and various correlation times of the first inhibitory domain of calpastatin (CSD1), MAP2c, ERD10, and bovine serum albumin (BSA) as reference, which are all interpreted in terms of the structure and function of these IUPs.
Theoretical background
Protein solutions are composed of protein molecules, a hydration shell, and unbound water as main components. The protein molecules have rapidly and slowly movable protons, which act as relaxation centers of all the magnetic dipoles. Whereas the rapidly moving centers originate from side chains with rotatable groups or any other flexible parts of the protein, the slowly moving centers correspond to the more rigid backbone. The spin polarization of the water protons is transferred first by rapid material exchange from the bulk solvent to the hydration shell and from there to the relaxation centers by dipolar interaction and spin diffusion. Also, there exists a slight material exchange between the water and protein protons (Noack, 1971). To separate the various water phases present in the samples, the protein solutions are frozen down: the narrow proton signal (time-domain FID) of hydrogen atoms in the ice phase falls within the dead time of the spectrometer. The phases of ice protons, protein protons, and (unfrozen) water protons are clearly separated in the FID signal by virtue of large differences in the spin-spin relaxation rate R2. Ice protons yield a solid signal fraction with a typical value of R2 > 200,000 s−1, which is completely buried in the dead time of the spectrometer. Protein protons yield a solidlike signal fraction also with a large value of R2 > 20,000 s−1. The water signal has a much smaller spin-spin relaxation rate, typically R2 < 2000 s−1. This enables specific recording of FIDs that belong to the hydrate layer of proteins (Fig. 1). In terms of the behavior of water molecules, four distinct temperature regions (a–d) could be identified, within which the contribution of water phases to the NMR signal show characteristic differences.
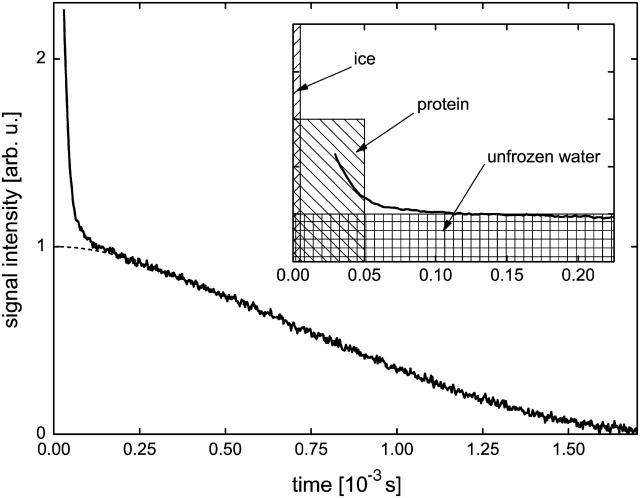
FIGURE 1.
Illustration of the method applied to measure the fraction of the unfrozen water component xunfrozen (MAP2c solution, T = −10.4°C). The slowly decaying part of the time-domain FID signal was extrapolated to t = 0 by applying Lorentzian approximation (dashed line). The extrapolated signal intensity was then compared to the signal intensity measured above 0°C when the whole sample is in liquid state, to get the xunfrozen value. The inset shows the typical spread in time of FID signals produced by ice protons, protein protons, and unfrozen water protons.
Region a
Above the freezing point, all the water of the protein solution is in liquid state and contributes to the NMR signal. A mean activation energy value related to the water dynamics can be calculated from the temperature dependence of the measured spin-lattice relaxation rates.
Region b
The process of freezing initially acts only upon bulk water outside the hydration shell. Thus, the protein solution can be treated as a two-phase (hydration shell and free water) spin system at about the freezing point, with molecular exchange between the phases. The case of rapid exchange applies if the lifetime of spins in each phase is much shorter than their relaxation times in the given phase. In this case, the relaxation of nuclear magnetization is exponential and
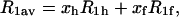 |
(1) |
where R1av is the measured spin-lattice relaxation rate measured just before freezing, whereas ‘h’ and ‘f’ stand for the hydration shell and free water, respectively. x denotes the probability density of the spins in a phase and 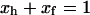 (Zimmerman and Brittin, 1957). In this picture, the relative amount of unfrozen water just below the freezing temperature is equal to xh and the spin-lattice relaxation rate at this temperature is taken as R1h. The fraction xh results directly from the FID intensity measurements (see Experimental). The spin-lattice relaxation rate in the free water phase, R1f, at about the freezing point can then be calculated from the equation above.
(Zimmerman and Brittin, 1957). In this picture, the relative amount of unfrozen water just below the freezing temperature is equal to xh and the spin-lattice relaxation rate at this temperature is taken as R1h. The fraction xh results directly from the FID intensity measurements (see Experimental). The spin-lattice relaxation rate in the free water phase, R1f, at about the freezing point can then be calculated from the equation above.
Region c
The temperature range from freezing point down to ∼250 K is characterized by rapidly changing spin-lattice relaxation rates and amounts of unfrozen water in the hydrate layer. The measured spin-lattice relaxation rates can be interpreted as weighted averages of the relaxation rates in different unfrozen water fractions. The measurable relaxation rate R1M of an n-fraction sample is (Noack, 1971)
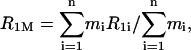 |
(2) |
where mi is the number of protons in the ith fraction and R1i is the relaxation rate of the ith fraction. The original theoretical description (Noack, 1971) was developed for vacuum dehydration of biological samples at room temperature. It can be applied without change to our case with the only difference that instead of the dehydration process, the freezing process was used to separate the contributions of the different water fractions to R1M. When the first hydration-shell phase freezes, the first term disappears from the summation. Upon further cooling, the next term becomes zero and so on until only one, quasi-rigid phase is left, which is analyzed in the next section.
Region d
The unfrozen water fraction can be treated as a system with identical 1H nuclei below 250 K. The amount of unfrozen water changes only slightly in this temperature range. The relaxation rate R1 is described by the Redfield-Slichter model (Racz et al., 1983). The spin-lattice relaxation in an applied magnetic field B0 is interpreted as the statistical ensemble average of local field Bloc induced transitions between two Zeeman levels. The dipoles interact with the fluctuating magnetic field Bloc. The spin-lattice relaxation rate formula of this model with isotropic fluctuations is
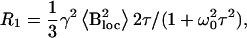 |
(3) |
where γ is the gyromagnetic ratio of the 1H nuclei, ω0 is the Larmor frequency, and τ is the mean jump time characteristic to the fluctuation of Bloc. The mean jump time obeys the Arrhenius law: 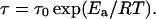 The mean squared amplitude (variance) of the isotropic fluctuations is
The mean squared amplitude (variance) of the isotropic fluctuations is 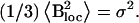 The spin-spin relaxation rate R2 is given by the equation
The spin-spin relaxation rate R2 is given by the equation 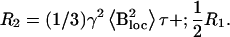
The relative magnitudes of the spin-spin (R2) and the rotating-frame spin-lattice (R1ρ) relaxation rates compared to the spin-lattice relaxation rate at the temperature of the R1 maximum is informative with respect to the nature of the correlation time(s). When R2 and R1ρ are greater by one or two orders of magnitude than that predicted by the Redfield-Slichter or the Bloembergen-Purcell-Pound (Bloembergen et al., 1948) model fitted to the R1 values and 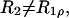 the relaxation cannot be described by a single correlation time. Two distinct molecular motions characterized by two different correlation times τs and τr can be assumed to account for the measured relaxation rates. The relaxation rates at the temperature of the R1 maximum then can be expressed as given in Slichter (1990):
the relaxation cannot be described by a single correlation time. Two distinct molecular motions characterized by two different correlation times τs and τr can be assumed to account for the measured relaxation rates. The relaxation rates at the temperature of the R1 maximum then can be expressed as given in Slichter (1990):
 |
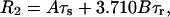 |
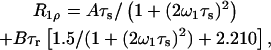 |
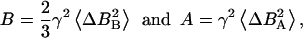 |
where A and B are the strengths of interactions responsible for the relaxation and τs and τr are the correlation times for slow molecular motion and reorientational fast motions, respectively. The parameters A, B, τs, and τr can be calculated from the above equations at the temperature of the R1 maximum.
Region b–d
The frozen water (ice) fraction acts as a rigid spin system. Obviously, it is present in increasing amounts when cooling the sample below TFP. Its NMR signal is characterized by a rapid decay (Barnaal and Lowe, 1967) lost in the dead time of the spectrometer and by a very small spin-lattice relaxation rate. The contribution of ice therefore cannot be detected directly, only its absence is experienced.
EXPERIMENTAL
DNA constructs
The clones of human calpastatin domain 1 (CSD1, Ala137-Lys277 of CST) and rat MAP2c were kindly provided by Prof. M. Maki from Nagoya University (Nagoya, Japan) and Prof. A. Matus from the Friedrich Miescher Institut (Basel, Switzerland). The coding regions were amplified by PCR and subcloned into the NdeI-XhoI sites of the expression vector pET22b (Novagene, Darmstadt, Germany). The gene of ERD10, including an intron, was amplified from Arabidopsis thaliana genomial DNA, with Erd10F (GGAATTCCATATGGCAGAAGAGTACAAGAACAC) and Erd10R (ATAGTTTAGCGGCCGCATCAGACACTTTTTCTTTCTTCTCTTCC) primers. The exons from this primary product were amplified with Erd10F and Erd10R, and two inner primers (Erd10endoF, CCAACAGCTCTTCTTCCTCT TCGAGTGATGAAGAAGGTGAAG and Erd10endoR, CTTCACCTTCTTCATCACTCGAAGAGGAAGAAGAGCTGTTGG, which encode for an overlap in the middle). The exons were then joined by simple annealing PCR, to produce ERD10 cDNA, which was then ligated into pET22b. All the constructs were checked by sequencing at MWG-AG Biotech (Ebersberg, Germany).
Protein purification
CSD1 was expressed in the Escherichia coli strain BL21 and purified on diethylaminoethyl-cellulose anion exchange column as described in (Yang et al., 1994). MAP2c was prepared according to (Ferralli et al., 1994). ERD10 was also expressed in E. coli BL21 and purified by Ni-nitrilotriacetic acid affinity chromatography. Purified proteins were dialyzed into distilled water and dried down by lyophilization. For NMR experiments, the proteins were dissolved on the day of use in buffer 20 mM Tris + 150 mM NaCl + 0.1 mM EDTA at 50 mg/ml (CSD1, MAP2c, and BSA) or 25 mg/ml (ERD10).
NMR spectroscopy
The measurements and data acquisition were accomplished by a Bruker (Rheinstetten, Germany) SXP 4-100 pulse NMR spectrometer of ∼10−6 resolution at ω0/2π = 44.14 and 82.57 MHz. Spin-lattice relaxation rates were measured by the inversion-recovery method (Fig. 2). As an addition to the known NMR methods, the standard in situ measurement of 1H concentration was also solved (Tompa et al., 2003), which is operative in the time range of minutes and enables the simultaneous measurement of NMR relaxation rates and hydrogen content. Determination of the unfrozen water fraction (xunfrozen) is based on the comparison of the FID signal intensity extrapolated to t = 0 (Fig. 1) with that measured at a temperature where the whole sample is in liquid state. The zero-time FID signal intensity is proportional to the number of resonant nuclear spins in the sample.
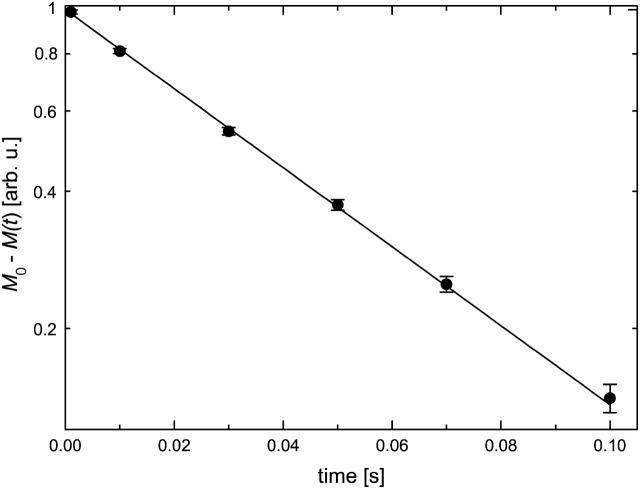
FIGURE 2.
Typical magnetization versus pulse spacing curve of π-t-π/2 experiments (inversion-recovery method). Spin-lattice relaxation rate R1 was obtained by fitting the equation M0 − M(t) = 2 M0 × exp(−t × R1) (line) to the experimental data (circles).
The measurements were made by a variable-temperature probe. The temperature was controlled to ±0.1°C.
MATERIALS
For DNA purification, the Nucleo-Spin extract kit (Macherey-Nagel, Püren, Germany) was used. BSA and all other chemicals were purchased from Sigma Chemical, St. Louis, MO. Buffers were made in Millipore (Billerica, MA) MilliQ water.
RESULTS AND DISCUSSION
Region a spin-lattice relaxation above freezing point
Spin-lattice relaxation was exponential in the studied samples above the freezing point. R1 data showed simple Arrhenius type behavior, and approximate activation energies (Ea) were calculated (Table 1). The results can be interpreted as mean activation energy values characteristic to the dynamics of water molecules. As seen, the values are within experimental error for the solutions of IUPs and the globular control, i.e., the proteins have a negligible effect on bulk water. The 18–20 kJ/mol value accords with the typical 20 kJ/mol energy of a hydrogen bond in water.
TABLE 1.
Mean activation energy values related to the water dynamics in liquid state solutions determined from spin-lattice relaxation rates
| Sample | Ea [kJ/mol] |
|---|---|
| MAP2c | 18 ± 5 |
| CSD1 | 19 ± 3 |
| ERD10_a | 18 ± 5 |
| ERD10_b | 20 ± 7 |
| BSA | 18.4 ± 0.7 |
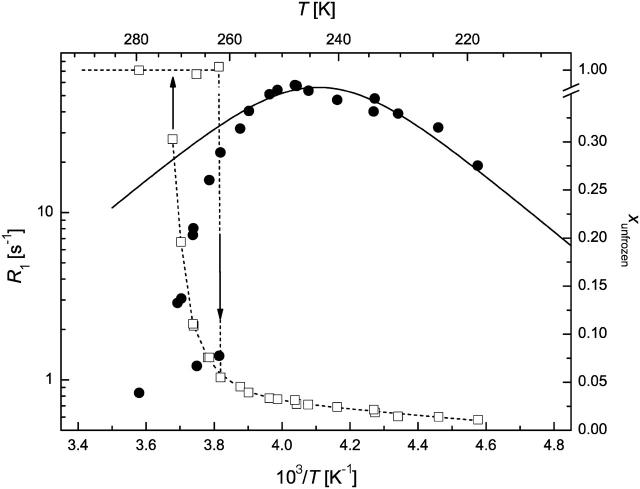
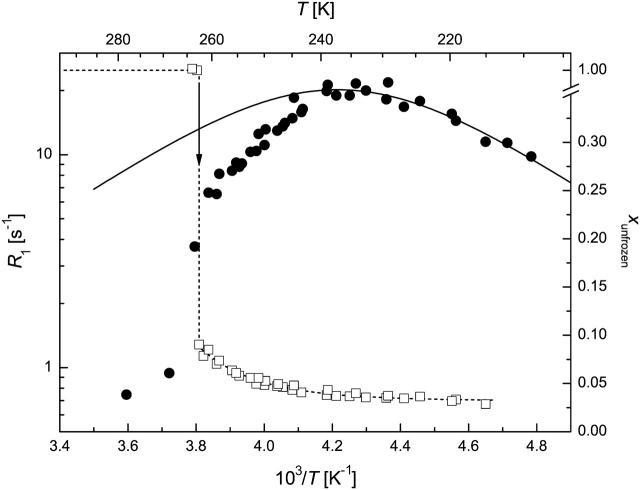
Region b freezing
The FID intensity measurements allowed the monitoring of the freezing process. Supercooling was experienced, and a sharp freezing point could be detected at 261–263 K (Table 2 and Figs. 3–6). A two-phase analysis of the FID intensity and spin-lattice relaxation rate values measured just above and below the freezing point (TFP) was made to get a simple description of the hydration shell and the free-water content. R1av = 1.1–1.4 s−1 was obtained just above TFP, indicative of the fast relaxation of water bound to proteins in comparison to pure water, which is due to the solutes acting as relaxation centers. As slowly relaxing free water that accounts for ∼80%–90% of total water freezes out, the relaxation rates measured just below TFP become significantly higher. The calculated R1f values (Eq. 1) show that 1H nuclei in the free-water phase relax much more slowly than in the hydration shell. Further, CSD1 and ERD10 are more effective at promoting relaxation than BSA, which might be due to their special amino acid composition (Dunker et al., 2002; Tompa, 2002; Uversky et al., 2000) or enhanced flexibility (Tompa, 2002; Wright and Dyson, 1999). When the fraction of bound water, xh, is considered, IUPs bind significantly more water than globular proteins, in agreement with their unfolded, mostly solvent-exposed character. To our knowledge, this is the first quantitative assessment of the hydration capacity of IUPs. An interesting further observation is that IUPs display marked hysteresis in terms of freezing/melting point and the amount of water they bind. This effect is most conspicuous with ERD10, which appears to bind a very large amount of water at its melting temperature, i.e., under equilibrium conditions. This finding is in perfect agreement with the proposed physiological function of ERD10 as a water-binding dehydration stress protein (Alsheikh et al., 2003; Kiyosue et al., 1994).
TABLE 2.
Freezing point (TFP), measured (R1av, R1h), and calculated (R1f) spin-lattice relaxation rates and bound water fraction (xh) for various proteins
| MAP2c | CSD1 | ERD10 | BSA | |
|---|---|---|---|---|
| TFP [K] | 261 ± 1 | 261 ± 1 | 262 ± 1 | 263 ± 1 |
| ω0/2π [MHz] | 44.14 | 44.14 | 82.57 | 44.14 |
| R1av [s−1] | 1.07 ± 0.05 | 1.10 ± 0.06 | 1.41 ± 0.07 | 1.11 ± 0.06 |
| R1h [s−1] | 3.3 ± 0.2 | 6.1 ± 0.3 | 5.8 ± 0.3 | 3.7 ± 0.2 |
| R1f [s−1] | 0.7 ± 0.1 | 0.3 ± 0.1 | 0.7 ± 0.1 | 0.85 ± 0.09 |
| xh | 0.142 ± 0.005 | 0.135 ± 0.005 | 0.110 ± 0.005* | 0.090 ± 0.005 |
Calculated for 50 mg/ml concentration.
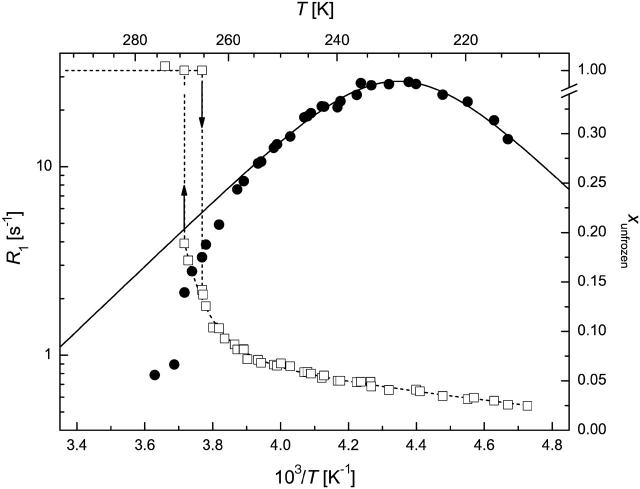
FIGURE 3.
1H spin-lattice relaxation rate (circles) and unfrozen water fraction (squares) in MAP2c solution (50 mg/ml) at ω0/2π = 44.14 MHz. (Solid line) Redfield-Slichter model fitted to R1 data; dotted lines are guides to the eye. SEs are represented by the size of the symbols.
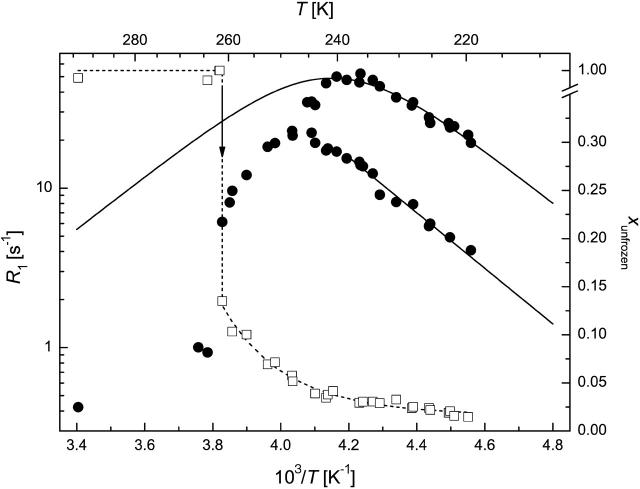
FIGURE 4.
1H spin-lattice relaxation rate (circles) and unfrozen water fraction (squares) in CSD1 solution (50 mg/ml) at ω0/2π = 44.14 MHz. (Solid line) Redfield-Slichter model fitted to R1 data; dotted lines are guides to the eye. SEs are represented by the size of the symbols.
FIGURE 5.
1H spin-lattice relaxation rate (circles) and unfrozen water fraction (squares) in ERD10 solution (25 mg/ml) at ω0/2π = 82.57 MHz. (Solid line) Redfield-Slichter model fitted to R1 data; dotted lines are guides to the eye. SEs are represented by the size of the symbols.
FIGURE 6.
1H spin-lattice relaxation rate (circles) and unfrozen water fraction (squares) in BSA solution (50 mg/ml) at ω0/2π = 44.14 MHz. (Solid line) Redfield-Slichter model fitted to R1 data; dotted lines are guides to the eye. SEs are represented by the size of the symbols.
Region c hydration water below freezing
The temperature range of TFP > T > 250 K is characterized by the amount of frozen water increasing fast at the expense of the hydrate layer, as witnessed by unusually steep ln(R1) versus 1/T curves (Figs. 3–6). The outer hydration shell of proteins can be considered in this region as a multiphase system at the first approximation, with phases of different “freezing points”. The measured spin-lattice relaxation rate R1M just below TFP is then an average value according to Eq. 2. This approximation can principally reproduce the temperature dependence of R1M, but more extensive measurements are required to make quantitative statements.
In qualitative terms, the behavior of this “loose” hydration layer of IUPs correlates with their unstructured nature. First, IUPs seem to have a spatially more extended outer hydration layer, as they lose significantly more of their bound water in this phase (between 260 and 250 K), than BSA (∼5%–7% vs. 3.5%). This may suggest that their effect on water structure and dynamics extends deeper into space than that of globular proteins. The rapid interaction of IUPs with their partners may stem from their enhanced capture radius (Pontius, 1993; Shoemaker et al., 2000; Tompa, 2002) but also from an increased electrostatic screening effect (Schreiber and Fersht, 1996) that results from their high net charge (Uversky et al., 2000). This screening effect may be related to the observed hydration pattern. The second point of note is that the behavior of CSD1 differs significantly from the other proteins studied. This deviation becomes most apparent below 240 K but is also unmistakable in this range. In effect, the hydrate layer of CSD1 is more heterogeneous than that of the other proteins, as the R1 value is worst fitted by the Redfield-Slichter model and becomes clearly two componential below 250 K.
In principle, this inhomogeneity might result simply from the heterogeneity of the protein surface, spotted with hydrophobic patches and charged clusters. This, however, would apply equally to the other IUPs studied and would not explain the difference observed. Rather, our contention is that the inhomogeneity reflects residual structure, as it competes with water binding. This is most characteristic of CSD1, shown to have a significant residual structure apparent by local structural preferences (Mucsi et al., 2003; Todd et al., 2003), suboptimal hydration (Konno et al., 1997), and also transient long-range interactions shown by circular dichroism and limited proteolysis (P. Tompa, unpublished observations). This feature appears to ensure that calpastatin is structurally primed for very rapid and effective interaction with calpain (Emori et al., 1988; Maki et al., 1989). Such a structural organization and priming for fast interaction is not required in the case of MAP2c, the major function of which is to ensure spacing in the cytoskeleton (Mukhopadhyay and Hoh, 2001) or ERD10, which is mostly implicated in water binding (Alsheikh et al., 2003; Kiyosue et al., 1994). A final note on this section is that lowering the temperature to 250 K leaves about one equivalent of hydrate water (w/w) mobile, which supports our assertion that from here on only water molecules directly in contact with the protein remain.
Region d “frozen solution”
Below 240–250 K, the amount of unfrozen water changes only slightly and behaves single phased. This region is the most informative when studying the hydration properties since this is the innermost water fraction bound to the protein. Further, the spin-lattice relaxation rates show a maximum in this low-temperature region (Figs. 3–6), and the Redfield-Slichter model can be used for the quantitative evaluation of water dynamics. By fitting Eq. 3 to the measured R1 data (Table 3), the dynamics of water molecules in the hydration shell of BSA can be characterized by significantly lower activation energy (20 kJ/mol) than that of the IUPs (32 kJ/mol). Thus, IUPs not only bind more water, they also bind the inner hydration layer much more strongly. The related correlation time constants also differ according to the intrinsically unstructured (10−16 s) or globular (10−13 s) nature of the protein, perhaps due to more rotational freedom of water molecules on the surface of IUPs than on globular proteins. This marked difference may be of physiological relevance in terms of the rapid rearrangements of hydrate layer that IUPs have to undergo, as outlined in the introduction. This is most apparent with CSD1 (Emori et al., 1988; Maki et al., 1989) but may also apply to MAP2c that binds microtubules (Matus, 1994; Sanchez et al., 2000) and ERD10, in light of its possible membrane association (Bussell and Eliezer, 2003; Koag et al., 2003). The inhomogeneity of CSD1 hydration, pointed out in the previous section, is relevant here, due to the significant residual structure and rapid binding function of this protein.
TABLE 3.
Activation energy (Ea), correlation time constant (τ0), and average local field 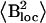 values obtained from the Redfield-Slichter model fitted to R1 data below 240–250 K
values obtained from the Redfield-Slichter model fitted to R1 data below 240–250 K
| Sample | Ea [kJ/mol] | τ0 [s] |
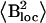 [10−8 T2] [10−8 T2] |
|---|---|---|---|
| MAP2c | 31 ± 2 | 10−15 | 32 ± 2 |
| CSD1 (rapid R1) | 32 ± 4 | 10−16 | 57 ± 4 |
| CSD1 (slow R1) | 33 ± 3 | — | — |
| ERD10 | 32 ± 3 | 10−16 | 66 ± 8 |
| BSA | 20 ± 2 | 10−13 | 23.4 ± 0.7 |
Correlation time constants could be determined to a precision of one order of magnitude.
The average local field obtained for the hydration shell of BSA (23.4 ± 0.7 × 10−8 T2) equals the value of 23.2 × 10−8 T2 calculated for a water molecule. The much greater values obtained for the hydration shells of the IUPs indicate that further relaxation mechanisms should be taken into account. This assumption is also supported by the facts that the measured R2 values are greater by one or two orders of magnitude than predicted by the Redfield-Slichter model fitted to the R1 values and that R2 is not equal to R1ρ. The predicted value of the R2/R1 ratio is 2:3 in the Redfield-Slichter and 0.6158 in the Bloembergen-Purcell-Pound model. Our results give 10–100 times larger values than the models (Table 4). It can be concluded therefore that the relaxation for nonfreezable water protons cannot be approached by a single correlation time even in this temperature region.
TABLE 4.
Spin-spin (R2) and rotating-frame spin-lattice (R1ρ) relaxation rates and correlation time values (τr, τs) obtained from the Redfield-Slichter model fitted to R1 data below 240−250 K
| MAP2c | CSD1* | ERD10 | BSA | ||
|---|---|---|---|---|---|
| ω0/2π [MHz] | 44.14 | 44.14 | 82.57 | 44.14 | |
| T(R1 = R1max) [K] | 229 ± 1 | 241 ± 1 | 243 ± 1 | 237 ± 1 | |
| R1max [s−1] | 28 ± 1 | 50 ± 2 | 57 ± 3 | 19 ± 1 | |
| R1ρ [103 s−1] | 0.17 ± 0.03 | 0.8 ± 0.1 | 0.11 ± 0.02 | 0.7 ± 0.1 | 0.22 ± 0.03 |
| R2 [103 s−1] | 0.34 ± 0.03 | 1.5 ± 0.2 | 0.58 ± 0.06 | ≈ R1ρ | 0.60 ± 0.06 |
| R2/R1 | 12 ± 2 | 53 ± 8 | 12 ± 2 | 7 ± 2 | 32 ± 5 |
| τr [10−9 s] | 2.2204 ± 0.0004 | 2.2204 ± 0.0004 | 1.1870 ± 0.0002 | 2.2204 ± 0.0004 | |
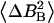 [10−8 T2] [10−8 T2] |
11.5 ± 0.6 | 20 ± 1 | 44 ± 2 | 7.7 ± 0.4 | |
| τs [10−7 s] | 9 ± 2 | 7 ± 1 | 30 ± 3 | — | 11 ± 1 |
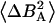 [10−8 T2] [10−8 T2] |
0.47 ± 0.08 | 3.0 ± 0.5 | 0.24 ± 0.03 | — | 0.8 ± 0.1 |
Rapid R1 branch.
CONCLUSIONS
We have demonstrated that quantitative information on the hydration of proteins can be obtained by solid-state relaxation NMR. The method provides data on both the amount and dynamics of bound water, the heterogeneity of which is unveiled by progressive freezing out. The technique enables the characterization of IUPs, showing their significantly larger hydration than globular proteins. The activation energy obtained for the dynamics of the most strongly bound part of the hydration shell is 50% larger for IUPs than for a globular protein. The correlation time constants also markedly differ by orders of magnitude between the two types of proteins. To get additional information and more precise data on the outer parts of the hydration shell and on its behavior at ambient temperatures, more extensive measurements are needed at T > 240 K.
Acknowledgments
This work was supported by the Hungarian Scientific Research Fund (OTKA; grant T34255) and by the International Senior Research Fellowship GR067595 from the Wellcome Trust. Monika Bokor and Peter Tompa acknowledge the support of the Bolyai János Scholarship.
References
- Alsheikh, M. K., B. J. Heyen, and S. K. Randall. 2003. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 278:40882–40889. [DOI] [PubMed] [Google Scholar]
- Antzutkin, O. N. 2002. Molecular structure determination: applications in biology. In Solid-State NMR Spectroscopy. M. J. Duer, editor. Blackwell Science, Oxford.
- Barnaal, D. E., and I. J. Lowe. 1967. Experimental free-induction-decay shapes and theoretical second moments for hydrogen in hexagonal ice. J. Chem. Phys. 46:4800–4809. [Google Scholar]
- Bloembergen, N., E. M. Purcell, and R. V. Pound. 1948. Relaxation effects in nuclear magnetic resonance absorption. Phys. Rev. 73:679–712. [Google Scholar]
- Bochicchio, B., and A. M. Tamburro. 2002. Polyproline II structure in proteins: identification by chiroptical spectroscopies, stability, and functions. Chirality. 14:782–792. [DOI] [PubMed] [Google Scholar]
- Bussell, R. Jr., and D. Eliezer. 2003. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329:763–778. [DOI] [PubMed] [Google Scholar]
- Chen, J., Y. Kanai, N. J. Cowan, and N. Hirokawa. 1992. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 360:674–677. [DOI] [PubMed] [Google Scholar]
- Demchenko, A. P. 2001. Recognition between flexible protein molecules: induced and assisted folding. J. Mol. Recognit. 14:42–61. [DOI] [PubMed] [Google Scholar]
- Dunker, A. K., C. J. Brown, J. D. Lawson, L. M. Iakoucheva, and Z. Obradovic. 2002. Intrinsic disorder and protein function. Biochemistry. 41:6573–6582. [DOI] [PubMed] [Google Scholar]
- Dyson, H. J., and P. E. Wright. 2002. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12:54–60. [DOI] [PubMed] [Google Scholar]
- Dyson, H. J., and P. E. Wright. 2004. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 104:3607–3622. [DOI] [PubMed] [Google Scholar]
- Emori, Y., H. Kawasaki, S. Imajoh, Y. Minami, and K. Suzuki. 1988. All four repeating domains of the endogenous inhibitor for calcium-dependent protease independently retain inhibitory activity. Expression of the cDNA fragments in Escherichia coli. J. Biol. Chem. 263:2364–2370. [PubMed] [Google Scholar]
- Ferralli, J., T. Doll, and A. Matus. 1994. Sequence analysis of MAP2 function in living cells. J. Cell Sci. 107:3115–3125. [DOI] [PubMed] [Google Scholar]
- Fuxreiter, M., I. Simon, P. Friedrich, and P. Tompa. 2004. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338:1015–1026. [DOI] [PubMed] [Google Scholar]
- Goyal, K., L. Tisi, A. Basran, J. Browne, A. Burnell, J. Zurdo, and A. Tunnacliffe. 2003. Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J. Biol. Chem. 278:12977–12984. [DOI] [PubMed] [Google Scholar]
- Iakoucheva, L., C. Brown, J. Lawson, Z. Obradovic, and A. Dunker. 2002. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323:573–584. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., K. Yamaguchi-Shinozaki, and K. Shinozaki. 1994. Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol. 35:225–231. [PubMed] [Google Scholar]
- Koag, M. C., R. D. Fenton, S. Wilkens, and T. J. Close. 2003. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 131:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno, T., N. Tanaka, M. Kataoka, E. Takano, and M. Maki. 1997. A circular dichroism study of preferential hydration and alcohol effects on a denatured protein, pig calpastatin domain I. Biochim. Biophys. Acta. 1342:73–82. [DOI] [PubMed] [Google Scholar]
- Leulliot, N., and G. Varani. 2001. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 40:7947–7956. [DOI] [PubMed] [Google Scholar]
- Lisse, T., D. Bartels, H. R. Kalbitzer, and R. Jaenicke. 1996. The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineum displays no defined three-dimensional structure in its native state. Biol. Chem. 377:555–561. [DOI] [PubMed] [Google Scholar]
- Maki, M., H. Bagci, K. Hamaguchi, M. Ueda, T. Murachi, and M. Hatanaka. 1989. Inhibition of calpain by a synthetic oligopeptide corresponding to an exon of the human calpastatin gene. J. Biol. Chem. 264:18866–18869. [PubMed] [Google Scholar]
- Matus, A. 1994. MAP2. In Microtubules. J. S. Hyams and V. W. Lloyd, editors. Wiley-Liss, New York. 155–166.
- Mucsi, Z., F. Hudecz, M. Hollosi, P. Tompa, and P. Friedrich. 2003. Binding-induced folding transitions in calpastatin subdomains A and C. Protein Sci. 12:2327–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, R., and J. H. Hoh. 2001. AFM force measurements on microtubule-associated proteins: the projection domain exerts a long-range repulsive force. FEBS Lett. 505:374–378. [DOI] [PubMed] [Google Scholar]
- Noack, F. 1971. Nuclear Magnetic Relaxation Spectroscopy. NMR Basic Principles and Progress. Springer Verlag, Berlin .
- Otting, G. 1997. NMR studies of water bound to biological molecules. Progr. NMR Spectr. 31:259–285. [Google Scholar]
- Pontius, B. W. 1993. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem. Sci. 18:181–186. [DOI] [PubMed] [Google Scholar]
- Racz, P., K. Tompa, I. Pocsik, and P. Banki. 1983. Water fractions in normal and senile cataractous eye lenses studied by NMR. Exp. Eye Res. 36:663–669. [DOI] [PubMed] [Google Scholar]
- Sanchez, C., J. Diaz-Nido, and J. Avila. 2000. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 61:133–168. [DOI] [PubMed] [Google Scholar]
- Schreiber, G., and A. R. Fersht. 1996. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 3:427–431. [DOI] [PubMed] [Google Scholar]
- Shoemaker, B. A., J. J. Portman, and P. G. Wolynes. 2000. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. USA. 97:8868–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slichter, C. P. 1990. Principles of Magnetic Resonance. Springer Verlag, Berlin .
- Syme, C. D., E. W. Blanch, C. Holt, R. Jakes, M. Goedert, L. Hecht, and L. D. Barron. 2002. A Raman optical activity study of rheomorphism in caseins, synucleins and tau. Eur. J. Biochem. 269:148–156. [DOI] [PubMed] [Google Scholar]
- Todd, B., D. Moore, C. C. Deivanayagam, G. D. Lin, D. Chattopadhyay, M. Maki, K. K. Wang, and S. V. Narayana. 2003. A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor. J. Mol. Biol. 328:131–146. [DOI] [PubMed] [Google Scholar]
- Tompa, P. 2002. Intrinsically unstructured proteins. Trends Biochem. Sci. 27:527–533. [DOI] [PubMed] [Google Scholar]
- Tompa, P. 2003. The functional benefits of protein disorder. J. Mol. Struct. THEOCHEM. 666– 667:361–371. [Google Scholar]
- Tompa, K., P. Banki, M. Bokor, and G. Lasanda. 2001. Hydrogen spectroscopy of Pd0.9Ag0.1-H alloys on NMR scales. Europhys. Lett. 53:79–85. [Google Scholar]
- Tompa, K., P. Banki, M. Bokor, G. Lasanda, and L. Vasaros. 2003. Diffusible and residual hydrogen in amorphous Ni(Cu)-Zr-H alloys. J. Alloys Comp. 350:52–55. [Google Scholar]
- Tompa, P., and P. Csermely. 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 18:1169–1175. [DOI] [PubMed] [Google Scholar]
- Uversky, V. N. 2002a. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11:739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky, V. N. 2002b. What does it mean to be natively unfolded? Eur. J. Biochem. 269:2–12. [DOI] [PubMed] [Google Scholar]
- Uversky, V. N., J. R. Gillespie, and A. L. Fink. 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 4:415–427. [DOI] [PubMed] [Google Scholar]
- Uversky, V. N., S. Winter, O. V. Galzitskaya, L. Kittler, and G. Lober. 1998. Hyperphosphorylation induces structural modification of tau-protein. FEBS Lett. 439:21–25. [DOI] [PubMed] [Google Scholar]
- Ward, J. J., J. S. Sodhi, L. J. McGuffin, B. F. Buxton, and D. T. Jones. 2004. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337:635–645. [DOI] [PubMed] [Google Scholar]
- Wider, G. 1998. Technical aspects of NMR spectroscopy with biological macromolecules and studies of hydration in solution. Progr. NMR Spectr. 32:193–275. [Google Scholar]
- Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, P. E., and H. J. Dyson. 1999. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293:321–331. [DOI] [PubMed] [Google Scholar]
- Yang, H. Q., H. Ma, E. Takano, M. Hatanaka, and M. Maki. 1994. Analysis of calcium-dependent interaction between amino-terminal conserved region of calpastatin functional domain and calmodulin-like domain of mu-calpain large subunit. J. Biol. Chem. 269:18977–18984. [PubMed] [Google Scholar]
- Zimmerman, J. R., and W. E. Brittin. 1957. Nuclear magnetic resonance studies in multiple phase systems: lifetime of a water molecule in an adsorbing phase on silica gel. J. Phys. Chem. 61:1328–1333. [Google Scholar]