Abstract
Negative cofactor 2 (NC2) is an evolutionarily conserved transcriptional regulator that was originally identified as an inhibitor of basal transcription. Its inhibitory mechanism has been extensively characterized; NC2 binds to the TATA-binding protein (TBP), blocking the recruitment of TFIIA and TFIIB, and thereby inhibiting preinitiation complex assembly. NC2 is also required for expression of many yeast genes in vivo and stimulates TATA-less transcription in a Drosophila in vitro transcription system, but the mechanism responsible for the NC2-mediated stimulation of transcription is not understood. Here we establish that yeast NC2 can directly stimulate activated transcription from TATA-driven promoters both in vivo and in vitro, and moreover that this positive role requires the same surface of TBP that mediates the NC2 repression activity. On the basis of these results, we propose a model to explain how NC2 can mediate both repression and activation through the same surface of TBP.
Keywords: BUR6‖yeast
The binding of TATA-binding protein (TBP) to the TATA box is the first step in the assembly of the RNA polymerase II preinitiation complex at the promoter, triggering the subsequent recruitment of the remaining general transcription factors and polymerase II (1). As the first step in promoter-specific transcription, TBP recruitment is a frequent target for regulation. Several proteins that stimulate formation of the TBP-TATA complex have been identified (2), including promoter-bound activators, TFIIA, and IIB (3–7), but TBP-TATA complex formation is also inhibited through distinct mechanisms by histones (8), Mot1 (9), dTAFII230 (10), and TBP dimerization (11).
TBP is also regulated after it is bound to the TATA box, by negative cofactor 2 (NC2). NC2 was initially identified as an activity in human nuclear extracts that binds to TBP, inhibiting the recruitment of TFIIA and TFIIB (12), and thus the assembly of the preinitiation complex. NC2 consists of two subunits (13–15) that are highly conserved in eukaryotes; in Saccharomyces cerevisiae the NC2α subunit is encoded by BUR6/NCB1, and the NC2β subunit by YDR1/NCB2 (15–18). Consistent with its biochemical characterization, the genes encoding both NC2 subunits were identified by genetic selections that were expected to reveal general repressors of transcription. Mutations in BUR6 increase transcription from a UAS-less SUC2 promoter (19), whereas both bur6 and ydr1 mutations were found to suppress mutation in SRB4, which encodes a subunit of the polymerase II holoenzyme (17, 20).
NC2 function in vivo is likely to be more complex than implied by this simple model, because bur6 mutations reduce transcription from many promoters in yeast (16, 21) and ydr1 mutations reduce transcription from the HIS3 and HIS4 TATA-less promoters (22). These observations suggested that NC2 can both inhibit and stimulate transcription in vivo, although it was not known whether its positive role was direct or indirect. NC2 was subsequently purified from a Drosophila nuclear extract as an activity required for transcription from TATA-less promoters containing a downstream promoter element (23), providing biochemical evidence that NC2 can directly stimulate transcription under certain circumstances. The relevance of this finding for the more typical TATA-driven promoters remains unclear, however, because that same study detected no stimulatory effect of NC2 on TATA-containing promoters, and downstream promoter elements have been extensively characterized only in Drosophila (24, 25). Another indication that the positive role of NC2 might be direct came from chromatin immunoprecipitation studies showing that the Bur6 (21) and Ydr1 (26) subunits of NC2 are associated with active promoters and that Bur6 is recruited to those promoters under inducing conditions. These chromatin immunoprecipitation studies provide a correlation between the presence of both NC2 subunits at active promoters, but do not address the mechanistic role of NC2 during activation. The target of NC2 for stimulating transcription remains unknown, and detailed studies of the mechanism of NC2-mediated stimulation at TATA-containing promoters would be greatly facilitated by an in vitro system that accurately reproduces all of its in vivo characteristics.
Here we investigate the mechanistic basis for the positive role of NC2 during transcription. We found that NC2 directly stimulates activator-dependent transcription from TATA-driven promoters both in vivo and in vitro, and that its stimulatory effect is mediated by contacts with TBP. These results lead to a model to explain the dual regulation of TBP by NC2.
Materials and Methods
Strains and Plasmids.
For the altered specificity experiments the GAL1 TATA box was mutated to TGTA by oligo-directed mutagenesis of pYC60, which contains a 1.9-kb GAL1-GAL10 EcoRI fragment in pRS406. The genomic GAL1 promoter in strain GY281 (MATα ura3–52 trp1Δ63) was then replaced with the mutant TGTA-GAL1 allele by standard two-step transplacement procedures, creating the reporter strain YY96. The TGTA-HIS3 strain has been described (27). NC2 binding-defective TBP m3 mutants m3–601 (pYC58) and m3–644 (pYC59) were created by incorporating the spt15–601 (F182V) and spt15–644 (F82V H179Q) mutations into plasmid m3IIDy/y (CEN spt15-m3 URA3) (27) by oligo-directed mutagenesis of TBP m3 and were confirmed by DNA sequencing.
Whole-Genome Transcriptional Analysis.
Strains GY473 (BUR+), YY83 (bur6–1), and YY94 (spt15–601) were grown in 200 ml of yeast extract/peptone/dextrose at 30°C to 1 × 107 cells per ml. RNA was isolated as described (28). Poly(A)+ selection, cDNA synthesis, in vitro transcription, and hybridizations were all performed essentially as described (29). Expression analysis was performed by using Affymetrix (Santa Clara, CA) genechip software. For details on the whole-genome expression results and data analysis, see http://steelhead.aecom.yu.edu/expression.html.
RNA Analysis.
Northern blots were performed by using RNA isolated as described (16) from cells grown under the following conditions. For GAL1-GAL10 induction, cells were grown in synthetic complete (SC) medium containing 2% raffinose, followed by 3 h of induction in SC medium containing 5% galactose. For HSP12 and CUP1, cells were grown in glucose minimal (SD) medium at 24°C and subjected to either a 30-min heat shock at 39°C (HSP12) or a 20-min treatment with 1 mM cupric sulfate (CUP1). For PHO5 induction, cells were grown in SD medium containing 7.5 mM or 0.1 mM phosphate. For HO induction, cells were grown in SD medium. The probe for GAL1 and GAL10 analysis was a 1.9-kb EcoRI fragment from p4812. The probes for all other transcripts were PCR fragments amplified from the coding regions of the respective genes.
In Vitro Transcription Reactions.
Whole-cell extract preparation from BUR6+ (GY480) and bur6–1 (GY565) strains and transcription assays were performed essentially as described (30, 31) with minor modifications. Recombinant NC2 (1:1 molar ratio of recombinant His-6-Bur6 and His-6-Ydr1) (32) was preincubated with whole-cell extract on ice for 10 min. A premix containing template DNA pGAL4CG- in the presence or absence of 100 ng of purified Gal4-VP16 (gift from V. Palham, The Rockefeller University, New York) was incubated on ice for 5 min before addition into the NC2/extract mixture. After 5 min at room temperature, reactions were started by adding ATP, CTP, [α-32P]UTP, and phosphoenolpyruvate and incubated at 25°C for 30 min; the products were examined by using 5% urea-polyacrylamide gels and autoradiography.
β-Galactosidase Assays.
BUR6+ and bur6–1 strains were transformed with a LacZ reporter driven by a LexA binding site (pYEp21-SC3423) (33) and a plasmid expressing either the LexA DNA-binding domain or the LexA DNA-binding domain fused to the activation domains of Hap4, Gal4, and Gcn4 (34). β-Galactosidase assays were performed as described (16).
Results
Similar Transcriptional Defects in bur6 and TBP NC2-Defective Mutants.
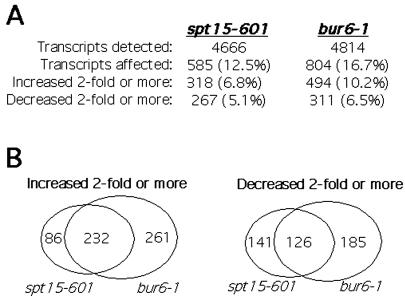
Recent genomewide expression analysis with a bur6 temperature-sensitive strain revealed effects on 17% of yeast transcripts, with a roughly equivalent number of transcripts increasing as decreasing (21). To determine whether the observed changes are caused by the loss of NC2–TBP interactions, we performed similar whole-genome analysis to examine transcript levels in bur6–1 and spt15–601 strains. The bur6–1 allele was isolated in the original Bur selection as a mutation that increased transcription from the suc2Δuas UAS-less promoter (19), whereas the spt15–601 allele is representative of a cluster of TBP mutants that are defective for binding NC2, but still bind DNA, TFIIA, TFIIB, and Mot1 (32). In broad agreement with the results of Geisberg et al. (21) with a conditional bur6 mutant, 17% of the detected transcripts were affected in our bur6–1 strain. Not surprisingly, on the basis of their relative growth rates, slightly less (12%) of the transcripts were affected in the spt15–601 missense mutant (Fig. 1A). The transcripts affected by bur6–1 and spt15–601 mutations display considerable overlap, because 61% of the transcripts affected 2-fold or more in the spt15–601 strain were also affected 2-fold or more in the bur6–1 strain (Fig. 1B); by using a more restrictive 3-fold cutoff, 67% of the affected transcripts were shared between the bur6–1 and spt15–601 strains, indicating that the extensive overlap is independent of the chosen cutoff point. By comparison, only 19% of the transcripts affected at least 3-fold in the spt15–601 strain are also affected in a bur1 mutant strain (data not shown). BUR1 encodes a protein kinase that was identified by the same selection as BUR6, yet affects transcription by a different mechanism, mediated through the RNA polymerase II carboxyl-terminal domain. The extensive overlap between bur6–1 and spt15–601 suggests a shared mechanistic defect, namely the inability of NC2 to bind to TBP. Seventy-three percent of the transcripts with decreased expression in the spt15–601 strain also decreased expression more than 2-fold in the bur6–1 strain (Fig. 1B), indicating that most of the transcripts that require BUR6 for activation also require the NC2-interacting surface of TBP. Although these results do not distinguish direct from indirect effects, they provide a valuable catalog of the transcripts that are most sensitive to disruption of the NC2–TBP interaction and implicate the NC2–TBP interaction in stimulating transcription from a significant number of promoters in vivo.
Figure 1.
Comparison of transcriptional effects in bur6–1 and spt15–601 strains. Whole-genome expression analysis was performed by using Affymetrix YE6100 arrays on RNA prepared from strains containing bur6–1 or spt15–601 mutations. BUR6 encodes the yeast NC2 α subunit, whereas the spt15–601 TBP mutation is defective for binding NC2. (A) The number of transcripts that are detected in the spt15–601 and bur6–1 mutant strain and the number of transcripts that are affected, increase, or decrease 2-fold or greater relative to a wild-type strain are presented. The percent of transcripts that are affected, increase, or decrease relative to wild type are shown in parentheses. (B) The extent of overlap between the affected transcripts in the bur6–1 and spt15–601 strains are depicted diagrammatically, with the overlap between transcripts that increase shown on the left, and overlap between transcripts that decrease shown on the right.
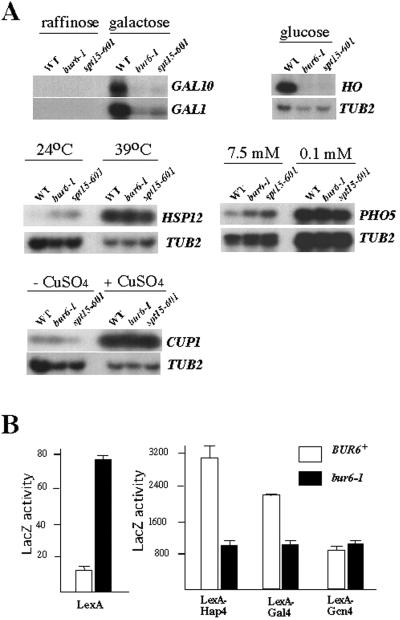
The whole-genome analysis approach determines transcript levels under a single growth condition; to examine whether the NC2–TBP interaction affects levels of specific transcripts under both induced and uninduced conditions, several representative well-studied genes were chosen and their RNA levels analyzed by Northern blotting (Fig. 2A). Transcriptional induction of GAL1, GAL10, and HO was impaired in both bur6–1 and spt15–601 strains, but uninduced GAL1 and GAL10 mRNA levels were essentially unchanged. The identical results observed by using bur6–1 and spt15–601 strains strongly suggest that binding of NC2 to TBP is required to stimulate transcription from these promoters. By contrast, induced levels of HSP12 and PHO5 mRNA were unaffected in bur6–1 and spt15–601 strains, whereas their basal expression was increased. Other transcripts, such as CUP1 and TUB2, were unaffected in either strain. BUR6 therefore has complex, promoter-specific roles, selectively repressing basal transcription from some promoters and stimulating activated transcription from others, and both effects require the TBP–NC2 interaction surface.
Figure 2.
Promoter- and activator-specific requirements for BUR6. (A) Northern blot analysis of selected transcripts from wild-type (WT), bur6–1, and spt15–601 strains grown under uninduced and induced conditions as indicated. (B) Activation domain-specific requirement for BUR6 with LexA-activation domain fusions. BUR6+ and bur6–1 strains containing a LacZ reporter driven by a LexA-binding site and a plasmid expressing either the LexA DNA-binding domain or the LexA DNA-binding domain fused to the activation domains of Hap4, Gal4, and Gcn4. β-Galactosidase assays were performed in triplicate, with the average LacZ activity and standard deviations shown.
Activator-Dependent Requirement for BUR6.
To investigate the basis for the promoter-specific role of NC2 on activated transcription, we tested whether different activation domains were equally dependent on BUR6 function (Fig. 2B). Expression from a LexAop-CYC1p-LacZ reporter driven by the LexA-Hap4 and LexA-Gal4 activators was reduced 3- and 2-fold, respectively, in a bur6–1 mutant, whereas the same reporter driven by LexA-Gcn4 was unaffected. This result agrees with our finding that the GAL1 UAS conferred BUR6 dependence (16), and more specifically demonstrates that the activation domain contributes to BUR6 dependence. By contrast, expression of the same reporter in the absence of an activation domain (the LexA DNA-binding domain alone) increases 6-fold in the bur6 mutant, as expected on the basis of the previous characterization of BUR6 as a repressor of UAS-less promoters (16, 32).
Direct Requirement for NC2–TBP Interaction for Activation in Vivo.
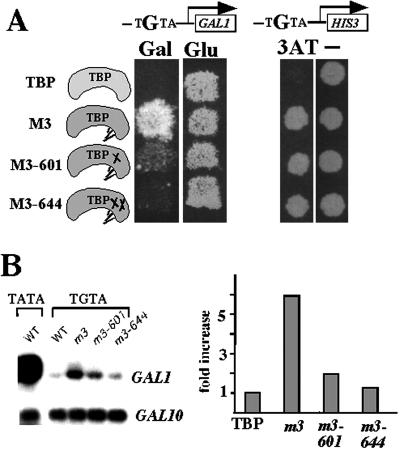
We have previously shown that the bur6–1 mutation and TBP mutants such as spt15–601 that are unable to bind NC2 confer defects in GAL1 induction. To determine whether the positive role of NC2 at the GAL1 promoter in vivo is direct, we took advantage of an altered DNA-binding specificity mutant of TBP (27) that had been used to examine direct TBP interactions in vivo (35, 36). The TBP m3 mutant recognizes the sequence TGTAAA in addition to the canonical TATAAA element (27). Replacement of the genomic GAL1 TATA box with TGTA greatly reduced GAL1 induction, resulting in a Gal− phenotype, and as expected, introduction of TBP m3 into this strain on a low-copy-number (CEN) plasmid partially restored growth on galactose-containing medium (Fig. 3A) and increased transcription from the TGTA-GAL1 promoter (Fig. 3B). To determine whether the NC2–TBP interaction is directly required for activation, we then constructed two TBP m3 derivatives containing single (m3–601) or double (m3–644) mutations on the TBP surface that reduce binding of NC2 (32). Because TBP m3 is unable to support viability in the absence of wild-type TBP, TBP m3 and its NC2-defective mutants were tested for activity in a strain that also expressed wild-type TBP from its normal chromosomal location. In this genetic background, transcription from TGTA-GAL1 is directly regulated by TBP m3 or its NC2-defective derivatives, whereas other genes are still transcribed normally by the wild-type TBP; therefore, any effects observed on TGTA-GAL1 transcription in the TBP m3 NC2-defective mutants are likely to result directly from loss of NC2 binding. TGTA-GAL1 transcription decreased in the TBP m3 mutants unable to bind NC2, consistent with a direct role for the NC2–TBP interaction in stimulating GAL1 transcription (Fig. 3B). The reduced transcription from TGTA-GAL1 in the m3 NC2-defective mutant strain was not due to a general activation defect or a defect in GAL gene induction, because the adjacent GAL10 gene, which still contains its TATA box and shares the same UAS with GAL1, was transcribed normally. In contrast, both TBP m3 and the NC2-defective derivatives supported growth of a yeast strain containing a TGTA-HIS3 allele (27) on medium lacking histidine and containing 3-aminotriazole (Fig. 3A). This result is consistent with other results that BUR6 (Fig. 2B) and YDR1 (22) are not required for Gcn4-dependent transcriptional activation and suggests that introduction of these surface mutations into the m3 background did not reduce TGTA-GAL1 transcription simply by impairing recognition of the TGTA box. These results constitute strong genetic evidence that the direct positive role of NC2 at some promoters is mediated through contacts with TBP.
Figure 3.
Direct requirement of NC2 binding to TBP for activated transcription in vivo. (A) Gal− phenotype of TBP mutants defective for binding NC2. The TATA box of the endogenous GAL1 and HIS3 genes were replaced by TGTA, and the resulting strains (TGTA-GAL1 = YY96; TGTA-HIS3 = KY577) were then transformed with plasmids expressing wild-type TBP, TBP m3, or two TBP m3 derivatives that are defective for binding NC2 (32). Transformants were grown on either SC plates containing galactose (Gal) or glucose (Glu) or SD plates in the presence or absence (−) of 0.1 mM 3-aminotriazole (3AT). (B) Northern blot analysis (Left) and quantitation (Right) of induced GAL1 and GAL10 transcription from the strains described in A.
Stimulation of Activated Transcription by NC2 in Vitro.
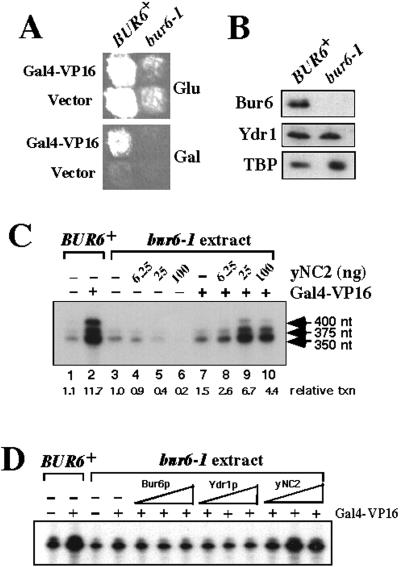
If NC2 directly stimulates transcription, then activation defects should be detectable in an in vitro transcription system that lacks NC2. A previous attempt at immunodepletion of NC2 by using a Ydr1 antibody column resulted in an activation defect, but that defect was due to codepletion of other general factors, including RNA polymerase II (37). To eliminate depletion of other factors, an NC2-deficient whole-cell extract was prepared from a bur6–1 strain, which produces undetectable amounts of Bur6 protein, at least 50 times lower than the level of expression in a BUR6+ strain. In contrast, the levels of other transcription factors tested, including Ydr1 and TBP were essentially unchanged relative to the BUR6+ extract (Fig. 4B). When assayed by using a promoter containing a Gal4-binding site and the CYC1 TATA box upstream from a G-less cassette, basal transcription with the bur6–1 extract was inhibited by addition of purified recombinant NC2 (Fig. 4C, lanes 3–6), demonstrating that the extract was NC2-deficient and that the recombinant NC2 was active for repression. This finding is consistent with the initial characterization of NC2 as a basal repressor and the genetic characterization of BUR6 as a repressor in vivo. In the presence of Gal4-VP16, strong activation was obtained by using extracts prepared from a BUR6+ strain (Fig. 4C, lanes 1 and 2), whereas only 1.5-fold activation was obtained with the bur6–1 extract (Fig. 4C, lanes 3 vs. 7). The reduced activation in the bur6–1 extract was due to the absence of NC2 activity, because addition of recombinant NC2 stimulated Gal4-VP16-dependent activation 4-fold more, equivalent to the level of activation obtained by using the Bur6+ extract (Fig. 4C, compare lanes 1 and 2 with lanes 5 and 9). Restoration of activation was due to NC2 activity, and not some contaminant or nonspecific mechanism, because both of the purified recombinant Bur6 or Ydr1 subunits were required for stimulatory activity (Fig. 4D). The amount of NC2 that stimulates transcription in these reactions approximates the amount of NC2 present in the wild-type extract; when added at concentrations exceeding that found in wild-type extracts (Fig. 4C, lane 10), NC2 began to repress activated transcription, as reported (38). Addition of recombinant TFIIA, by contrast, had no stimulatory effect on the bur6–1 extract (data not shown), indicating that TFIIA is not able to compensate functionally for the absence of NC2. The results observed with Gal4-VP16 in vitro accurately reproduce in vivo effects, because activation by both the endogenous Gal4 (16) and Gal4-VP16 (Fig. 4A) require BUR6+ function in vivo. Finally, addition of NC2 to the wild-type extract had no effect on activation, indicating that saturating amounts of NC2 are present and that stimulation is not caused by a nonspecific mechanism. Purified recombinant NC2 thus inhibited basal transcription and stimulated Gal4-VP16-dependent activation in bur6 mutant extracts, precisely reproducing the results observed in vivo and demonstrating that NC2 can have a direct role in stimulating activated transcription.
Figure 4.
NC2 inhibits basal transcription and stimulates activated transcription in vitro. (A) BUR6+ gal4Δ and bur6–1 gal4Δ strains transformed with either Gal4-VP16 or vector CEN plasmids were replica plated to SC-Leu glucose (Glu) or SC-Leu galactose (Gal) plates. As observed previously with GAL4, GAL4-VP16 is defective for activation in the bur6–1 strain. (B) Western blot detection of Bur6, Ydr1, and TBP levels in whole-cell extracts prepared from BUR6+ and bur6–1 strains. (C) In vitro transcription assays with wild-type (lanes 1 and 2) or bur6–1 (lanes 3–10) whole-cell extract with purified Gal4-VP16 and yNC2 proteins added as indicated at the top. Transcription levels are indicated at the bottom, relative to basal transcription from the bur6–1 extract. (D) Reactions were assembled as in C, and either 1, 4, or 16 ng of recombinant Bur6, Ydr1, or Bur6 + Ydr1 (NC2) were added as shown.
Discussion
Many gene-specific transcriptional regulatory proteins that were originally characterized as repressors were subsequently found to have additional roles as activators (and vice versa), with the phage λcI protein serving as a prototype (39). Dual positive and negative roles have also been observed for factors that have more general roles as transcriptional regulators. Mutations in the genes encoding histones and Mot1, for example, cause some transcripts to decrease (16, 40, 41), contrary to their original characterizations as repressors, whereas a snf5 mutation causes more transcripts to increase than decrease, contrary to its originally perceived role in facilitating activation (42, 43). The challenge is to determine which of the transcriptional effects are direct and which are indirect, and to understand the mechanisms that result in both activation and repression.
Stimulatory effects on transcription have recently been attributed to NC2 (16, 21–23, 37), in contrast to its initial characterization as a basal repressor. In particular, the detection of stimulatory activity of NC2 in vitro on Drosophila TATA-less promoters (23) and the finding that NC2 is present at transcriptionally active promoters in vivo (21) are strong indications that NC2 has direct positive roles during transcription. Although the mechanism of repression by NC2 has been well characterized, much less is known regarding the mechanism responsible for its stimulatory role. The results presented here substantially expand our current understanding of the positive roles of this essential transcription factor. First, whole-genome analysis revealed that the Bur6 subunit of NC2 and the NC2-binding domain of TBP are required for both the stimulatory and inhibitory functions of NC2 in vivo. Second, by using an altered-specificity TBP mutant that is defective for binding NC2, we found that the NC2–TBP interaction is directly required at an affected promoter. Combined, the results from these two approaches indicate that the positive effects of NC2 are mediated through the same TBP domain that is required for its repression activity. Third, NC2 can repress basal transcription and stimulate activation from TATA-containing promoters by the model transactivator Gal4-VP16 in vitro. Transcriptional stimulation and inhibition by NC2 in this in vitro system requires both subunits and is observed by using amounts of NC2 that approximate its in vivo level. Most importantly, both the stimulatory and inhibitory effects of NC2 observed in this naturally depleted in vitro system faithfully reflect both of its characterized in vivo functions. This system will be invaluable for further studies on the stimulatory role of NC2 and should provide an interesting counterpoint to the Drosophila NC2-dependent TATA-less transcription system (23).
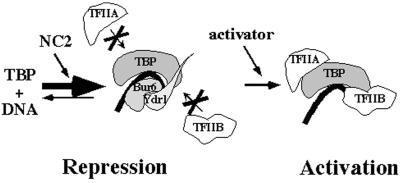
These results expand on an emerging body of data on the roles of NC2, leading us to propose a model to explain how the same TBP-NC2 contacts can mediate both activation and repression (Fig. 5). The model comprises two steps. In the first step NC2 stimulates TBP binding to the TATA box through formation of a TBP-NC2-DNA complex. Stimulation of TBP-TATA binding by NC2 has been observed (12–14, 32, 44), although it has not been extensively characterized or quantitated. Our equilibrium-binding studies using a gel mobility-shift assay revealed approximately 30-fold stimulation of TBP binding to the adenovirus major late promoter by purified NC2 (data not shown). This TBP-NC2-DNA complex is transcriptionally inactive, because NC2 sterically blocks the recruitment of TFIIA and TFIIB (12, 32, 45, 46). The basis for simultaneous inhibition of TFIIA and TFIIB was provided by the recent TBP-NC2-DNA crystal structure, with NC2 binding to the underside of the TBP-DNA surface, blocking contacts of TFIIA and TFIIB with TBP (46). This inactive NC2-TBP intermediate must be overcome in an activator-dependent fashion in the second step, allowing TFIIA and TFIIB recruitment before transcription initiation can proceed. Existing evidence suggests that the ability to counteract NC2-mediated repression is limited to specific activation domains; the activation domains of VP16 and E1A were able to overcome repression caused by overexpression of NC2 in human cells, whereas those of Sp1 and CTF were not (47), and the adenovirus E1A activation domain was able to dissociate physically the NC2-TBP complex in vitro (48). The basis for the activator specificity and its exact mechanism are not known, but these published data are consistent with our LexA-activation domain fusion results suggesting that activation domains are a major determinant of NC2-dependent stimulation. Other determinants are likely to influence NC2 responsiveness further, such as the presence of a consensus TATA box, local chromatin accessibility, and other factors that independently influence TBP occupancy. In summary, in our model NC2 inhibits TBP interactions with TFIIA and TFIIB as originally described, whereas its activation function is mediated by stimulating TBP binding. We therefore consider the NC2-TBP-DNA complex as an intermediate state that can have net positive or negative effects, depending on the presence or absence of an appropriate activator.
Figure 5.
A two-step model for NC2 as a repressor of basal transcription and stimulator of activated transcription. NC2 stimulates the binding of TBP to DNA in the first step, yet its continued presence inhibits subsequent recruitment of TFIIA and TFIIB. Activators then counteract NC2 either directly or through co-activator intermediates, resulting in greater net stimulation by the combination of NC2 and activator due to the enhanced binding of TBP in the initial step. Activators can either physically remove NC2 from the TBP complex as shown, or inactivate its repression activity while allowing NC2 to remain bound to TBP.
The functions of NC2 need to be coordinated and balanced with those of other global factors that might counteract or overlap with NC2. The inhibitory function of NC2, for example, partially overlaps with that of Mot1, which also has direct inhibitory and stimulatory roles in vivo (32). Similarly, the positive role of NC2 might partially overlap with that of TFIIA, because TFIIA also stabilizes the TBP-TATA complex (3–5), it can overcome NC2-mediated repression at the AdML and E4 promoters in vitro (44), and a mutation in the TOA1 subunit of TFIIA can suppress the inviability caused by bur6Δ or ydr1Δ mutations (49). The positive functions of TFIIA and NC2 in vivo are not completely redundant, however, because addition of purified recombinant TFIIA cannot restore activation in the bur6 mutant extract (data not shown). This finding was not surprising, because the Gal− phenotype caused by bur6–1 or spt15–601 mutations is suppressed by the combined overexpression of the BUR6 and YDR1 NC2 subunits, but not by overexpression of TFIIA subunits (32). In fact, TFIIA overexpression exacerbates the bur6–1 and spt15–601 Gal− phenotypes and causes those strains to grow extremely slowly (data not shown). Finally, the activity of still other general regulatory factors is likely to overlap with NC2, because loss of NC2 activity can be compensated for by mutations in the SIN4 holoenzyme subunit (22, 50), and mutations in either BUR6 or YDR1 suppress mutations in the SRB4 subunit of holoenzyme (17, 20).
Our model predicts that promoters with weak TATA boxes or that are limited for TBP occupancy by nucleosomes, Mot1, or other inhibitory factors will depend more on NC2 for activation. In support of this idea, NC2 is required for transcription from TATA-less promoters in vivo in yeast (22) and has been purified from Drosophila extracts as being required for TATA-less downstream promoter element-mediated transcription (23). The observation by Willy et al. (23) that dNC2 had no effect on TATA-driven promoters might either be due to a requirement for other factors that are present in our whole-cell extract but absent from the fractionated Drosophila system, or due to the limited number of TATA-driven promoters that were only examined in that study. In particular, the experiments presented here were initiated because the unselected bur6 Gal− phenotype implicated NC2 in GAL1 induction; analogous methods to those used here, such as whole-genome transcript analysis or informative phenotypes caused by NC2 mutations will be necessary to identify NC2-responsive promoters in other organisms. Here we demonstrate that NC2 can directly stimulate transcription from a model TATA-driven promoter and that this positive effect is achieved through interactions with TBP. On the basis of the broad effects of NC2 on transcription and the evolutionary conservation of NC2 and the TBP residues required for interaction with NC2, this model will have general implications for transcriptional regulation in other eukaryotes, including humans.
Acknowledgments
We thank Karen Arndt, Scott Emmons, and Ian Willis for reading versions of the manuscript, and Michael Brenowitz, David Stillman, Judith Jaehning, and Vikas Palham for plasmids, reagents, and technical advice. This work was supported by National Institutes of Health Grant GM52486 (to G.P.).
Abbreviations
- NC2
negative cofactor 2
- TBP
TATA-binding protein
- SC
synthetic complete
- SD
glucose minimal
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 2.Lee T I, Young R A. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 3.Yokomori K, Zeidler M P, Chen J L, Verrijzer C P, Mlodzik M, Tjian R. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 4.Weideman C A, Netter R C, Benjamin L R, McAllister J J, Schmiedekamp L A, Coleman R A, Pugh B F. J Mol Biol. 1997;271:61–75. doi: 10.1006/jmbi.1997.1152. [DOI] [PubMed] [Google Scholar]
- 5.Imbalzano A N, Zaret K S, Kingston R E. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 6.Zhao X, Herr W. Cell. 2002;108:615–627. doi: 10.1016/s0092-8674(02)00648-7. [DOI] [PubMed] [Google Scholar]
- 7.Hisatake K, Roeder R G, Horikoshi M. Nature (London) 1993;363:744–747. doi: 10.1038/363744a0. [DOI] [PubMed] [Google Scholar]
- 8.Workman J L, Roeder R G. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 9.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 10.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 11.Jackson-Fisher A J, Chitikila C, Mitra M, Pugh B F. Mol Cell. 1999;3:717–727. doi: 10.1016/s1097-2765(01)80004-6. [DOI] [PubMed] [Google Scholar]
- 12.Meisterernst M, Roeder R G. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 13.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 14.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 15.Goppelt A, Meisterernst M. Nucleic Acids Res. 1996;24:4450–4455. doi: 10.1093/nar/24.22.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prelich G. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Na J G, Hampsey M, Reinberg D. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prelich G, Winston F. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbois E L, Young R A. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisberg J V, Holstege F C, Young R A, Struhl K. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaire M, Xie J, Meisterernst M, Collart M A. Mol Microbiol. 2000;36:163–173. doi: 10.1046/j.1365-2958.2000.01839.x. [DOI] [PubMed] [Google Scholar]
- 23.Willy P J, Kobayashi R, Kadonaga J T. Science. 2000;290:982–984. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 24.Burke T W, Willy P J, Kutach A K, Butler J E, Kadonaga J T. Cold Spring Harbor Symp Quant Biol. 1998;63:75–82. doi: 10.1101/sqb.1998.63.75. [DOI] [PubMed] [Google Scholar]
- 25.Burke T W, Kadonaga J T. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christova R, Oelgeschlager T. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 27.Strubin M, Struhl K. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 28.Carlson M, Botstein D. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 29.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 30.Woontner M, Jaehning J A. J Biol Chem. 1990;265:8979–8982. [PubMed] [Google Scholar]
- 31.Woontner M, Wade P A, Bonner J, Jaehning J A. Mol Cell Biol. 1991;11:4555–4560. doi: 10.1128/mcb.11.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cang Y, Auble D T, Prelich G. EMBO J. 1999;18:6662–6671. doi: 10.1093/emboj/18.23.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hope I A, Struhl K. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 34.Pina B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tansey W P, Herr W. Science. 1997;275:829–831. doi: 10.1126/science.275.5301.829. [DOI] [PubMed] [Google Scholar]
- 36.Tansey W P, Ruppert S, Tjian R, Herr W. Genes Dev. 1994;8:2756–2769. doi: 10.1101/gad.8.22.2756. [DOI] [PubMed] [Google Scholar]
- 37.Castano E, Gross P, Wang Z, Roeder R G, Oelgeschlager T. Proc Natl Acad Sci USA. 2000;97:7184–7189. doi: 10.1073/pnas.140202297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T K, Zhao Y, Hui G, Bernstein R, Roeder R G. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 39.Ptashne M. A Genetic Switch. 2nd Ed. Cambridge, MA: Cell Press & Blackwell Scientific; 1992. [Google Scholar]
- 40.Wyrick J J, Holstege F C, Jennings E G, Causton H C, Shore D, Grunstein M, Lander E S, Young R A. Nature (London) 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 41.Dasgupta A, Darst R P, Martin K J, Afshari C A, Auble D T. Proc Natl Acad Sci USA. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudarsanam P, Iyer V R, Brown P O, Winston F. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Parvin J D, Shykind B M, Sharp P A. J Biol Chem. 1996;271:18405–18412. doi: 10.1074/jbc.271.31.18405. [DOI] [PubMed] [Google Scholar]
- 45.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 46.Kamada K, Shu F, Chen H, Malik S, Stelzer G, Roeder R G, Meisterernst M, Burley S K. Cell. 2001;106:71–81. doi: 10.1016/s0092-8674(01)00417-2. [DOI] [PubMed] [Google Scholar]
- 47.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 48.Kraus V B, Inostroza J A, Yeung K, Reinberg D, Nevins J R. Proc Natl Acad Sci USA. 1994;91:6279–6282. doi: 10.1073/pnas.91.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie J, Collart M, Lemaire M, Stelzer G, Meisterernst M. EMBO J. 2000;19:672–682. doi: 10.1093/emboj/19.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, Cabane K, Hampsey M, Reinberg D. Mol Cell Biol. 2000;20:2455–2465. doi: 10.1128/mcb.20.7.2455-2465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]