Abstract
HemAT from Bacillus subtilis is a new type of heme protein responsible for sensing oxygen. The structural and functional properties of the full-length HemAT protein, the sensor domain (1–178), and Tyr-70 mutants have been characterized. Kinetic and equilibrium measurements reveal that both full-length HemAT and the sensor domain show two distinct O2 binding components. The high-affinity component has a Kdissociation ≈ 1–2 μM and a normal O2 dissociation rate constant, kO2 = 50–80 s−1. The low-affinity component has a Kdissociation ≈ 50–100 μM and a large O2 dissociation rate constant equal to ∼2000 s−1. The low n-value and biphasic character of the equilibrium curve indicate that O2 binding to HemAT involves either independent binding to high- and low-affinity subunits in the dimer or negative cooperativity. Replacement of Tyr-70(B10) with Phe, Leu, or Trp in the sensor domain causes dramatic increases in kO2 for both the high- and low-affinity components. In contrast, the rates and affinity for CO binding are little affected by loss of the Tyr-70 hydroxyl group. These results suggest highly dynamic behavior for the Tyr-70 side chain and the fraction of the “up” versus “down” conformation is strongly influenced by the nature of the iron-ligand complex. As a result of having both high- and low-affinity components, HemAT can respond to oxygen concentration gradients under both hypoxic (0–10 μM) and aerobic (50–250 μM) conditions, a property which could, in principle, be important for a robust sensing system. The unusual ligand-binding properties of HemAT suggest that asymmetry and apparent negative cooperativity play an important role in the signal transduction pathway.
INTRODUCTION
Bacterial chemotaxis utilizes a network of sensing and regulatory proteins to change bacterial swimming behavior in response to fluctuations of environmental conditions. Nearly all of the major protein components in this signaling transduction system have been identified (Djordjevic and Stock, 1998; Falke et al., 1997; Manson et al., 1998; Stock and Mowbray, 1995). Six methyl-accepting chemotactic proteins in Escherichia coli and 10 in Bacillus subtilis have been isolated, nearly all of which are transmembrane proteins (Grebe and Stock, 1998; Le Moual and Koshland, 1996). Crystal structures of periplasmic binding proteins and the ligand-binding domains of Tar, CheA, CheB, and CheR have been determined and provide a molecular understanding of this signaling transduction pathway (Bilwes et al., 1999; Djordjevic and Stock, 1997; Milburn et al., 1991; Sharff et al., 1992; West et al., 1995; Yeh et al., 1993).
A newly discovered protein family of heme-based sensors appears to be responsible for bacterial sensing of diatomic gases (Chan, 2001; Rodgers, 1999). These heme proteins play important roles in the signaling transduction pathway of aerotaxis and are also involved in other cellular activities such as nitrogen fixation and cellulose synthesis (Chang et al., 2001; Gilles-Gonzalez et al., 1991). Proteins in this family include HemAT, guanylyl cyclase, and CooA, which are able to detect O2, NO, and CO, respectively (Shelver et al., 1997; Stone and Marletta, 1995). These sensors normally have two domains. The heme-containing domain detects the presence of the gas by coordination to the iron atom and then transmits the conformational changes induced by ligand binding to the second domain. The latter is either a kinase itself or is coupled to other enzymes that activate a downstream signal transduction cascade.
Our understanding of how the chemoreceptors detect external molecules and transmit signals to their regulatory proteins remains incomplete (Falke and Hazelbauer, 2001). The major difficulty is the complex nature of membrane proteins and the highly dynamic behavior of their individual domains (Bass and Falke, 1999; Seeley et al., 1996). Soluble chemotactic receptors have been constructed by deleting the transmembrane fragment from its native protein (Ottemann and Koshland, 1997). A Tar/Tsr chimera protein was used to demonstrate that attachment of the complementary domains of two different types of receptors still allows chemotactic function (Krikos et al., 1985; Weerasuriya et al., 1998). The latter result provides strong evidence to support the hypothesis that the signaling relay of chemotactic receptors has adopted a similar molecular mechanism for a variety of external signals.
Discovery of the soluble HemAT protein from B. subtilis provides a unique opportunity to elucidate a more detailed molecular mechanism for signal transduction in aerotaxis (Aono et al., 2002; Hou et al., 2000; Zhang et al., 2003). The signaling domain of HemAT has a protein architecture similar to that of the cytoplasmic domains of other chemotactic receptors. Knowledge gained from studying HemAT can be applied not only to the specific oxygen-sensing mechanism in B. subtilis, but also to a more general understanding of signal transduction by transmembrane chemoreceptors. The crystal structures of the reduced unliganded and cyanomet forms of the HemAT sensor domain have shown that the protein is a homodimer with a four-helical bundle as a core (Zhang et al., 2003). Each subunit preserves a classic globin fold with an extra helix at the N-terminus, named the Z helix, and is missing a D helix. The structure of unliganded HemAT is more asymmetrical than the liganded form. The opposite situation occurs for most E. coli chemotaxis receptors where the liganded receptor is asymmetrical (Milburn et al., 1991). Symmetry disruption in HemAT is caused by a unique conformational change in the B subunit involving an outward and upward movement of Tyr-70 away from the heme iron atom. Our current work shows that this residue plays a key role in regulating O2 binding, presumably by forming a hydrogen bond with the bound ligand. A hydrogen bonding interaction with bound cyanide is seen in the ferric structure of HemAT.
Bacterial chemotaxis is an excellent model for investigating the adaptation of cellular networks (Barkai and Leibler, 1997; Yi et al., 2000). Both experimental evidence and theoretical models suggest that the adaptation of bacterial chemotaxis is robust, responding to concentrations of O2 ranging from ∼40 to 1000 μM (Wong et al., 1995; Yu et al., 2002). This property is an inherent consequence of the protein network itself (Alon et al., 1999; Barkai and Leibler, 1997; Jasuja et al., 1999). In this study, we have characterized functionally the full-length HemAT protein, its sensor domain, and different mutants from B. subtilis using spectroscopic, biophysical, and kinetics techniques. The results are interpreted in terms of the previously published structures. In contrast to the initial work of Aono et al. (2002), we observed markedly biphasic O2 dissociation time courses and equilibrium curves for the wild-type proteins, indicating high- and low-affinity components within the homodimer. The unusual ligand binding properties of HemAT suggest that asymmetry and apparent negative cooperativity play an important role in the signal transduction pathway for chemotaxis.
EXPERIMENTAL PROCEDURES
Growth of bacteria
The wild-type HemAT gene from B. subtilis was commercially cloned into pET29b expression plasmid by polymerase chain reaction (ATG, Eden Prairie, MN). E. coli (BL21(DE3)) cells were transformed with the pET29b-HemAT plasmid and then grown overnight at 37°C on 2×YT agar plates with addition of 30 μg/ml kanamycin. A single colony was picked to inoculate 50 ml of 2×YT media with antibiotics and grown overnight. This 50-ml culture was poured into 1 L 2×YT media. Cell growth was monitored by absorbance of the cell suspension at 600 nm. Protein expression was induced by addition of 1 mM isopropylthio-β-galactoside when the absorbance at 600 nm reached 0.5∼0.6. The temperature was decreased to 28°C, and expression and growth was continued overnight. Cells were harvested by centrifugation at 4000 × g, 4°C, and stored at −80°C for later use.
In addition to full-length HemAT, the sensor domain of HemAT (1–178 residues) and three mutants (Y70F, Y70L, and Y70W) were cloned. Sequences encoding a six-histidine “tag” and one Factor Xa cleavage site (Ile-Glu-Gly-Arg) were added to the sensor domain to facilitate purification. Cell growth, induction, and storage conditions were quite similar to those used for the full-length protein.
Protein purification
Cell paste was resuspended in 50 mM NaCl, 50 mM Tris HCl, pH 7.5. Cells were broken either by a French cell press or lysozyme with a small amount of DNase 1. The cellular debris was removed by centrifugation at 28,000 × g, 4°C, for 30min. For wild-type full-length HemAT protein, solid ammonium sulfate was gradually added to the supernatant to a final concentration of 1 M. The resulting suspension was centrifuged to remove the precipitate. The clear orange-colored supernatant was loaded onto a phenyl Sepharose column (Pharmacia, Piscataway, NJ) preequilibrated with 1 M (NH4)2SO4 in 50 mM Tris.HCl, pH 8.0. The recombinant HemAT protein was eluted with a linear gradient of (NH4)2SO4 from 1 to 0 M. Fractions with red color were pooled, and dialyzed against 50 mM Tris.HCl, pH 8.0, 20 mM NaCl, and 1 mM EDTA at 4°C overnight. The dialysate was concentrated using a Centriprep (Millipore, Billerica, MA). The partially purified HemAT protein was loaded onto a Source 15Q column (Pharmacia) preequilibrated with 50 mM Tris.HCl, pH 8.0, 20 mM NaCl, and 1 mM EDTA. Fractions containing HemAT were eluted around 200–300 mM NaCl with a salt gradient and then concentrated by using a VIVAspin6 filtration device (VIVA Life Science, Costa Mesa, CA). For the polishing step, the samples were passed through a Superose6 gel filtration column (Pharmacia) preequilibrated with 50 mM Tris.HCl, pH 8.0, 200 mM NaCl, and 1 mM EDTA. Protein peaks were well resolved, and the ratio of absorbance at 410:280 nm for the purified HemAT was >3. Fractions containing HemAT from the gel filtration column were pooled and concentrated to final protein concentration of 10 mg/ml.
For the truncated sensor domains, purification was facilitated by a His-tag. Cells were broken as described for the full-length protein, and centrifugation was carried out to remove cell debris. The clear supernatant was loaded onto a Ni-chelating column, and bound protein was eluted with 500 mM imidazole. The His-tag was removed by digestion with Factor Xa (Novagen, Madison, WI). HemAT sensor domains were purified further by anion exchange and hydroxyapatite chromatography (Pharmacia and BioRad (Cambridge, MA), respectively). The buffer for anion exchange (Source 15Q) is 20 mM NaCl, 50 mM Tris.Cl, pH 8.5, and 1 M NaCl, 50 mM Tris.Cl, pH 8.5. The eluted HemAT containing fractions from salt gradient was concentrated to small volume and was passed through desalting column preequilibrated with 10 mM NaPOi, pH 6.8. The sample was loaded to hydroxyapatite column preequilibrated with 10 mM NaPOi, pH 6.8, and eluted with gradient of 500 mM NaPOi, pH 6.8. The final wild-type HemAT sensor domain was examined by gel electrophoresis to be >90% pure and the mass verified with matrix assisted laser desorption ionization mass spectrometry.
Ultraviolet-visible absorption measurements
Spectra of purified full-length HemAT were recorded on a Shimadzu UV-2401 PC spectrophotometer (Columbia, MD). Deoxygenated HemAT was obtained by adding a few grains of solid dithionite to the sample. Fully oxygenated HemAT was prepared by passing the dithionite-reduced HemAT protein through a small Sephadex G-25 column, and then flushing the sample with pure oxygen gas in a stoppered cuvette to obtain 100% saturation. The carbon monoxide derivative was obtained by flushing the gas space above the sample with pure carbon monoxide after reduction with dithionite.
Circular dichroism spectral measurements
Circular dichroism (CD) spectra of purified full-length HemAT were collected in an AVIV model 62A DS spectrophotometer (Lakewood, NJ) at 22°C. A quartz cuvette with 1 cm pathlength was used in the experiment. Protein was diluted to a concentration of 50 μg/ml in 100 mM sodium phosphate buffer, pH 7.0. Circular dichroism wavelength scans were taken from 190 to 250 nm with a bandwidth of 1 nm and 1 min averaging time. The final spectrum was reported as extinction coefficient difference (Δɛ) versus wavelength after subtracting a baseline spectrum and dividing the measured CD signal by the protein concentrations. Secondary structure analysis was performed using the software provided by AVIV.
Analytical ultracentrifugation experiments
Sedimentation velocity and sedimentation equilibrium experiments were performed on a Beckman (Fullerton, CA) model XL-A analytical ultracentrifuge at 10°C. The purified full-length HemAT was prepared in 50 mM Tris.HCl buffer, pH 7.0, 5 mM NaCl, and 4% glycerol. The sedimentation velocity experiment was carried out at rotor speed of 60,000 rpm, and protein concentration was monitored by the absorbance at 415 nm. The second-moment method was used to analyze the sedimentation velocity data. The weight average sedimentation and diffusion coefficients were calculated from the slope of the plot ln(r) versus ω2t and the plot of the boundary spreading, z, versus t (correlation coefficient 0.99 for both values; data not shown).
Sedimentation equilibrium experiments were carried out at 8000, 11,000, and 14,000 rpm using six-sector cells at three different protein concentrations. The partial specific volume of HemAT was computed based on its amino acid composition. Solvent density was calculated based on the CRC handbook. A global fitting algorithm provided by Beckman was used to determine the molecular weight of purified HemAT using combined data at three different concentrations and three different rotor speeds.
Oxygen equilibrium determination
Oxygen equilibrium curves were measured using a Shimadzu spectrophotometer equipped with an Imai cell and an O2 electrode (Imai, 1981). The system was calibrated with 100% nitrogen, 20% oxygen, and 100% oxygen before each measurement. Purified full-length HemAT (50 μM) in 100 mM sodium phosphate, pH 7.0, was injected into the cuvette and the formation of deoxygenated protein was monitored by absorbance changes at 560 nm at 25°C. Absorbance data were collected and stored at preset changes in PO2 as measured by an electrode/picoammeter system. Spectra from 400 to 700 nm were collected at 100% and 0% oxygen concentrations to monitor the extent of oxygenation. Reoxygenation experiments were also carried out to confirm the reversibility of the deoxygenation experiments.
Kinetic experiments
The association rates (k′) for oxygen and carbon monoxide binding to full-length HemAT and the sensor domain proteins were measured by flash photolysis techniques using a pulsed dye laser as described previously (Rohlfs et al., 1990). Deoxygenated sample was diluted to a final concentration of 50 μM in 100 mM sodium phosphate buffer, pH 7.0, equilibrated with either 1 atm of oxygen or carbon monoxide gas. The sample was injected into a stoppered 1-mm pathlength cuvette and converted to fully liganded form by flushing with pure oxygen or carbon monoxide gas. Ligand rebinding was measured by absorbance changes that were monitored at 436 or 412 nm. For each time course, the experiment was repeated at least three times.
Rapid mixing techniques were carried out on a Gibson-Dionex stopped-flow apparatus equipped with an On-Line Instrument Systems (Bogart, GA) model 3820 data collection system at 20°C. The dissociation rates (k) for oxygen and carbon monoxide were determined by using ligand replacement techniques as described previously (Olson, 1981). Fully oxygenated HemAT was rapidly mixed with a stock solution of 1 mM carbon monoxide in 100 mM sodium phosphate buffer, pH 7.0. Replacement rate constants were obtained by fitting the experimental time courses to a double exponential expression. O2 dissociation rate constants were calculated from replacement rates, rO2, by compensating for the competition between O2 and CO rebinding (kO2 = rO2(1 + k′O2[O2]/k′CO[CO]). The rate constant for CO dissociation from HemAT was determined by mixing the CO derivative at ∼100 μM free carbon monoxide with a 2-mM solution of NO (Olson, 1981). Under these conditions, the observed rate is directly equal to the rate of CO dissociation.
FTIR spectroscopic measurements
The infrared spectral data of full-length HemAT and the sensor domain proteins were carried out using a Nicolet Nexus 470 (Thermo Electron-Nicolet, Waltham, MA) Fourier transform infrared spectrometer with the assistance of OMNIC software. The samples were concentrated to ∼2 mM, reduced with dithionite, and equilibrated with 1 atm CO gas in a small tube in 10 mM sodium phosphate buffer, pH 7.0. An airtight syringe, equilibrated with nitrogen gas, was then used to draw CO-HemAT from the tube. The protein sample was rapidly added to a CaF2 BioCell (Rancho Dominguez, CA) IR cuvette (5 mm thickness × 50 mm diameter, separated by a 40-μm spacer; BioTools, Wauconda, IL) to obtain a uniform, bubble-free film. Then the windows of the cuvette were quickly sealed. The cuvette was placed in the sample chamber of the spectrometer that was purged with nitrogen gas 1 h before, and then during data collection. A baseline spectrum with buffer was collected for water vapor correction. The absorbance spectra of the HemAT samples were recorded as a function of wavenumber with a resolution of 2 cm−1 at 25°C. A total 64 scans were collected and averaged and then the water vapor background spectrum was subtracted to give the final FTIR spectrum of the iron-carbonyl complex.
RESULTS
Protein purification and spectroscopic characterization
Both recombinant full-length HemAT and its sensor domain proteins were successfully expressed in E. coli cells. The harvested cell pellet was red, which suggested that the prosthetic group heme was synthesized by the bacterium. However, most of the full-length HemAT protein was expressed in inclusion bodies if the normal induction procedure under the control of T7 promoter was used. Lowering the growth temperature to 28°C and prolonging the induction time were essential to obtain more soluble full-length holoprotein. Under these conditions, the yield of soluble full-length HemAT was 5∼7 mg/l of cell culture.
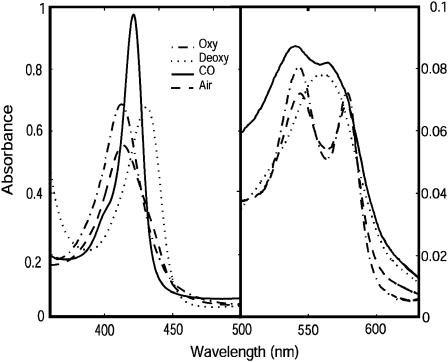
The spectra of both full-length HemAT and the sensor domains indicated that the proteins were purified in a partially oxygenated state. This finding supports the view that HemAT can sense O2 gradients even at high absolute concentrations. As was reported by Aono et al. (2002), three major diagnostic peaks are observed in the spectrum of fully oxygenated HemAT: 412 nm (Soret), 544 nm (β-band), and 578 nm (α-band), which compare well with the peaks at 415–418, 540–543, and 576–581 nm for sperm whale MbO2 and human HbO2. The spectrum of deoxygenated HemAT, produced by reduction with dithionite, has peaks at 434 and 556 nm (Fig. 2). The sensor domain proteins have spectra almost identical to those of the full-length protein. Our spectra and those of Aono's group for HemAT are different from those of Hou et al. (Aono et al., 2002; Hou et al., 2000), who reported absorption peaks at 406 nm (Soret), 538 nm (β-band), and 578 nm (α-band) for the oxygenated form of HemAT and 425 nm (Soret) and 555 nm (β-band) for the deoxygenated form. This discrepancy is due to significant amounts of oxidized protein in Hou et al.'s first preparation of HemAT (Hou et al., 2000).
FIGURE 2.
Ultraviolet-visible spectra of the purified full-length HemAT protein. Comparison of absorption spectra of the CO-, deoxy-, oxy-forms of HemAT in 100 mM sodium phosphate buffer, pH 7.0, at 22°C. The spectrum of HemAT exposed in air is also shown.
Secondary and tertiary structure of full-length HemAT protein
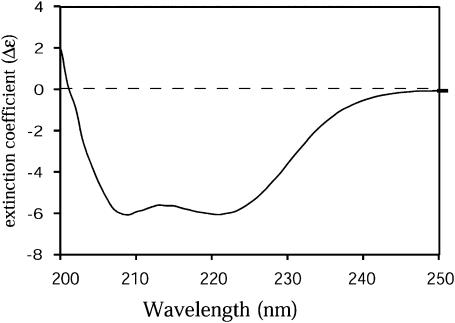
Protein sequence alignments reveal that the N-terminus of HemAT (residues 1∼170) exhibits limited homology to sperm whale myoglobin and that the C-terminus of HemAT (residues 198–432) shares 30% sequence identity to E. coli serine receptor Tsr and other methyl-accepting chemotactic proteins (Hou et al., 2000). Modeling of the cytoplasmic domains of 20 different chemotactic receptors indicate that this region is mainly helical structure with characteristic heptad repeats (Le Moual and Koshland, 1996). The crystal structure of the cytoplasmic domain of E. coli Tsr was determined and revealed that Tsr is a dimeric protein with an elongated coiled coil of antiparallel helix (Kim et al., 1999). The secondary structure of HemAT was examined using circular dichroism spectroscopy to estimate the content of α-helical and β-sheet structure. As shown in Fig. 3, the CD spectrum of full-length HemAT indicates a completely helical protein based on the characteristic double minima at 218 and 208 nm. The secondary structure composition was estimated to be 67% α-helix, 6.9% β-sheet, 12.5% turn, and 15.9% random coil using software and standard spectra provided by AVIV. The helical composition of HemAT based on this analysis is slightly less than that of sperm whale myoglobin (78.0% α-helix content).
FIGURE 3.
Circular dichroism spectrum of the purified full-length HemAT protein. The protein sample was at a concentration of 50 μg/ml in 100 mM sodium phosphate buffer, pH 7.0, at 22°C. The y axis is extinction coefficient difference (Δɛ) after correcting baseline spectrum and the protein concentrations. A very highly α-helical structure is indicated.
In contrast to the sensor domain, full-length HemAT does not behave like a globular protein during purification. We were unable to determine its molecular weight accurately using size exclusion chromatography, and the column elution profiles showed a single, early eluting species suggesting an elongated shape. In contrast, Aono et al. (2002) reported chromatographic data suggesting that HemAT is a homotetrameric protein. The cytoplasmic domain of E. coli Tar receptor also displays complex molecular weight behavior at different pH conditions indicating odd shapes and/or dissociating oligomeric structures (Seeley et al., 1996). To resolve the discrepancy between our initial observations and those of Aono et al. (2002), analytical ultracentrifugation was used to determine more directly the molecular weight of our preparation of full-length HemAT.
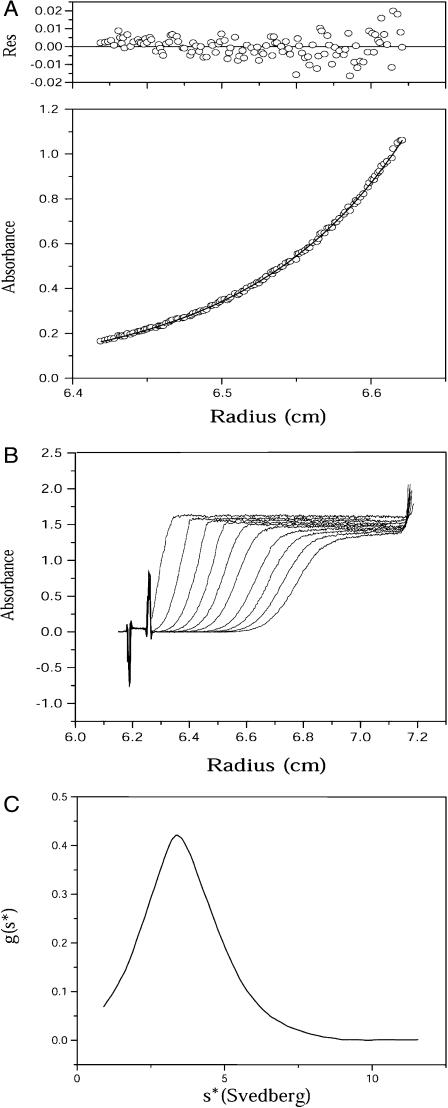
Sedimentation equilibrium experiments were carried out to obtain the molecular weight of full-length HemAT without having to determine its frictional coefficient. A molecular weight of 98,292 was obtained from these analyses (Fig. 4 A) and is remarkably close to the theoretical value of an intact HemAT dimer based on its sequence (98,766 Da including the heme moiety). Thus, the native form of HemAT forms a stable homodimer without significant dissociation to monomers or further aggregation in the concentration range used in our experiments (2–20 μM).
FIGURE 4.
Analytical ultracentrifugation experiments. (A) Sedimentation equilibrium curve of the purified full-length HemAT. Data shown here were recorded at rotor speed 11,000 rpm and the absorbance was monitored at 415 nm. The residual distributions are shown at the top panel. (B) Sedimentation velocity experiment of the full-length HemAT, which were carried out at rotor speed of 60,000 rpm and the absorbance was monitored at 415 nm with protein concentration of ∼15 μM at 10°C. (C) Sedimentation coefficient distribution g(s*) plot. The peak of the curve corresponds to sedimentation coefficient 2.63 s corresponding to a dimer.
Results from sedimentation velocity experiments are shown in Fig. 4, B and C, and also suggest that the protein is a single molecular species in solution. However, the sedimentation coefficient, 2.63 s, is very small, in comparison to that expected value of 7–8 s for a globular protein with a molecular weight of ∼98,000. This low sedimentation coefficient shows that HemAT has an extremely large frictional coefficient. The overall shape of full-length HemAT can be deduced from the results of both analytical ultracentrifugation experiments. The molecular weight determined by equilibrium sedimentation and the measured sedimentation coefficient was used to calculate the frictional coefficient of HemAT (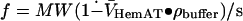 ). The ratio of the experimentally determined frictional coefficient (f) to the theoretical frictional coefficient for a spherical particle (fo) is 2.5, which demonstrates that the full-length HemAT has a highly asymmetric rodlike shape. Thus, our centrifugation results show that in solution full-length HemAT is an elongated homodimer with axial ratio >10, consistent with the structure of all other receptors involved in bacterial chemotaxis.
). The ratio of the experimentally determined frictional coefficient (f) to the theoretical frictional coefficient for a spherical particle (fo) is 2.5, which demonstrates that the full-length HemAT has a highly asymmetric rodlike shape. Thus, our centrifugation results show that in solution full-length HemAT is an elongated homodimer with axial ratio >10, consistent with the structure of all other receptors involved in bacterial chemotaxis.
Equilibrium O2 binding
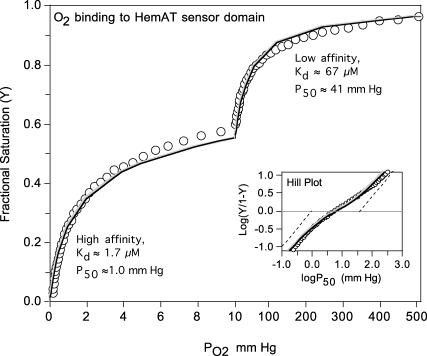
Equilibrium oxygen binding to full-length HemAT shows two distinct components, one with high affinity and a P50 ≈ 1 mm Hg (Kd ≈ 1.7 μM) and one with very low affinity and a P50 ≈ 40 mm Hg (Kd ≈ 70 μM) (Fig. 5). Biphasic binding curves are not unique to HemAT and have also been observed for ligand binding to the Tar and Tsr chemoreceptors (Biemann and Koshland, 1994; Lin et al., 1994). The overall P50 (concentration of free O2 at 50% saturation) for full-length HemAT is ∼10 μM, and the n-value (Hill coefficient) at 50% saturation is ∼0.45. The low n-value and biphasic character indicate that O2 binding to HemAT involves either independent binding to high and low subunits in the dimer or negative cooperativity. The data in Fig. 5 could be fitted to either mechanism using Eqs. 1 or 2, respectively.
FIGURE 5.
Equilibrium O2 binding to 50 μM full-length HemAT at 25°C in 0.1 M phosphate, pH 7.0. Data were collected using an Imai apparatus in which PO2 is recorded with an oxygen electrode and absorbance changes are measured in a spectrophotometer and then converted to fractional saturation, Y. The inset shows a Hill plot of the observed data, where the dashed lines show theoretical plots for the high- and low-affinity binding components. The observed partial pressure at 50% saturation was ∼6 mm Hg (∼10 μM) and the n-value is very low, ∼0.45 at 50% saturation. The gray line represents a fit to two independent high- and low-affinity binding components with the Kd and P50 values shown beside each hyperbolic region in the curve. The black line represents a fit to a two-step Adair equation with the high- and low-affinity Kd values representing 1/K1 and 1/K2 in this sequential scheme. The PO2 scale for the left portion of the main figure is 0–10 mm Hg and defines the high-affinity binding phase and the scale for the right portion is 10–510 mm Hg, defining the low-affinity binding phase. The analyses assume equal absorbance changes at 560 nm for the two components (i.e., the two subunits in the HemAT dimer). The deviations seen at 50% saturation may reflect slight differences in the extinction coefficient changes for O2 binding the two components.
Two independent components:
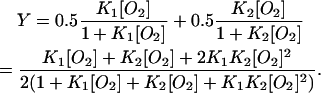 |
(1) |
Two consecutive reaction Adair scheme:
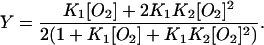 |
(2) |
When K1 ≫ K2, Eq. 1 reduces to Eq. 2 and the Adair scheme becomes indistinguishable from the two independent binding components scheme because the K2[O2] terms become negligible compared to the K1[O2] and K1K2[O2]2 terms. Thus, significant negative cooperativity, 0.5K1 ≫ 2K2, cannot be distinguished from binding to two independent components with widely different affinities.
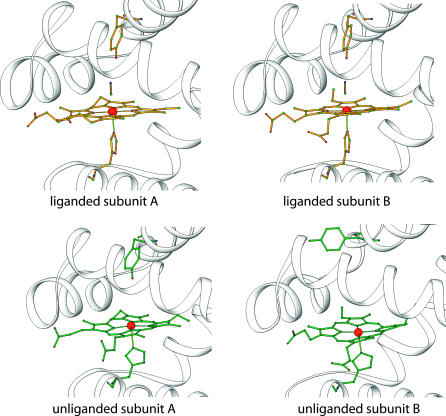
The fitted K1 and K2 values for the high- and low-affinity components of HemAT are ∼0.6 and ∼0.015 μM−1, respectively. The structures of unliganded HemAT in Fig. 1 also suggest that the two ligand binding sites in HemAT are not equivalent initially and that O2 may bind preferentially at equilibrium to the subunit (molecule A) that allows stabilization of bound O2 by Tyr-70 in the “down” conformation. At low [O2], ligands would bind only to the A subunit, with the remaining heme group unoccupied until the concentration of ligand is increased to ≥100 μM to allow binding in the absence of hydrogen-bond stabilization by Tyr-70. The net result is that HemAT shows changes in saturation over almost three decades of O2 concentration from 0.2 to ≥200 μM (Fig. 5).
FIGURE 1.
Diagrams of the heme regions of the HemAT sensor domain. The proteins are drawn as ribbons and residues Tyr-70, His-123, heme groups, and ligands are represented as ball/stick models. The top panels show liganded HemAT sensor domain and the bottom panels show unliganded structure. The Protein Data Bank access codes for the two structures are 1OR4 and 1OR6.
Rates of O2 binding
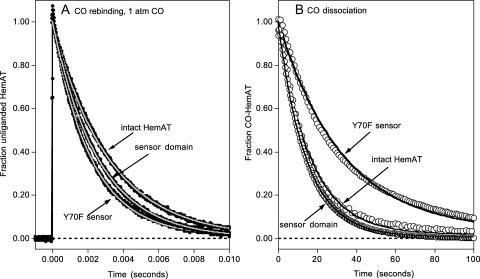
Kinetic parameters for O2 binding to full-length HemAT, wild-type and mutant HemAT sensor domains, and wild-type and mutant sperm whale myoglobin are compared in Table 1. After laser photolysis, O2 rebinding to full-length HemAT and the sensor domain shows a major rapid phase followed by a very slow process with a small amplitude (Figs. 6 A and 7).
TABLE 1.
Rate and equilibrium constants for ligand binding to HemAT proteins
| Protein | k′O2μM−1s−1 | kO2 s−1 | KO2μM−1 | k′COμM−1s−1 | kCO s−1 | KCOμM−1 | KCO/KO2 |
|---|---|---|---|---|---|---|---|
| Wild-type SwMb* | 17 | 15 | 1.1 | 0.51 | 0.019 | 27 | 25 |
| SwMb H64L* | 98 | 4100 | 0.023 | 26 | 0.024 | 1100 | 48,000 |
| Full-length HemAT | 19 | 1900 | 0.010 | 0.34 | 0.067 | 5.1 | 510 |
| 87 | 0.22 | (23) | |||||
| Sensor wild type | 19 | 1800 | 0.011 | 0.43 | 0.070 | 6.1 | 550 |
| 50 | 0.37 | (16) | |||||
| Sensor Y70F | 53 | 19,000 | 0.003 | 0.47 | 0.030 | 16 | 5300 |
| 1800 | 0.030 | (530) | |||||
| Sensor Y70L | 89 | 70,000 | 0.001 | 0.16 | 0.029 | 5.5 | 5500 |
| 3500 | 0.026 | (210) | |||||
| Sensor Y70W | 57 | 22,000 | 0.003 | 0.28 | 0.043 | 6.5 | 2200 |
| 3100 | 0.019 | (340) |
Data for sperm whale myoglobin taken from Springer et al. (1994).
FIGURE 6.
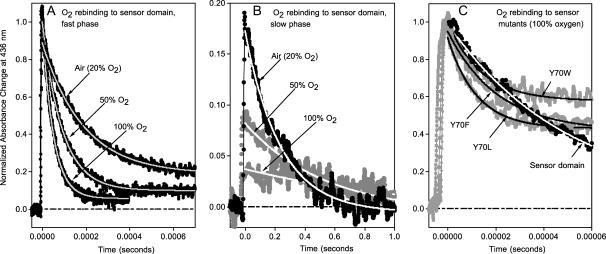
Normalized time courses for O2 rebinding to the sensor domain of HemAT at pH 7.0, 25°C, after laser photolysis with a 300-ns excitation pulse. Conditions were: ∼50 μM heme and three different concentrations of O2, 250 μM (air), 625 μM (50% O2), and 1250 μM (100% O2) in 0.1 M sodium phosphate buffer, pH 7. The time courses were measured at 436 nm (peak for deoxygenated HemAT) and identical results were obtained at 412 nm (peak for oxygenated HemAT). (A) Dependence of the time courses for the major, fast bimolecular phase on [O2]. As the O2 concentration decreases from 1250 μM (100% O2) to 250 μM (air), the final equilibrium saturation decreases from 1.0 to ∼0.9 (see Fig. 5). As result the amplitude of the normalized absorbance change decreases in air. (B) The minor, very slow phase shows the opposite dependence on [O2] with the rate and amplitude decreasing with increasing ligand concentration. The signal/noise level is high because: 1), the data were collected using the same small time constant used to collect data for the 1000 times more rapid bimolecular phase; and 2), the amplitude of the slow phase is small. (C) The time courses for O2 rebinding to the wild-type sensor domain and Tyr-70 mutants in buffer equilibrated with 100% oxygen (1250 μM).
FIGURE 7.
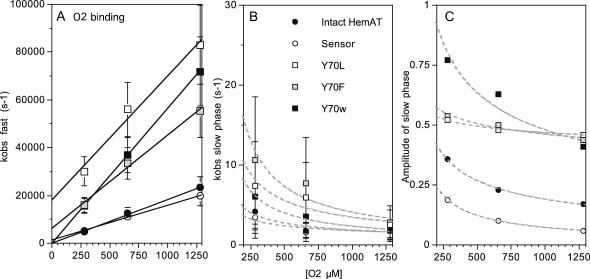
Dependence of the fast and slow observed rates for O2 rebinding on total [O2] for full-length HemAT, wild-type sensor domain, and Y70 mutants of the sensor domain at pH 7, 25°C. (A) kobs for the fast phase depends linearly on [O2], and the k′O2 values in Table 1 were taken from the average of slopes determined in these types of experiments. The intercepts provide estimates of the rate of O2 dissociation in the initial rapid binding process and are similar to the values of kO2 obtained for the slow phases seen in direct O2 dissociation experiments (Table 1; Fig. 7). The errors for the fast rate constants were estimated to be ±20% based on multiple experiments with the wild-type sensor protein. (B and C) The rates and amplitudes of the slow phases seen in O2 rebinding experiments show a complex inverse dependence on [O2], with both the rate and amplitude decreasing to 0 at high ligand concentration. This pattern suggests that a slow conformational transition can occur after the high-affinity site is filled, enhancing O2 binding to the low-affinity site. Thus, as [O2] increases less of this change will occur and it will vanish when 100% saturation is achieved in the initial rapid bimolecular phase. The errors in kobs,slow and the fraction of slow phase were estimated to be ±50% and ±0.1 based on multiple experiments with the wild-type sensor domain.
The observed pseudo-first-order rate constants (kobs) for the fast phases show linear dependences on [O2] for all of the HemAT derivatives examined (Figs. 6 A and 7 A). Thus, the fast phase represents a simple bimolecular process with a second-order rate constant, k′O2, equal to the slope of these plots (Fig. 7 A). The association rate constants for both full-length HemAT and the sensor domain are nearly identical, 19 μM−1s−1 at pH 7.0, 20°C, and comparable to that of sperm whale myoglobin (Springer et al., 1994). Aono et al. (2002) observed a similar value of k′O2 = 32 μM−1s−1 at pH 8.0 and room temperature, however, the dependence of the observed time courses on [O2] was not reported. O2 binding to the Tyr-70 mutants is approximately three- to fivefold faster than that to the wild-type proteins (Fig. 6 C; Table 1), and the amplitude of the slow O2 rebinding phase is much larger, ≥50%. A minor slow phase is observed during O2 rebinding to full-length HemAT and the wild-type sensor (Fig. 6 B). Both the rate and extent of this process decrease with increasing [O2] and are ≤2 s−1 and 10%, respectively, when ≥90% saturation is achieved in buffer equilibrated with 1 atm of pure oxygen (Figs. 6 B and 7 C). The structural cause and mechanism of this slow phase are not apparent, but it is clear that the process is not bimolecular because both kobs,slow and the amplitude of the slow phase decrease with increasing [O2] (Fig. 7 C).
The unusual dependence of the rebinding time course on [O2] can also be rationalized in terms of high- and low-affinity components. If the association rate constants of both components are roughly the same, but the low-affinity component has an extremely large dissociation rate constant, then simple monophasic time courses would be observed at high [O2]. However at low [O2], when incomplete saturation occurs, complex biphasic time courses would be observed. Under the latter conditions, the binding of the first ligand may allow a fraction of the partially liganded dimers to isomerize to a new conformation in which the second site can bind O2 more readily, presumably by downward movement of Tyr-70 and hydrogen binding to bound O2. If the conformational transition were slow (≤10 s−1) and incomplete, additional ligand binding would be slow and first order. In addition, the amplitude of this phase would decrease at high O2 where binding can occur to the low-affinity subunit without the need for isomerization to allow hydrogen bonding by Tyr-70.
This slow conformational transition mechanism assumes that the differences in affinity are due solely to differences in the O2 dissociation rate constants at the two binding sites. This interpretation is supported by the markedly biphasic time courses for O2 dissociation from HemAT (Fig. 8 and results below) and the observation that the major difference between the asymmetric subunits in unliganded HemAT is the proximity of the polar Tyr-70 side chain to the site for O2 binding.
FIGURE 8.
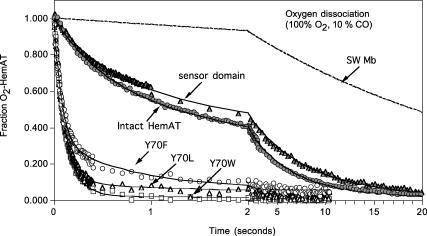
Time courses for O2 dissociation from full-length HemAT, wild-type sensor domain, and Y70F, Y70L, and Y70W mutant sensor domains at pH 7, 20°C, 0.1 M sodium phosphate buffer. Reduced HemAT (3–8 μM) was injected into buffer equilibrated with 1 atm of O2 (1250 μM before mixing) and then rapidly mixed with 100 μM CO. Oxygen displacement was monitored by absorbance increases at 421 nm (peak of CO-HemAT) and absorbance decreases at 412 nm (peak of O2-HemAT). Data were collected on two timescales, 0–2 s (100 points) and then 2–10 s or 20 s (100 points) to define the fast and slow phases. Similar time courses were obtained at either wavelength; the data shown are for 421 nm. The dashed time course was that expected for sperm whale myoglobin under these high O2 and low CO conditions. The time courses were fitted to two exponential expressions and the observed replacement rates (robs) for the fast phases were 1.8, 1.9, and 13 s−1 for full-length HemAT, wild-type sensor domain, and the Y70F mutant. The calculated value of robs for sperm whale MbO2 under these conditions is 0.36 (dashed line). The fitted values for the slow phases are 0.22, 0.16, and 1.3 s−1, respectively, for the same set of HemAT proteins. Reactions had to be carried out at ≥1000 μM [O2] to ensure complete saturation of the protein, and low [CO] was required because of the large values of kO2 for the fast phase components, from ∼2000 to ≥20,000 s−1 (Table 1).
Rates of O2 release
Time courses for the dissociation of O2 from HemAT are also markedly biphasic (Fig. 8). Aono et al. (2002) examined the reaction of oxygenated HemAT with excess sodium dithionite and reported a single phase with kO2 ≈ 20 s−1 at pH 8.0, 25°C. We obtained similar results when the HemAT-O2 forms of full-length and sensor domain proteins were mixed with dithionite alone or dithionite in CO saturated buffer. However, over half the expected absorbance change was “lost” in the dead time of stopped flow device. To examine the ultrafast O2 absorbance phase, we measured time courses for O2 replacement by CO. In these experiments, HemAT-O2 in 100% O2 was mixed with a low concentration of CO to decrease the replacement rate to examine the entire reaction. The rate of O2 replacement is given by
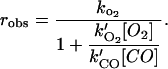 |
(3) |
By keeping the ratio [O2]/[CO] high, the observed rate can be reduced to a value accessible in rapid mixing experiments, t1/2 ≤ 300 s−1. The true value of kO2 can be calculated from robs = (1 + k′O2[O2]/k′CO[CO]), where the bimolecular rates of O2 and CO binding are determined independently in laser photolysis experiments (Figs. 6 and 9).
FIGURE 9.
Time courses for CO association with and dissociation from full-length HemAT, wild-type sensor domain, and the Y70F sensor domain mutant at pH 7.0, 25°C. (A) CO rebinding after laser photolysis at [CO] = 1000 μM. Time courses were measured at 436 nm (black lines and circles) and fitted to single exponential expressions (gray lines) with observed rates of 35 s−1, 38 s−1, and 47 s−1 for full-length HemAT, wild-type sensor domain, and the Y70F mutant, respectively. (B) Time courses for CO dissociation were obtained by mixing 3–8 μM protein in 100 μM CO with anaerobic buffer containing 2000 μM NO. Under these conditions, the observed replacement rate is directly equal to the rate of CO dissociation, kCO. Time courses were recorded at 422 nm (○) and fitted to single exponential expressions (solid lines) with observed rates equal to the kCO values in Table 1. As shown in panel B, the time course for CO dissociation from the Y70F (and the other mutants (Y70L and Y70W) not shown due to superimposition on these for Y70F) is better fit to a multiple exponential expression. However the differences in the fast and slow component rates are less than a factor of 3 and very small compared to the 20-fold differences seen in the O2 dissociation time courses (Fig. 7 and Table 1).
The dissociation rate constants (kO2) for the fast phases of the HemAT proteins are very large, ranging from 70,000 to 2,000 s−1 (Table 1). The computed kO2 value for the fast phase of full-length HemAT (1900 s−1) is ∼100 times faster than that observed for sperm whale myoglobin and indicates a relatively apolar distal pocket with the Tyr-70 side chain primarily in the “up” conformation. Similar high oxygen dissociation rate constants are observed for Glycera dibranchiata Hb II (1800 s−1), which has a completely apolar distal pocket with a Leu at the E7 position (Mims et al., 1983; Rohlfs et al., 1990), and sperm whale myoglobin mutants in which His-E7 is replaced with apolar amino acids (2000 ∼ 10,000 s−1; see entry for H64L Mb in Table 1; Springer et al., 1994). In both cases, the high values of kO2 have been shown to be due to the lack of stabilization of bound O2 by hydrogen-bonding donors (Rohlfs et al., 1990).
We estimated the values of kO2 for the slow dissociation phases of all the HemAT derivatives by assuming that the measured bimolecular rate constants for O2 and CO binding, k′O2 and k′CO, apply to both dissociation components. The O2 dissociation rate constants for the slow phases of full-length HemAT and the sensor domain are only four- to sixfold greater than that of mammalian myoglobins (Table 1). More importantly, both the fast and slow kO2 values of the full-length protein and the wild-type sensor domain are almost identical, supporting the conclusion that the ligand binding properties of HemAT are independent of the C-terminal domain.
Oxygen affinity constants (KO2) for the fast and slow dissociating components can be calculated from the ratio of the rate constants (k′O2/kO2) and were estimated to ∼0.2–0.4 and ∼0.01 μM−1, respectively, in good agreement with the fitted values of K1 and K2 from analyses of the oxygen equilibrium curve shown in Fig. 5 (0.6 and 0.015 μM−1, respectively). Both of these KO2 values are less than that of sperm whale myoglobin (Table 1). The higher K2 value for the slow phase component is close to the O2 affinity of HemAT reported previously by Hou et al. and Aono et al. (Aono et al., 2002; Hou et al., 2001).
Replacement of Tyr-70 with Leu, Phe, and Trp causes dramatic 10- to 30-fold increases in rates of O2 dissociation and the time courses become almost monophasic compared to those observed for the full-length HemAT and the wild-type sensor domain (Fig. 8; Table 1). As a result, the O2 affinities of all three mutants are lowered 3- to 10-fold by the loss of hydrogen-bonding potential at position 70. In the crystal structure of the cyanomet form of the wild-type sensor domain, bound cyanide is stabilized by a hydrogen bond from the side chain of Tyr-70 (Fig. 1). The mutations made in sensor domain (Y70F, Y70L, and Y70W) remove the hydrogen-bonding capability at position 70, and the resultant lack of stabilization by Tyr-70 appears to be the underlying cause of the dramatic increases in kO2 and decreases in KO2 caused by the substitutions.
Carbon monoxide binding
The kinetic parameters for CO binding to all five HemAT proteins are also listed in Table 1. The time courses for CO rebinding after laser photolysis are monophasic (Fig. 9 A), and the observed association rate constants are all similar to each other (k′co = 0.2–0.5 μM−1s−1) and to that of sperm whale myoglobin (k′co = 0.51 μM−1s−1; Springer et al., 1994). The time courses for the dissociation of CO from the HemAT derivatives are also monophasic (Fig. 9 B). The Tyr-70 mutations have little effect on the observed rates (0.03–0.07 s−1), which in this case are ∼2–3 times greater than those seen for mammalian MbCOs. The calculated CO affinities of full-length HemAT and the wild-type sensor domain are low, 5–6 μM−1, which is ∼5 times smaller than that for sperm whale myoglobin. In both proteins, in-plane iron movement and coordination with exogenous ligands is probably restricted by the large rotations required for ligand binding and the eclipsed orientations of the proximal imidazole ring in both the liganded and unliganded structure of HemAT (Fig. 1). Aono et al. (2002) came to similar conclusions based on comparisons of the resonance Roman spectra of HemAT and Mb complexes.
In contrast to O2 binding, the three Tyr-70 mutants show CO binding behavior, which is similar to that of full-length HemAT and the wild-type sensor domain. This lack of effect indicates that the observed changes in KO2 and kO2 are due primarily to the loss of electrostatic interactions between Tyr-70 and bound O2. Because the FeCO complex is apolar, loss of hydrogen-bonding potential at this position is expected to have only small effects on the kinetic and equilibrium parameters for CO binding.
IR spectra of CO-HemAT complexes
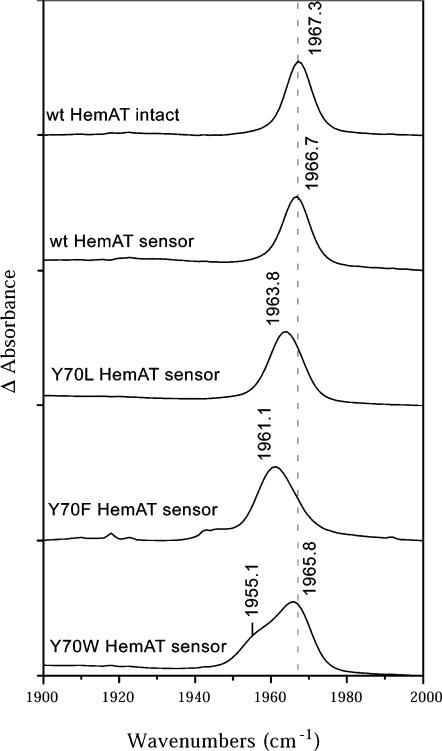
FTIR spectra of full-length and sensor domain CO-HemAT complexes were recorded to examine more thoroughly hydrogen bonding and electrostatic fields in the distal pockets of these proteins. The stretching frequency of bound CO, νCO, has been shown to correlate inversely with the calculated electrostatic field in the immediate vicinity of the bound ligand in 20 different mutants of sperm whale myoglobin (Li et al., 1994; Phillips et al., 1999). These results and those with model hemes and other heme proteins have demonstrated that νCO can serve as a sensitive probe of the electrostatic potential in the ligand binding site (Chu et al., 2000; Kaposi et al., 2001a,b; Thomas et al., 2001). The presence of strong or multiple hydrogen-bond donors correlates with νCO peaks in the 1910–1930 cm−1 region. Moderate hydrogen-bonding interactions give rise to peaks between 1940 and 1950 cm−1. Neutral (apolar) and negative fields shift the νCO peaks to 1960–1970 and 1970–1990 cm−1, respectively.
As shown in Fig. 10, both full-length CO-HemAT and the sensor domain show a single peak centered at 1967 cm−1, which indicates an apolar environment and little or no hydrogen bonding between Tyr-70 and the bound ligand. Aono et al. reported a similar νCO peak at 1964 cm−1 in the resonance Roman spectra of HemAT-CO complex (Aono et al., 2002). The Tyr-70 to Leu and Phe mutations produce only small changes in νCO from 1967 cm−1 to 1964 and 1961 cm−1, respectively. Mutation of Tyr-70 to Trp causes a further small decrease and splitting into bands at 1966 and 1955 cm−1, indicating multiple conformations of the bulky side chain around the buried CO.
FIGURE 10.
FTIR spectra of full-length HemAT protein and the sensor domain. These data were measured at protein concentration of 2 mM in 10 mM sodium phosphate buffer, pH 7.0. The absorbance change was recorded as a function of wavenumber with the resolution of 2 cm−1 at 25°C. The results show that the environments around the CO ligand in the intact protein and isolated domains are quite similar and that the Tyr-70 side chain likely points away from the CO ligand.
All of the FTIR data suggest that, if anything, the nonbonded electron pair of the Tyr-70 hydroxyl O atom is interacting weakly and unfavorably with bound CO in the wild-type protein, presumably because the side chain is primarily in the “up” conformation. The presence of an unfavorable interaction is supported by the decrease in kCO from 0.07 to 0.03 s−1 when Tyr-70 is replaced with Phe or Leu (Table 1). This result is surprising because the crystal structure of the cyanomet form of the sensor domain shows a strong interaction between the hydroxyl group of Tyr-70 and bound cyanide. Although the Fe(III)CN complex is isoelectronic with Fe(II)CO, the cyanide complex is highly charged, with a significant partial negative charge on the ligand atoms and a partial positive charge on the iron. The negative charge on the cyano group would appear to attract the Tyr-70 side chain into the “down” conformation and, more importantly, induce donation of a hydrogen bond from the Tyr-70 hydroxyl, whereas the apolar iron-carbonyl complex does not, as judged by the νCO peak at 1967 cm−1. These results predict that the Tyr-70 side chain probably has an “up” conformation in both subunits of the CO-HemAT dimer, which is analogous to that seen for the B subunit in the unliganded dimer structure (Fig. 1). The large increases in kO2 caused by the Tyr-70 mutations indicate that the partial negative charge on bound O2 can induce the “down” conformation and hydrogen bonding but to markedly different extents in the A and B subunits. Similar differential effects of FeCO and FeO2 complexes on the conformation of polar amino acid side chains have been reported for leghemoglobin (Kundu et al., 2004).
DISCUSSION
Structural mechanism for biphasic O2 binding
Both full-length HemAT and the sensor domain show two distinct O2 binding components. The high-affinity component has a Kd ≈ 1–2 μM and a normal O2 dissociation rate constant, 50–80 s−1. The low-affinity component has a Kd ≈ 50–100 μM and a large O2 dissociation rate constant equal to ∼2000 s−1. The Kd for the high-affinity component is similar to that reported previously (Aono et al., 2002; Hou et al., 2001). Biphasic equilibrium binding attributed to negative cooperativity has been reported for the aspartate receptor from E. coli and Salmonella typhimurium and for the serine receptor from E. coli, and thus, this unusual binding phenomenon may be an intrinsic behavior of chemotactic sensor proteins (Biemann and Koshland, 1994; Lin et al., 1994).
As described in Eqs. 1 and 2, it is impossible to distinguish between negative cooperativity and two independent components if the observed rate and equilibrium constants for the first and last steps in binding are widely different. The simplest interpretation is to assume that the A and B subunits in the unliganded HemAT dimer have widely different ligand binding properties and behave as independent components. As shown in Fig. 1 (bottom panels), there is strong structural evidence for asymmetric conformations and nonidentical ligand binding sites in the reduced unliganded HemAT homodimer. In molecule A, the side chain of Tyr-70 is primarily in the “down” position with the aromatic hydroxyl group in a position to interact strongly with bound ligands. In molecule B, the opposite situation occurs, the Tyr-70 side chain is pointing out into solvent, and the distal pocket appears to be complete apolar. Thus, the molecule A conformer can be assigned to the high-affinity O2 binding component with a low rate of oxygen dissociation and the B conformer can be assigned to the low-affinity component that behaves like naturally occurring or mutant Mbs and Hbs with completely apolar distal pockets.
The effects of the Tyr-70 mutations support this interpretation. Removal of the phenolic hydroxyl group causes dramatic 30- to 70-fold increases in kO2 and >10-fold decreases in KO2 for the high-affinity component, presumably subunit A. The Tyr-70 replacements also affect the ligand binding parameters of the low-affinity component, causing kO2 to increase and KO2 to decrease significantly. The latter results suggest that in the B subunit, Tyr-70 is in equilibrium between the up and down conformation. Thus, a significant fraction (10–20%) of the Tyr-70 side chains in this subunit do rotate downward and form a stabilizing hydrogen bond, being attracted by the partial negative charge on the bound O2 molecule. In the A subunit, the Tyr-70 side chain is already ≥90% in the down position in the unliganded state and thus can form a strong hydrogen bond with bound ligands without the free energy cost of altering its conformation.
O2 rebinding to the Y70F, L and W mutants is markedly biphasic (Fig. 6 C), with large slow phases. These results imply a rapid equilibration of O2 with one subunit followed by slow first-order conformational changes that result in further O2 binding to the remaining subunit in the HemAT dimer. In these cases, enhancement of O2 binding cannot occur by hydrogen binding to Tyr-70(B10). The slow conformational changes may be caused by alterations in proximal coordination geometry that enhance iron reactivity or to the slow interconversion of open and closed conformations of the active site as is observed for plant nonsymbiotic hemoglobins (Trent et al., 2001). Such changes are probably also occurring in the wild-type proteins.
In contrast to O2 binding, CO binding to HemAT shows simple monophasic behavior (Figs. 8 and 9; Table 1). These observations support the view that the heterogeneous O2 binding is primarily due to different electrostatic interactions in the asymmetric subunits of the HemAT homodimer. The stability of the apolar FeCO complex should be little affected by the presence or absence of the Tyr-70 hydroxyl group. In fact, the FTIR results suggest that CO binding induces an “up” conformation of the Tyr-70 side chain because only a single peak at 1967 cm−1 is observed implying apolar distal pockets in both subunits of the fully saturated CO-HemAT dimer. This contrasts with the crystal structure of the cyanomet HemAT dimer where the both Tyr-70 side chains are in the down position, interacting with the highly polar Fe(III)CN complex.
Together, the O2, CO, and cyanide binding results suggest highly dynamic behavior for the Tyr-70 side chain. The fraction of the “up” versus “down” conformation is strongly influenced by the nature of the iron-ligand complex. In sperm whale myoglobin, small movements (≤0.3 Å) of His-E7 away from the iron are seen when comparing the average position of the imidazole side chain in MbO2 versus MbCO at room temperature (Quillin et al., 1993). In Mb, the distal histidine is held in place by strong steric interactions with the side chain of Phe-CD4 and weakly by electrostatic interactions with the heme propionates and Arg or Lys-CD3 (Lai et al., 1995). Thus, the position of the imidazole side chain is only weakly affected by the nature of the bound ligand. In HemAT, the polar Tyr-70 side chain appears to be much more flexible and thus more greatly influenced by the polarity of the iron-ligand complex. In the CO complexes, the Tyr-70 hydroxyl group appears to move away from the heme group and interact with solvent water molecules, whereas in cyanomet-HemAT, it is attracted to and interacts strongly with the zwitterionic Fe(III)+1CN−1 complex. Intermediate behavior is probably occurring for the partially charged FeO2 complexes. The observed heterogeneity in O2 binding appears to result from the differences in the thermodynamic stability of the “down” conformation in the asymmetric A and B subunits in the HemAT dimer.
Physiological significance
Regardless of the exact structural interpretation, the results in Fig. 5 show that HemAT can respond to oxygen concentration gradients under both hypoxic (0–10 μM) and aerobic (50–250 μM) conditions, as result of binding to the high- and low-affinity components, respectively. Thus, the dynamic range of the chemotactic response induced by HemAT can cover three decades of O2 concentration. A systemic study of the O2 concentration dependence of the aerotaxis response of B. subtilis has not been reported. However, Taylor's group (Wong et al., 1995) has shown smooth swimming responses in both air and 100% O2, and Alam's group (Yu et al., 2002) has shown that B. subtilis can respond rapidly and strongly to photochemically produced pulses of O2 at concentrations on the order of 20∼50 μM. This behavior is in contrast to most other O2 sensors, particularly those involved in transcriptional regulation, which show high affinity and respond to low levels of O2 (Gilles-Gonzalez et al., 1991).
The N-terminus of HemAT has a myoglobin-like fold that distinguishes it from the heme-based oxygen sensors, FixL and Dos, whose heme-containing domains have a PAS fold (Delgado-Nixon et al., 2000; Gong et al., 2000). Thus, different molecular mechanisms have evolved for detecting oxygen. HemAT-like sensors bind O2 in a manner analogous to Hbs and Mbs, causing conformational changes that forward structural information to the C-terminal signaling domain. Homologous HemAT-like proteins have been found in archaea genomes, which suggest that oxygen sensing could be an ancient function of proteins with myoglobin-like folds. The primary function of these ancient Mb and Hbs may have been to detect environmental diatomic oxygen, and later this function evolved to remove oxygen by reaction with NO or to store O2 and transport it for aerobic metabolism.
Biphasic ligand binding is also seen for the Tar and Tsr chemotactic receptors (Biemann and Koshland, 1994; Lin et al., 1994). The appearance of half-the-site reactivity is often seen in enzymatic catalysis. Various molecular mechanisms have been proposed with some structural investigation (Anderson et al., 1999; Koshland, 1996; Peterson and Smith, 1999). Site-directed mutagenesis and structure studies have been carried out to investigate the origin of heterogeneity and/or negative cooperativity for the aspartate receptor of S. typhimurium and E. coli (Kolodziej et al., 1996; Yeh et al., 1996; Yu and Koshland, 2001). These studies reveal that Ser-68 plays a crucial role in the regulating of aspartate binding. Ligand binding at one site appears to trigger a downward shift of one α-helix that breaks the symmetry of whole structure and blocks ligand binding to the second site (Biemann and Koshland, 1994; Yu and Koshland, 2001). Thus, in the case of the Asp receptor, negative cooperativity provides the simplest structural explanation of the biphasic equilibrium binding curves. Again, regardless of exact interpretation, the biphasic nature of the ligand binding curves for the receptors examined explains how bacteria can respond to gradients over a wide range of absolute concentrations. The “design” of the HemAT structure seems to satisfy both sensitivity and robustness requirements for chemotaxis.
Acknowledgments
The authors thank Drs. Yi Dou and Antony J. Mathews for their helpful discussions on kinetics experiments and protein purification and Mr. George Blouin for assisting FTIR data collection.
This work was supported by the Robert A. Welch Foundation C-1142 (G.N.P.), by Robert A. Welch Foundation grant C-612 (J.S.O.), National Institutes of Health grants AR40252 (G.N.P.), HL47020 (J.S.O.), and GM35649 (J.S.O.), the Wisconsin Alumni Research Foundation, and the W. M. Keck Center for Computational Biology.
Abbreviations used: Tar, aspartate receptor; HemAT, heme-based aerotactic transducer; SwMb, sperm whale myoglobin; Hb, hemoglobin; Tsr, serine receptor; CheA, histidine kinase; CheB, methylesterase; CheR, methyltransferase.
References
- Alon, U., M. G. Surette, N. Barkai, and S. Leibler. 1999. Robustness in bacterial chemotaxis. Nature. 397:168–171. [DOI] [PubMed] [Google Scholar]
- Anderson, A. C., R. H. O'Neil, W. L. DeLano, and R. M. Stroud. 1999. The structural mechanism for half-the-sites reactivity in an enzyme, thymidylate synthase, involves a relay of changes between subunits. Biochemistry. 38:13829–13836. [DOI] [PubMed] [Google Scholar]
- Aono, S., T. Kato, M. Matsuki, H. Nakajima, T. Ohta, T. Uchida, and T. Kitagawa. 2002. Resonance Raman and ligand binding studies of the oxygen-sensing signal transducer protein HemAT from Bacillus subtilis. J. Biol. Chem. 277:13528–13538. [DOI] [PubMed] [Google Scholar]
- Barkai, N., and S. Leibler. 1997. Robustness in simple biochemical networks. Nature. 387:913–917. [DOI] [PubMed] [Google Scholar]
- Bass, R. B., and J. J. Falke. 1999. The aspartate receptor cytoplasmic domain: in situ chemical analysis of structure, mechanism and dynamics. Structure. 7:829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemann, H. P., and D. E. Koshland, Jr. 1994. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry. 33:629–634. [DOI] [PubMed] [Google Scholar]
- Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell. 96:131–141. [DOI] [PubMed] [Google Scholar]
- Chan, M. K. 2001. Recent advances in heme-protein sensors. Curr. Opin. Chem. Biol. 5:216–222. [DOI] [PubMed] [Google Scholar]
- Chang, A. L., J. R. Tuckerman, G. Gonzalez, R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. A. Gilles-Gonzalez. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry. 40:3420–3426. [DOI] [PubMed] [Google Scholar]
- Chu, G. C., K. Katakura, T. Tomita, X. Zhang, D. Sun, M. Sato, M. Sasahara, T. Kayama, M. Ikeda-Saito, and T. Yoshida. 2000. Histidine 20, the crucial proximal axial heme ligand of bacterial heme oxygenase Hmu O from Corynebacterium diphtheriae. J. Biol. Chem. 275:17494–17500. [DOI] [PubMed] [Google Scholar]
- Delgado-Nixon, V. M., G. Gonzalez, and M. A. Gilles-Gonzalez. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 39:2685–2691. [DOI] [PubMed] [Google Scholar]
- Djordjevic, S., and A. M. Stock. 1997. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 5:545–558. [DOI] [PubMed] [Google Scholar]
- Djordjevic, S., and A. M. Stock. 1998. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J. Struct. Biol. 124:189–200. [DOI] [PubMed] [Google Scholar]
- Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13:457–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez, M. A., G. S. Ditta, and D. R. Helinski. 1991. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 350:170–172. [DOI] [PubMed] [Google Scholar]
- Gong, W., B. Hao, and M. K. Chan. 2000. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry. 39:3955–3962. [DOI] [PubMed] [Google Scholar]
- Grebe, T. W., and J. Stock. 1998. Bacterial chemotaxis: the five sensors of a bacterium. Curr. Biol. 8:R154–R157. [DOI] [PubMed] [Google Scholar]
- Hou, S., T. Freitas, R. W. Larsen, M. Piatibratov, V. Sivozhelezov, A. Yamamoto, E. A. Meleshkevitch, M. Zimmer, G. W. Ordal, and M. Alam. 2001. Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria. Proc. Natl. Acad. Sci. USA. 98:9353–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, S., R. W. Larsen, D. Boudko, C. W. Riley, E. Karatan, M. Zimmer, G. W. Ordal, and M. Alam. 2000. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature. 403:540–544. [DOI] [PubMed] [Google Scholar]
- Imai, K. 1981. Measurement of accurate oxygen equilibrium curves by an automatic oxygenation apparatus. Methods Enzymol. 76:438–449. [DOI] [PubMed] [Google Scholar]
- Jasuja, R., Y. Lin, D. R. Trentham, and S. Khan. 1999. Response tuning in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 96:11346–11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaposi, A. D., J. M. Vanderkooi, W. W. Wright, J. Fidy, and S. S. Stavrov. 2001a. Influence of static and dynamic disorder on the visible and infrared absorption spectra of carbonmonoxy horseradish peroxidase. Biophys. J. 81:3472–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaposi, A. D., W. W. Wright, J. Fidy, S. S. Stavrov, J. M. Vanderkooi, and I. Rasnik. 2001b. Carbonmonoxy horseradish peroxidase as a function of pH and substrate: influence of local electric fields on the optical and infrared spectra. Biochemistry. 40:3483–3491. [DOI] [PubMed] [Google Scholar]
- Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 400:787–792. [DOI] [PubMed] [Google Scholar]
- Kolodziej, A. F., T. Tan, and D. E. Koshland, Jr. 1996. Producing positive, negative, and no cooperativity by mutations at a single residue located at the subunit interface in the aspartate receptor of Salmonella typhimurium. Biochemistry. 35:14782–14792. [DOI] [PubMed] [Google Scholar]
- Koshland, D. E., Jr. 1996. The structural basis of negative cooperativity: receptors and enzymes. Curr. Opin. Struct. Biol. 6:757–761. [DOI] [PubMed] [Google Scholar]
- Krikos, A., M. P. Conley, A. Boyd, H. C. Berg, and M. I. Simon. 1985. Chimeric chemosensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. USA. 82:1326–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu, S., G. C. Blouin, S. A. Premer, G. Sarath, J. S. Olson, and M. S. Hargrove. 2004. Tyrosine B10 inhibits stabilization of bound carbon monoxide and oxygen in soybean leghemoglobin. Biochemistry. 43:6241–6252. [DOI] [PubMed] [Google Scholar]
- Lai, H. H., T. Li, D. S. Lyons, G. N. Phillips Jr., J. S. Olson, and Q. H. Gibson. 1995. Phe-46(CD4) orients the distal histidine for hydrogen bonding to bound ligands in sperm whale myoglobin. Proteins. 22:322–339. [DOI] [PubMed] [Google Scholar]
- Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568–585. [DOI] [PubMed] [Google Scholar]
- Li, T., M. L. Quillin, G. N. Phillips, Jr., and J. S. Olson. 1994. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 33:1433–1446. [DOI] [PubMed] [Google Scholar]
- Lin, L. N., J. Li, J. F. Brandts, and R. M. Weis. 1994. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry. 33:6564–6570. [DOI] [PubMed] [Google Scholar]
- Manson, M. D., J. P. Armitage, J. A. Hoch, and R. M. Macnab. 1998. Bacterial locomotion and signal transduction. J. Bacteriol. 180:1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn, M. V., G. G. Prive, D. L. Milligan, W. G. Scott, J. Yeh, J. Jancarik, D. E. Koshland, Jr., and S. H. Kim. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 254:1342–1347. [DOI] [PubMed] [Google Scholar]
- Mims, M., A. Porras, J. Olson, R. Noble, and J. Peterson. 1983. Ligand binding to heme proteins. An evaluation of distal effects. J. Biol. Chem. 258:14219–14232. [PubMed] [Google Scholar]
- Olson, J. S. 1981. Stopped-flow, rapid mixing measurements of ligand binding to hemoglobin and red cells. Methods Enzymol. 76:631–651. [DOI] [PubMed] [Google Scholar]
- Ottemann, K. M., and D. E. Koshland, Jr. 1997. Converting a transmembrane receptor to a soluble receptor: recognition domain to effector domain signaling after excision of the transmembrane domain. Proc. Natl. Acad. Sci. USA. 94:11201–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, P. E., and T. J. Smith. 1999. The structure of bovine glutamate dehydrogenase provides insights into the mechanism of allostery. Structure. 7:769–782. [DOI] [PubMed] [Google Scholar]
- Phillips, G. N., Jr., M. L. Teodoro, T. Li, B. Smith, and J. S. Olson. 1999. Bound CO is a molecular probe of electrostatic potential in the distal pocket of myoglobin. J. Phys. Chem. B. 103:8817–8829. [Google Scholar]
- Quillin, M. L., R. M. Arduini, J. S. Olson, and G. N. Phillips Jr. 1993. High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin. J. Mol. Biol. 234:140–155. [DOI] [PubMed] [Google Scholar]
- Rodgers, K. R. 1999. Heme-based sensors in biological systems. Curr. Opin. Chem. Biol. 3:158–167. [DOI] [PubMed] [Google Scholar]
- Rohlfs, R. J., A. J. Mathews, T. E. Carver, J. S. Olson, B. A. Springer, K. D. Egeberg, and S. G. Sligar. 1990. The effects of amino acid substitution at position E7 (residue 64) on the kinetics of ligand binding to sperm whale myoglobin. J. Biol. Chem. 265:3168–3176. [PubMed] [Google Scholar]
- Seeley, S. K., R. M. Weis, and L. K. Thompson. 1996. The cytoplasmic fragment of the aspartate receptor displays globally dynamic behavior. Biochemistry. 35:5199–5206. [DOI] [PubMed] [Google Scholar]
- Sharff, A. J., L. E. Rodseth, J. C. Spurlino, and F. A. Quiocho. 1992. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 31:10657–10663. [DOI] [PubMed] [Google Scholar]
- Shelver, D., R. L. Kerby, Y. He, and G. P. Roberts. 1997. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl. Acad. Sci. USA. 94:11216–11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, B. A., S. G. Sligar, J. S. Olson, and G. N. Phillips, Jr. 1994. Mechanisms of ligand recognition in myoglobin. Chem. Rev. 94:699–714. [Google Scholar]
- Stock, A. M., and S. L. Mowbray. 1995. Bacterial chemotaxis: a field in motion. Curr. Opin. Struct. Biol. 5:744–751. [DOI] [PubMed] [Google Scholar]
- Stone, J. R., and M. A. Marletta. 1995. Heme stoichiometry of heterodimeric soluble guanylate cyclase. Biochemistry. 34:14668–14674. [DOI] [PubMed] [Google Scholar]
- Thomas, M. R., D. Brown, S. Franzen, and S. G. Boxer. 2001. FTIR and resonance Raman studies of nitric oxide binding to H93G cavity mutants of myoglobin. Biochemistry. 40:15047–15056. [DOI] [PubMed] [Google Scholar]
- Trent, J. T., 3rd, A. N. Hvitved, and M. S. Hargrove. 2001. A model for ligand binding to hexacoordinate hemoglobins. Biochemistry. 40:6155–6163. [DOI] [PubMed] [Google Scholar]
- Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, A. H., E. Martinez-Hackert, and A. M. Stock. 1995. Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB. J. Mol. Biol. 250:276–290. [DOI] [PubMed] [Google Scholar]
- Wong, L. S., M. S. Johnson, I. B. Zhulin, and B. L. Taylor. 1995. Role of methylation in aerotaxis in Bacillus subtilis. J. Bacteriol. 177:3985–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, J. I., H. P. Biemann, J. Pandit, D. E. Koshland, and S. H. Kim. 1993. The three-dimensional structure of the ligand-binding domain of a wild-type bacterial chemotaxis receptor. Structural comparison to the cross-linked mutant forms and conformational changes upon ligand binding. J. Biol. Chem. 268:9787–9792. [PubMed] [Google Scholar]
- Yeh, J. I., H. P. Biemann, G. G. Prive, J. Pandit, D. E. Koshland, Jr., and S. H. Kim. 1996. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol. 262:186–201. [DOI] [PubMed] [Google Scholar]
- Yi, T. M., Y. Huang, M. I. Simon, and J. Doyle. 2000. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. USA. 97:4649–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, E. W., and D. E. Koshland, Jr. 2001. Propagating conformational changes over long (and short) distances in proteins. Proc. Natl. Acad. Sci. USA. 98:9517–9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. S., J. H. Saw, S. Hou, R. W. Larsen, K. J. Watts, M. S. Johnson, M. A. Zimmer, G. W. Ordal, B. L. Taylor, and M. Alam. 2002. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol. Lett. 217:237–242. [DOI] [PubMed] [Google Scholar]
- Zhang, W., and G. N. Phillips Jr. 2003. Structure of the oxygen sensor in Bacillus subtilis: signal transduction of chemotaxis by control of symmetry. Structure. 11:1097–1110. [DOI] [PubMed] [Google Scholar]