Abstract
The plasma membrane H+-ATPase, Pma1, is an essential and long-lived integral membrane protein. Previous work has demonstrated that the Pma1-D378N mutant is a substrate for endoplasmic reticulum (ER)-associated degradation and causes a dominant negative effect on cell growth by preventing ER export of wild-type Pma1. We now show that Pma1-D378N is ubiquitylated, and it heterooligomerizes with wild-type Pma1, resulting in ubiquitylation and ER-associated degradation of wild-type Pma1. In temperature-sensitive lcb1-100 cells, defective in sphingoid base synthesis, Pma1 fails to oligomerize. At 30°C, lcb1-100 is a suppressor of pma1-D378N because wild-type Pma1 fails to heterooligomerize with Pma1-D378N; wild-type Pma1 moves to the cell surface, indicating that oligomerization is not required for delivery to the plasma membrane. Even in the absence of Pma1-D378N, wild-type Pma1 is ubiquitylated and it undergoes internalization from the cell surface and vacuolar degradation at 30°C in lcb1-100 cells. At 37°C in lcb1-100 cells, a more severe defect occurs in sphingoid base synthesis, and targeting of newly synthesized Pma1 to the plasma membrane is impaired. These data indicate requirements for sphingolipids at three discrete stages: Pma1 oligomerization at the ER, targeting to the plasma membrane, and stability at the cell surface.
The plasma membrane ATPase, encoded by PMA1, is a polytopic membrane protein whose essential physiological function is to pump protons out of the cell (1). Because Pma1 activity is critical for generating a membrane potential and regulating cytoplasmic pH, it is not surprising that numerous mechanisms exist to regulate Pma1 function at the plasma membrane (1), and to ensure efficient Pma1 delivery to the cell surface by way of the secretory pathway. Export of newly synthesized Pma1 from the endoplasmic reticulum (ER) is facilitated by a specialized COPII coat component, Lst1 (2, 3). En route to the plasma membrane, Pma1 associates with ergosterol- and sphingolipid-enriched microdomains called lipid rafts, which have been suggested to play a role in Pma1 targeting to the cell surface (4, 5). Pma1 association with lipid rafts is disrupted in temperature-sensitive lcb1-100 cells defective in sphingoid base synthesis (4). On arrival at the plasma membrane in wild-type cells, Pma1 remains stable with a half-life of ≈11 h (6). By contrast with cell surface proteins that undergo rapid turnover, Pma1 is not detectably ubiquitylated (7). The parameters that maintain Pma1 longevity are currently unknown, although a temperature-sensitive pma1 mutant with increased turnover at the plasma membrane has been recently characterized, which may provide insight into cell surface stability (8).
Because of its critical enzymatic function, PMA1 has been the focus of extensive mutagenesis studies (9). Many of these pma1 mutants are targeted for ER-associated degradation (ERAD) (10), making them a valuable resource for genetic approaches to understanding ER quality control (11). Of these mutants, Pma1-D378N is the best characterized. Pma1-D378N has increased sensitivity to trypsin, suggesting that it is misfolded (12), and it is retained in the ER and undergoes ERAD in a ubiquitin-dependent manner (11, 13, 14). Expression of Pma1-D378N causes a dominant-negative effect on growth, and in the presence of Pma1-D378N, wild-type Pma1 is also retained in the ER for degradation (11).
A variety of observations, including analysis of two-dimensional crystals, have suggested that Pma1 is oligomeric (5, 15). In this report, we demonstrate heterooligomerization of Pma1-D378N with wild-type Pma1. During ERAD, Pma1-D378N is ubiquitylated; as a result of its association with mutant Pma1, wild-type Pma1 is also ubiquitylated so that both proteins meet the same fate.
In lcb1-100 cells, defective in sphingoid base synthesis, Pma1 fails to oligomerize. At a semipermissive temperature of 30°C, lcb1-100 suppresses pma1-D378N because heterooligomerization of mutant and wild-type Pma1 does not occur. Wild-type Pma1 is allowed to move to the cell surface under these conditions, indicating that oligomerization is not essential for plasma membrane delivery. However, even in the absence of Pma1-D378N in lcb1-100 cells at 30°C, wild-type Pma1 becomes ubiquitylated and undergoes endocytosis and vacuolar degradation. At 37°C in lcb1-100 cells, Pma1 targeting to the plasma membrane becomes impaired. These data suggest a requirement for sphingolipid synthesis at several levels: Pma1 oligomerization, its targeting to the plasma membrane, and its stability at the cell surface.
Methods and Materials
Media, Strains, and Plasmids.
Standard yeast media and genetic manipulations were as described (16). F1105 is W303 (MATα ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100). RH3804 is MATα trp1 leu2 ura3 lys2 bar1-1 lcb1-100; RH3435 is the isogenic LCB1+ strain (MATa his4 leu2 ura3 lys2 bar1) (17). WQY7 (MATα trp1 leu2 ura3 lys2 bar1-1 lcb1-100 pep4∷URA3) was constructed by transformation of RH3804 with a PEP4 disruption construct [pAS173 (18)].
pND542 is a LEU2-marked centromeric plasmid bearing GAL-HA-PMA1; the hemagglutinin (HA) epitope is inserted after the second amino acid of Pma1 (19).
pWQ3 and pWQ4 are URA3-marked centromeric plasmids bearing GAL-myc-PMA1 and GAL-myc-PMA1-D378N, respectively. The plasmids were constructed by modifying pRN402U and pRN409U, URA3-marked centromeric plasmids containing PMA1 and the pma1-D378N mutation, respectively, within a 3.8-kb HindIII–SacI fragment from pPMA1.2 (12). A myc tag was introduced by replacing the 750-bp HindIII–BstEII fragment of pRN402U and pRN409U with that of pFT4; pFT4 carries PMA1 with a c-myc epitope introduced after the second amino acid. pRN402U, pRN409U, and pFT4 were obtained from Carolyn Slayman (Yale University, New Haven, CT).
Protein Induction, Immunoprecipitation, and Western Blotting.
Cells bearing epitope-tagged PMA1 under the control of GAL1 were grown at 25°C to midlogarithmic phase in synthetic complete medium with 2% raffinose. To induce synthesis of tagged Pma1, cells were shifted to medium containing 2% galactose for 4 h at 30°C. Phytosphingosine (50 μM) was added to lcb1-100 cells upon shift to galactose, and then cells were lysed. For coimmunoprecipitation, lysate (200 μg of protein) was solubilized in buffer containing 1% Nonidet P-40, 10 mM Tris⋅HCl at pH 7.5, 150 mM NaCl, and 2 mM EDTA. For ubiquitin blots, lysate was prepared in the presence of 5 mM N-ethylmaleimide, solubilized in 2% SDS, and then diluted 10-fold with buffer containing 1% Nonidet P-40 for immunoprecipitation. For coimmunoprecipitations, protein G-Sepharose and protein A-Sepharose were used to precipitate immune complexes with monoclonal anti-HA (Covance, Richmond, CA) and polyclonal anti-myc antibodies (Santa Cruz Biotechnology), respectively. For ubiquitin blots, filters were autoclaved (20) before incubating with monoclonal antibody (Zymed). Immune complexes were visualized by using an ECL Western blotting detection system (Amersham Pharmacia).
Ste3 endocytosis was assayed as described (21). Cells were transformed with a URA3-marked centromeric plasmid bearing GAL-STE3-myc (pSL2015; N. Davis, Wayne State University, Detroit, MI). Cells were grown at 30°C to midlogarithmic phase in synthetic medium without uracil containing 2% galactose. Glucose (3%) was added to terminate synthesis of Ste3-myc and lysate was prepared. Samples were normalized to protein for analysis by SDS/PAGE and Western blotting with anti-myc antibody.
Metabolic Labeling and Cell Fractionation.
Cultures were grown overnight at 25°C in minimal medium supplemented with required amino acids. Cells were resuspended at 1 OD600/ml in fresh medium and incubated at 30°C or 37°C for 10 min before adding Expre35S35S for 5 min. Synthetic complete medium with 20 mM methionine and 20 mM cysteine was added to start the chase. At various times of chase, cells were placed on ice with 10 mM azide. Lysate was prepared by vortexing with glass beads in the presence of a protease inhibitor mixture (22). Immunoprecipitates (IPs) with anti-Pma1 and anti-Gas1 antibodies (Tamara Doering, Washington University, St. Louis) were normalized to acid-precipitable cpm. Pma1 half-life in lcb1-100 cells was estimated by using fluorograms from six independent experiments for quantitation with NIH IMAGE.
Cell fractionation on Renografin density gradients was performed as described (23, 24). Fourteen fractions were collected. For analysis by IP, every two fractions were pooled and diluted into 150 mM NaCl/10 mM Tris⋅HCl, pH 7.5/2 mM EDTA/1% Triton X-100/1% sodium deoxycholate/0.1% SDS (RIPA buffer). For Western blotting, fractions were diluted 10-fold with Tris/EDTA buffer, and membranes were pelleted by centrifugation at 100,000 × g for 1 h. Polyclonal anti-Ras2 was a gift from James Broach (Princeton University, Princeton, NJ).
Indirect Immunofluorescence.
Indirect immunofluorescence was done essentially as described (25). Midlogarithmic-phase cultures in synthetic complete medium were fixed overnight with 4.4% formaldehyde in 0.1 M potassium phosphate, pH 6.5. Cells were spheroplasted and permeabilized with methanol and acetone before staining with affinity-purified rabbit anti-Pma1 and mouse monoclonal antibody against the 60-kDa subunit of V-ATPase (Molecular Probes). Primary antibody staining was detected with Cy3- and Cy2-conjugated secondary antibodies (Jackson ImmunoResearch). Images were visualized with an Olympus IX70 microscope (Lake Success, NY), collected digitally, and adjusted with Adobe PHOTOSHOP 4.0 (Adobe Systems, Mountain View, CA).
Results and Discussion
Oligomerization of Pma1 Requires Sphingoid Base Synthesis.
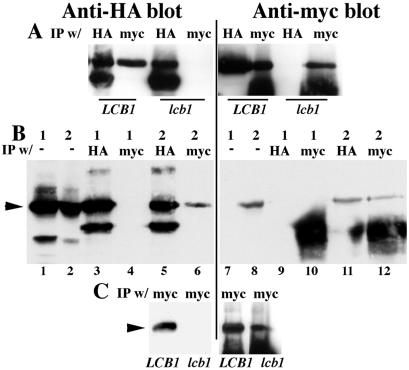
To determine whether Pma1-D378N exerts its dominant-negative effect by forming a complex with wild-type Pma1, we first established conditions for coimmunoprecipitation of Pma1. Homooligomerization of wild-type Pma1 was assayed by using constructs in which Pma1 expression is under the control of a GAL1 promoter, and independently tagged with HA and myc epitopes. Synthesis of tagged Pma1 was induced by shifting cells to medium containing galactose. Cells were lysed under nondenaturing conditions and Pma1 was immunoprecipitated with either anti-HA or anti-myc antibody. Western blot of the IPs shows that HA-Pma1 is present in an IP of myc-Pma1, and conversely, myc-Pma1 is precipitated with HA-Pma1 (Fig. 1A).
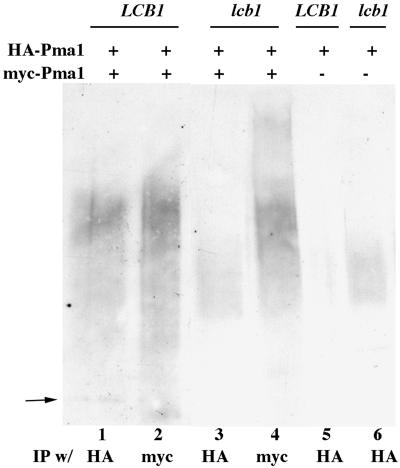
Figure 1.
Oligomerization of Pma1 is dependent on Lcb1. Cells were induced to synthesize tagged Pma1 by shifting to galactose medium for 4 h at 30°C, as described in Methods and Materials. Lysate was prepared, native IPs were performed with anti-HA and anti-myc antibodies and normalized to lysate protein, and were analyzed by Western blotting with the same antibodies. (A) Lysates were prepared from wild-type (F1105) and lcb1-100 (RH3804) cells expressing HA-Pma1 (pND542) and myc-wild-type Pma1 (pWQ3). (B) Lysates were prepared from F1105 cells bearing only GAL-HA-PMA1 (pND542) (1) or both GAL-HA-PMA1 and GAL-myc-pma1-D378N (pWQ4) (2). (C) Lysates were prepared from LCB1+ (F1105) and lcb1-100 (RH3804) cells expressing both HA-Pma1 (pND542) and myc-Pma1-D378N (pWQ4). Arrowheads indicate Pma1; IgG bands migrate below Pma1.
Fig. 1B shows heterooligomerization of Pma1-D378N and wild-type Pma1. In lysates containing both myc-tagged Pma1-D378N and HA-tagged wild-type Pma1, mutant and wild-type Pma1 were coimmunoprecipitated (Fig. 1B, lanes 6 and 11). The specificity of the coimmunoprecipitations is demonstrated by control experiments with lysate from cells expressing only HA-Pma1; these experiments show that anti-myc antibody does not bind HA-Pma1 (lanes 4 and 9).
Association of Pma1 with lipid rafts is disrupted in lcb1-100 cells (4) in which sphingolipid synthesis is reduced to 38% and 6% that of wild-type at 24°C and 37°C, respectively (17). To analyze further how Pma1 is affected by the composition of the lipid bilayer, we examined Pma1 oligomerization in lcb1-100 cells at 30°C, a semipermissive temperature. Western blots show that wild-type Pma1 tagged independently with HA and myc are expressed similarly in LCB1+ and lcb1-100 cells; however, homooligomerization of wild-type Pma1 is prevented in lcb1-100 cells (Fig. 1A). Fig. 1C shows that HA-wild-type Pma1 also fails to coimmunoprecipitate with myc-Pma1-D378N in lcb1-100 cells.
lcb1-100 Is a Suppressor of Pma1-D378N.
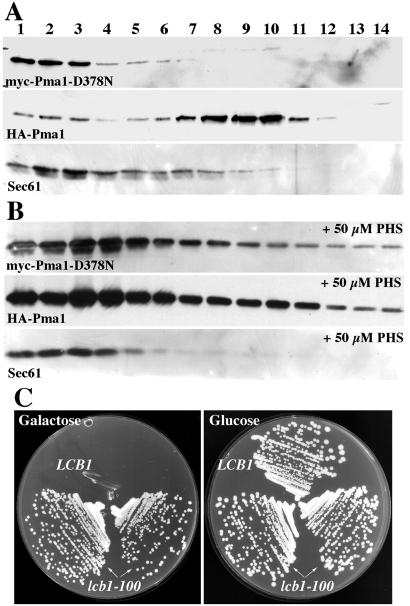
If ER retention of wild-type Pma1 is a consequence of heterooligomerization with Pma1-D378N, it seems possible that wild-type Pma1 might move to the plasma membrane when heterooligomerization is prevented. To test this hypothesis, membranes from lcb1-100 cells expressing both wild-type Pma1 and Pma1-D378N were fractionated. On Renografin density gradients, plasma membranes are found in dense fractions separated from intracellular membranes (23). Fig. 2A shows myc-Pma1-D378N localized predominantly to the lighter-density fractions 1, 2, and 3, which is where the ER membrane marker Sec61 is also concentrated. By contrast, HA-Pma1 is predominantly found in denser fractions 9 and 10. These dense fractions are enriched in endogenous (untagged) Pma1 (not shown), suggesting HA-Pma1 is delivered to the plasma membrane although it is not oligomeric in lcb1-100 (Fig. 1 A and C). Addition of the sphingoid base phytosphingosine to lcb1-100 cells restores colocalization of wild-type Pma1 with Pma1-D378N and Sec61 in the ER (Fig. 2B). Thus, defective sphingoid base synthesis in lcb1-100 cells results in failure of wild-type Pma1 to associate with Pma1-D378N, and independent localization of wild-type and mutant Pma1 to the plasma membrane and ER, respectively.
Figure 2.
Separation of wild-type Pma1 and Pma1-D378N in lcb1-100 cells. Cells (RH3804) bearing GAL-HA-PMA1 (pND542) and GAL-myc-pma1-D378N (pWQ4) were shifted to galactose-containing medium for 4 h. Lysates were prepared and fractionated. (A) Distribution of HA and myc epitopes and Sec61 was assayed by Western blotting. HA-Pma1 is predominantly found in fractions 9 and 10, whereas myc-Pma1-D378N is colocalized with Sec61 in fractions 1, 2, and 3. (B) Reversibility by phytosphingosine (PHS). Phytosphingosine (50 μM) was added on shift to galactose, and the distributions of HA and myc epitopes and Sec61 were assayed after fractionation. (C) lcb1-100 is a suppressor of pma1-D378N. Growth of cells bearing GAL-pma1-D378N (pWQ4) at 30°C on plates with synthetic complete medium without uracil containing glucose or galactose.
Because wild-type Pma1 moves to the plasma membrane despite ER retention of mutant Pma1, lcb1-100 cells can grow at 30°C in the presence of Pma1-D378N (Fig. 2C). By contrast, LCB1+ cells cannot grow when mutant Pma1-D378N is expressed, as described (11). Growth of lcb1-100 cells suggests that monomeric Pma1 at the plasma membrane is functional. These observations are in agreement with a previous report that Pma1 is active when reconstituted into liposomes as a monomer (26).
Vacuolar Degradation of Pma1 in lcb1-100.
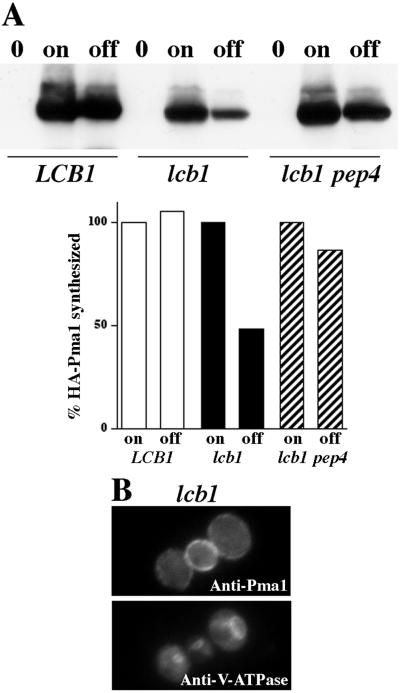
Wild-type Pma1 at the cell surface is normally long-lived (6). In lcb1-100 cells, wild-type Pma1 reaches the plasma membrane (Fig. 2 and see below). Nevertheless, Pma1 stability is decreased in lcb1-100 by comparison with that in LCB1+ cells (Fig. 3A). Pma1 turnover was examined by using the GAL-HA-PMA1 construct. HA-Pma1 synthesis was induced in the presence of galactose, and then glucose was added to shut off further expression. After a 4-h chase, HA-Pma1 levels remained constant in LCB1+ cells, but were diminished in lcb1-100 (Fig. 3A). HA-Pma1 was stabilized in lcb1-100 pep4Δ cells, indicating that Pma1 degradation is dependent on vacuolar proteinase A (Pep4) and a proteolytically active vacuole (27). These observations suggest that the stability of Pma1 is impaired in lcb1-100 cells at 30°C. Nevertheless, indirect immunofluorescence localization indicates that the steady-state distribution of Pma1 in lcb1-100 is at the plasma membrane (Fig. 3B).
Figure 3.
Stability of Pma1 in lcb1-100 cells. (A) Western blot of HA-Pma1. LCB1+ (F1105), lcb1-100 (RHY3804), and lcb1-100 pep4 (WQY7) cells bearing GAL-HA-PMA1 (pND542) were grown in medium with raffinose (0), shifted to galactose for 4 h at 30°C (on), and incubated for an additional 4 h after adding 3% glucose (off). Western blot was quantitated with NIH IMAGE and plotted as a percent of HA-Pma1 synthesized. Half-life of Pma1 is decreased in lcb1 cells; Pma1 degradation is dependent on Pep4. (B) Indirect immunofluorescence localization of Pma1. lcb1-100 (RH3804) growing at 30°C were fixed, permeabilized, stained with anti-Pma1 and V-ATPase, followed by Cy2- and Cy3-conjugated secondary antibodies.
Cell Surface Delivery and Endocytosis in lcb1-100.
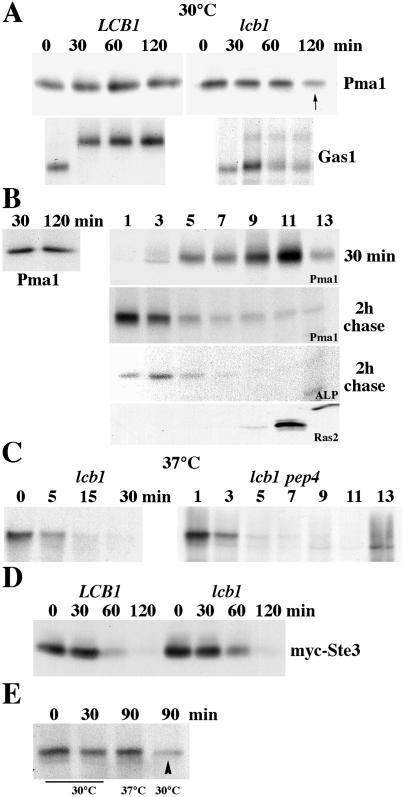
Stabilization of Pma1 in lcb1-100 pep4 cells (Fig. 3B), in conjunction with previous work suggesting that Pma1 targeting by lipid rafts is perturbed in lcb1-100 cells (4), prompted us to quantitate the fraction of newly synthesized Pma1 delivered to the cell surface. Cells were grown in minimal medium at 25°C, shifted to 30°C before pulse-labeling with [35S]methionine and cysteine for 5 min, and chased for various times. In wild-type cells newly synthesized Pma1 arrives at the plasma membrane by 30-min chase (not shown) and remains stable (Fig. 4A). ER-to-Golgi transport of the glycosylphosphatidylinositol-anchored plasma membrane protein Gas1 is rapid in wild-type cells; complete conversion of the ≈105-kDa precursor to the fully glycosylated ≈125-kDa mature form occurs within a 30-min chase (Fig. 4A) (28, 29). In lcb1-100 cells at 30°C, only a fraction of Gas1 matures by 30 min of chase, whereas the precursor persists (Fig. 4A); similar defects in Gas1 maturation in lcb1-100 cells have been reported (4, 29). In these cells, newly synthesized Pma1 has a half-life of ≈130 min (n = 6) (Fig. 4A).
Figure 4.
Cell surface delivery and endocytosis in lcb1-100 cells. (A) Pulse–chase analysis. LCB1+ (RH3435) and lcb1-100 cells (RH3804) were metabolically labeled at 30°C for 5 min and chased for various times. Immunoprecipitation of Pma1 and Gas1 was normalized to acid-precipitable cpm and was analyzed by SDS/PAGE and fluorography. lcb1-100 results are representative of six independent experiments. (B) Newly synthesized Pma1 is delivered to and removed from the plasma membrane at 30°C. lcb1-100 pep4 (WQY7) cells were pulse-labeled at 30°C for 5 min and chased for 30 min and 2 h. (Left) Pma1 was immunoprecipitated from total lysate. (Right) Cell lysate was fractionated on density gradients. Pma1 and alkaline phosphatase (ALP) were immunoprecipitated from gradient fractions. Ras2 localization was determined by Western blotting of gradient fractions. (C) Rapid vacuolar degradation of newly synthesized Pma1 at 37°C. lcb1-100 cells were metabolically labeled at 37°C for 5 min and chased for various times. (Left) Pma1 IP. (Right) lcb1-100 pep4 cells were pulse-labeled and chased for 30 min at 37°C. Lysate was fractionated on density gradients and Pma1 was immunoprecipitated from gradient fractions. (D) Endocytosis of Ste3. LCB1+ and lcb1-100 cells bearing GAL-STE3-myc (pSL2015) were grown in medium with galactose at 30°C. Cells were harvested at various times after addition of glucose (3%) and Ste3-myc was analyzed by Western blotting. (E) Stability of cell surface Pma1 in lcb1-100. Cells were pulse-labeled at 30°C for 5 min and chased for 30 min. Cells were then divided and the chase was continued for 60 min at 30°C or 37°C. Pma1 was immunoprecipitated from lysate. Cell surface Pma1 remains stable after chase at 37°C but not 30°C (arrowhead).
To examine cell surface delivery of Pma1, lcb1-100 pep4 cells were pulse-labeled and chased, and then fractionated on Renografin density gradients. At 30 min of chase, newly synthesized Pma1 is enriched in higher-density fractions 9 and 11 on Renografin gradients, coincident with the plasma membrane marker Ras2 (30) (Fig. 4B). After 2 h of chase in these cells, newly synthesized Pma1 remains stable (Fig. 4B Left) but is distributed predominantly in lighter-density fractions of the gradient, coincident with the vacuolar membrane marker alkaline phosphatase (Fig. 4B Right). These data suggest that newly synthesized Pma1 is delivered to the plasma membrane, and then undergoes internalization to the vacuole in lcb1-100 cells at 30°C.
At 37°C in lcb1-100 cells, degradation of newly synthesized Pma1 is faster than that seen at 30°C, so that by 30 min of chase, newly synthesized Pma1 is mostly degraded (compare Fig. 4C with Fig. 4A). Fractionation of lcb1-100 pep4 cells after 30 min of chase at 37°C reveals that newly synthesized Pma1 is localized only in light-density fractions. These data indicate that, by contrast with plasma membrane localization seen at 30°C in lcb1-100 cells, delivery of newly synthesized Pma1 to the cell surface is not detectable at 37°C. It is unlikely that Pma1 undergoes rapid internalization from the plasma membrane because lcb1 cells have a severe defect in endocytosis at 37°C; indeed, lcb1 cells are allelic with end8 (17). Thus, it seems that Pma1 is directed for vacuolar degradation without targeting to the plasma membrane in lcb1-100 cells at 37°C.
By contrast with the severe defect seen at 37°C, observations at 30°C support a model in which Pma1 arrives at the cell surface but fails to remain stable and undergoes endocytic transport to the vacuole. To reconcile an endocytic defect of lcb1-100 with increased turnover of cell surface Pma1, we examined endocytosis of the plasma membrane marker Ste3. Ste3, the a factor receptor, undergoes constitutive and rapid internalization and delivery for vacuolar degradation (21). Ste3 degradation in lcb1-100 cells was monitored by using a GAL-STE3-myc construct. Cells were shifted from growth in galactose to glucose to shut off Ste3-myc synthesis, and Ste3-myc remaining was measured by Western blot (Fig. 4D). At the semirestrictive temperature of 30°C, the rate of Ste3 degradation is not significantly affected in lcb1-100 cells (Fig. 4D). The endocytic defect of lcb1/end8 cells was then examined further by pulse-labeling cells at 30°C followed by chase for 30 min to allow newly synthesized Pma1 to move first to the plasma membrane. Cells were then shifted to 37°C or maintained at 30°C for an additional 60-min chase period. Under these conditions, newly synthesized Pma1 is unstable after extended chase at 30°C (Fig. 4E, arrowhead); however, Pma1 remains stable after chase at 37°C, consistent with a block in internalization at 37°C and stabilization of Pma1 at the plasma membrane.
Ubiquitylation of Pma1.
Because ERAD is mediated by ubiquitylation (10), it was of interest to determine whether association with mutant Pma1-D378N induces ubiquitylation of wild-type Pma1. Pma1 was immunoprecipitated under denaturing conditions from cell lysate containing both myc-Pma1-D378N and HA-wild-type Pma1. IPs were then examined by Western blotting with anti-ubiquitin. In agreement with reports (31), ubiquitylation of HA-wild-type Pma1 was virtually undetectable (Fig. 5, lane 5). However, in the presence of Pma1-D378N, ubiquitylation of wild-type Pma1 was observed as a high-molecular-mass smear, extending from ≥130 kDa to ≥205 kDa on an SDS/polyacrylamide gel, suggesting polyubiquitylation (Fig. 5, lane 1). Accordingly, wild-type Pma1 is an ERAD substrate in the presence of mutant Pma1 (11). In these samples, independent immunoprecipitation of myc-Pma1-D378N and HA-Pma1 was confirmed by Western blotting (not shown). Surprisingly, in lcb1-100 cells expressing wild-type and mutant Pma1, ubiquitylation of wild-type Pma1 was also detected (Fig. 5, lane 3). Because wild-type and mutant Pma1 do not heterooligomerize or even colocalize in lcb1-100 cells (Figs. 1 and 2), we reasoned that ubiquitylation of wild-type Pma1 is likely to occur independently from that of Pma1-D378N. Indeed, wild-type Pma1 is ubiquitylated in lcb1-100 cells in the absence of Pma1-D378N (Fig. 5, lane 6). The extent of polyubiquitylation of wild-type Pma1 in lcb1-100 seems less than that of Pma1-D378N (compare lanes 3 and 6 with lanes 1, 2, and 4), consistent with ubiquitylation occurring at a different cellular site. These data suggest that instability of wild-type Pma1 at the plasma membrane (Fig. 4) is accompanied by ubiquitylation in lcb1-100 cells.
Figure 5.
Ubiquitylation of Pma1. LCB1+ (F1105) and lcb1-100 (RH3804) cells bearing GAL1-HA-PMA1 (pND542) alone or with GAL1-myc-pma1-D378N (pWQ4) were shifted to galactose-containing medium for 4 h. IPs under denaturing conditions with anti-HA or anti-myc were assayed by Western blotting with anti-ubiquitin. Arrow shows position of Pma1.
Relationship Between Pma1 Stability, Sphingoid Base Synthesis, and Lipid Rafts.
At 37°C in lcb1 cells, the kinetics of Pma1 degradation (Fig. 4C) suggests that newly synthesized Pma1 is delivered to the vacuole before arrival at the plasma membrane. Moreover, because internalization from the cell surface is blocked at 37°C (17) (Fig. 4E), the possibility is unlikely that Pma1 arrives at the cell surface but has an exceedingly short residence time. Thus, severe depletion of sphingolipids impairs Pma1 targeting from Golgi to the plasma membrane, in agreement with Bagnat et al. (5).
At 30°C in lcb1-100 cells, Pma1 targeting to the plasma membrane remains essentially unperturbed (Fig. 4). By contrast with Pma1, Gas1 delivery to the cell surface is substantially delayed at 30°C, indicating intracellular transport of different proteins have different requirements for sphingolipid synthesis. In agreement with our results, export of Pma1 from the ER does not require sphingoid base synthesis and is not dependent on lipid rafts (5, 32). By contrast with reports that only a small fraction of Pma1 can reach the cell surface in lcb1-100 cells at 30°C (5, 32), our results indicate that the bulk of Pma1 is delivered to the plasma membrane. The reason for the discrepancy is not yet understood, but it seems that media differences and/or the cells' nutritional status may differentially affect sphingolipid pools in lcb1-100 cells. Indeed, media differences have been observed to affect the rate of endocytosis in lcb1-100 cells (17).
Analysis of lcb1 cells at 30°C reveals a sphingolipid requirement for cell surface stability and oligomerization of Pma1 more stringent than that for Pma1 targeting. Instability of Pma1 at the cell surface at 30°C is possibly a result of its being a monomer. Because the sphingolipid synthesis defect of lcb1-100 cells causes defective lipid raft assembly, it is also likely that rafts participate in maintaining Pma1 stability. We have recently characterized a temperature-sensitive pma1–10 mutant that fails to associate with a Triton-insoluble fraction and is also unstable at the cell surface at 37°C (8), lending support to the idea that raft association promotes Pma1 stability.
In the absence of Pma1-D378N, wild-type Pma1 is ubiquitylated in lcb1-100 cells at 30°C. Because wild-type Pma1 is accumulated at the plasma membrane in lcb1-100 at 30°C and vacuolar Pma1 is not detectable in these (PEP4+) cells (Fig. 3B), it seems plausible that ubiquitylation occurs at the cell surface to target Pma1 for internalization. In support of this idea, we have observed Pma1 ubiquitylation in lcb1-100 end3-1 cells at 30°C in which a constitutive block exists in internalization (unpublished observation). Further work is necessary to understand the relationship between membrane lipid composition and the potential for ubiquitination of wild-type Pma1.
Acknowledgments
We thank Randy Schekman and Marcus Lee for communicating their unpublished results, Cemile Guldal and James Broach for anti-Ras2, and Robert Dickson for helpful advice. This work was supported by National Institutes of Health Grant GM 58212.
Abbreviations
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- HA
hemagglutinin
- IP
immunoprecipitate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Portillo F. Biochim Biophys Acta. 2000;1469:31–42. doi: 10.1016/s0304-4157(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 2.Roberg K J, Crotwell M, Espenshade P, Gimeno R, Kaiser C A. J Cell Biol. 1999;145:659–672. doi: 10.1083/jcb.145.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimoni Y, Kurihara T, Ravazzola M, Amherdt M, Orci L, Schekman R. J Cell Biol. 2000;151:973–984. doi: 10.1083/jcb.151.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagnat M, Keranen S, Shevchenko A, Simons K. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagnat M, Chang A, Simons K. Mol Biol Cell. 2001;12:4129–4138. doi: 10.1091/mbc.12.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benito B, Moreno E, Lagunas R. Biochim Biophys Acta. 1991;1063:265–268. doi: 10.1016/0005-2736(91)90381-h. [DOI] [PubMed] [Google Scholar]
- 7.Hicke L. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 8.Gong X, Chang A. Proc Natl Acad Sci USA. 2001;98:9104–9109. doi: 10.1073/pnas.161282998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morsomme P, Slayman C W, Goffeau A. Biochim Biophys Acta. 2000;1469:133–157. doi: 10.1016/s0304-4157(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 10.Bonifacino J S, Weissman A M. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Chang A. EMBO J. 1999;18:5972–5982. doi: 10.1093/emboj/18.21.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamoto R K, Verjovski-Almeida S, Allen K E, Ambesi A, Rao R, Slayman C W. J Biol Chem. 1998;273:7338–7344. doi: 10.1074/jbc.273.13.7338. [DOI] [PubMed] [Google Scholar]
- 13.Harris S L, Na S, Zhu X, Seto-Young D, Perlin D, Teem J H, Haber J E. Proc Natl Acad Sci USA. 1994;91:10531–10535. doi: 10.1073/pnas.91.22.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeWitt N D, Tourinho dos Santos C F, Allen K E, Slayman C W. J Biol Chem. 1998;273:21744–21751. doi: 10.1074/jbc.273.34.21744. [DOI] [PubMed] [Google Scholar]
- 15.Auer M, Scarborough G A, Kuhlbrandt W. Nature (London) 1998;392:840–843. doi: 10.1038/33967. [DOI] [PubMed] [Google Scholar]
- 16.Sherman F, Hicks J B, Fink G R. Methods in Yeast Genetics: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 17.Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson B J, Riezman H. EMBO J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang A, Fink G R. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth A F, Sullivan D M, Davis N G. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swerdlow P S, Finley D, Varshavsky A. Anal Biochem. 1986;156:147–153. doi: 10.1016/0003-2697(86)90166-1. [DOI] [PubMed] [Google Scholar]
- 21.Davis N G, Horecka J L, Sprague G F., Jr J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang A, Slayman C W. J Cell Biol. 1991;115:289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo W-j, Chang A. Mol Biol Cell. 2000;11:579–592. doi: 10.1091/mbc.11.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenness D D, Li Y, Tipper C, Spatrick P. Mol Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 26.Goormaghtigh E, Chadwick C, Scarborough G A. J Biol Chem. 1986;261:7466–7471. [PubMed] [Google Scholar]
- 27.Jones E. J Biol Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- 28.Doering T L, Schekman R. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- 29.Sutterlin C, Doering T L, Schimmoller F, Schroder S, Riezman H. J Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharya S, Chen L, Broach J R, Powers S. Proc Natl Acad Sci USA. 1995;92:2984–2988. doi: 10.1073/pnas.92.7.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolling R, Losko S. EMBO J. 1997;16:2251–2261. doi: 10.1093/emboj/16.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M, Hamamoto S, Schekman R. J Biol Chem. 2002;277:22395–22401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]