Abstract
As a bacterial blue light sensor the photoactive yellow protein (PYP) undergoes conformational changes upon signal transduction. The absorption of a photon triggers a series of events that are initially localized around the protein chromophore, extends to encompass the whole protein within microseconds, and leads to the formation of the transient pB signaling state. We study the formation of this signaling state pB by molecular simulation and predict its solution structure. Conventional straightforward molecular dynamics is not able to address this formation process due to the long (microsecond) timescales involved, which are (partially) caused by the presence of free energy barriers between the metastable states. To overcome these barriers, we employed the parallel tempering (or replica exchange) method, thus enabling us to predict qualitatively the formation of the PYP signaling state pB. In contrast to the receptor state pG of PYP, the characteristics of this predicted pB structure include a wide open chromophore-binding pocket, with the chromophore and Glu46 fully solvent-exposed. In addition, loss of α-helical structure occurs, caused by the opening motion of the chromophore-binding pocket and the disruptive interaction of the negatively charged Glu46 with the backbone atoms in the hydrophobic core of the N-terminal cap. Recent NMR experiments agree very well with these predictions.
INTRODUCTION
The bacterial blue-light sensor photoactive yellow protein (PYP) originates from Halorhodospira halophila (Meyer et al.,1989; Sprenger et al., 1993). The light-sensitive part of PYP is the deprotonated para-hydroxy-coumaric acid, which is covalently attached to the protein via a thio-ester linkage to the unique cysteine at position 69 (van Beeumen et al., 1993). The buried negative charge of the chromophore is stabilized in its binding pocket by hydrogen bonds to the surrounding residues Glu46, Tyr42, Thr50, and the protein backbone (Borgstahl et al., 1995). Arg52 serves as the lid of the binding pocket, shielding the chromophore from contact with water molecules. Three-dimensional structure analysis identified two basic domains in PYP: a PER-ARNT-SIM (PAS) core (Pellequer et al., 1998), comprising amino acids 30–125, and an N-terminal cap containing the first 29 residues (Fig. 1). The chromophore is contained within the PAS-core (Borgstahl et al., 1995).
FIGURE 1.
Ribbon representation of PYP. The protein comprises two domains, the N-terminal domain (red) and the PAS-core (blue), including the chromophore binding pocket. The chromophore and its thio-ester linkage to the protein are shown in a yellow stick model. The shaded outline represents the molecular surface of PYP.
Absorption of a blue photon by the chromophore triggers a sequence of events occurring at various timescales and encompassing different parts of the protein (Hoff et al., 1994). After excitation the chromophore isomerizes from trans to cis (Kort et al., 1996), followed by the disruption of the hydrogen bond between the carbonyl oxygen of the chromophore and the protein backbone (Perman et al., 1998), resulting in the so-called pR state. Subsequently, on a microsecond timescale, a proton migrates from the Glu46 side chain to the chromophore causing a blue shift in its absorbance maximum (Meyer et al., 1993; Hendriks et al., 1999). The intermediate associated with this reversible process is denoted as pB′. Further proof of its existence was obtained with the deuterium isotope effect (Hendriks et al., 2003). More recently, resonance Raman spectroscopy has also confirmed the existence of this intermediate (Pan et al., 2004).
The proton transfer renders the protein metastable by leaving a negative charge at Glu46 and disrupting the stabilizing hydrogen-bonding network. The protonation of the chromophore is therefore also the trigger for the formation of pB, a process that occurs on a millisecond timescale (Hoff et al., 1994). The formation of pB is linked to large conformational rearrangements throughout the protein (Salamon et al., 1995; Hoff et al., 1999), sometimes even referred to as partial unfolding of the protein (van Brederode et al., 1996; Lee et al., 2001b). The observation that pB is the longest living state in the photocycle has led to the hypothesis that pB is, in fact, the signaling state of PYP (Hoff et al., 1994). The return to the ground state, completing the photocycle, is a subsecond process and includes the deprotonation and cis to trans re-isomerization of the chromophore.
Fig. 2 visualizes the chemical structure of the chromophore-binding pocket in the three states described above. In pG the chromophore is in a trans configuration, deprotonated, and hydrogen-bonded to the protonated Glu46. This hydrogen bond is retained in the pB′ configuration, whereas the proton on Glu46 has transferred to the chromophore in cis-configuration (Chen et al., 2003). The pB state has the same chemical structure, except for the disrupted hydrogen bond between the chromophore and Glu46. The hydrogen bond between the chromophore carbonyl oxygen and the protein backbone is disrupted in pB′ and reformed in pB (Pan et al., 2004).
FIGURE 2.
Structural differences among the pG, pB′, and the pB states. In pG the chromophore is deprotonated and in trans configuration, hydrogen-bonded to the protonated Glu46. In pB′, the chromophore is cis-isomerized and the proton has moved to the chromophore, leaving a negative charge on Glu46. The hydrogen bond between the groups is retained, but broken when the protein enters the pB state.
As to the nature and underlying mechanism of the changes upon the formation of pB, two areas in the protein are implicated: the chromophore-binding pocket and the N-terminal domain (van der Horst et al., 2001). CD-spectroscopy showed a loss of helical content in the protein upon activation (Lee et al., 2001a; Gensch et al., 2002), whereas the change in diffusion constants during the photocycle, as measured with transient grating spectroscopy (Takeshita et al., 2002), is best explained by conformational rearrangements and the increased exposure of protein hydrophobic interior. Results arising from the use of a hydrophobicity probe agree with the latter, specifying the chromophore-binding pocket as the region where the main rearrangements occur (Hendriks et al., 2002). Small angle x-ray scattering experiments showed an increase in the radius of gyration of N-terminal deletion mutants of PYP (Imamoto et al., 2002a; Harigai et al., 2003), confining the conformational changes to the PAS core and the first helix of the N-terminal domain. Finally, time-resolved fluorescence measurements on the unique tryptophan residue indicate that this residue has varying degrees of solvent contact during the photocycle (Kandori et al., 2000; Gensch et al., 2004).
Time-resolved crystallography on constantly illuminated crystals provided the first three-dimensional model of pB (Genick et al., 1997). In contrast to the results described above, this structure only shows differences in side-chain orientation in the chromophore binding pocket. More recent results imply, however, that the conformational changes occur throughout the protein (Schmidt et al., 2004; Ren et al., 2001). NMR spectroscopy indicated that the formation of a buried negative charge on Glu46 drives the conformational changes in the protein (Craven et al., 2000; Derix et al., 2003).
Molecular simulation methods, such as molecular dynamics (MD), can, in principle, provide a complementary atomistic picture of the changes occurring in PYP during its photocycle. For instance, by employing a combination of quantum mechanical and molecular mechanical calculations Groenhof and co-workers recently proposed a model describing the initial events, including the excitation and subsequent rearrangements (Groenhof et al., 2004). Groenhof and co-workers also used parameters from semi-empirical calculations for a protonated chromophore (Groenhof et al., 2002b), embedded in an equilibrated protein structure, to show initial rearrangements in the chromophore binding pocket and the N-terminal domain (Groenhof et al., 2002a). The proton transfer from Glu46 to the chromophore is assigned as the trigger for the conformational change to pB (Groenhof et al., 2002a). Simulation data on wild-type PYP and a mutant E46Q further elucidate this trigger as the weakening of the hydrogen bond between the chromophore and Glu46 (Antes et al., 2002).
In the first attempt to model the signaling state pB using MD, the ground-state chromophore vinyl bond was replaced with a single bond potential, to allow for faster rearrangements (van Aalten et al., 1998). In another simulation study starting from the crystal structure obtained from illuminated crystals, water molecules enter the chromophore binding pocket and hydrate Glu46 (Shiozawa et al., 2001). Both simulations show a slow drift away from the original crystal structure. Unfortunately, conventional MD is limited to relatively small timescales in the order of nanoseconds, whereas the formation of pB from pB′ is a millisecond process.
A recent study therefore used a coarse-grained Hamiltonian, which resulted in a description of pB as partially unfolded states stabilized by conformational entropy in the N-terminal domain and vibrational entropy around the chromophore (Itoh and Sasai, 2004). A disadvantage of using such coarse-grained models is that these are not able to resolve the atomic structure of the pB state. In conclusion, although previous studies investigated the initial conditions for pB formation, the extent of the conformational rearrangements, and the actual solution structure of pB, are still unknown.
The long timescales involved in the formation of pB are partly caused by free energy barriers between the several metastable states. One way to overcome the trapping of biomolecular systems in local minima between these barriers is to perform parallel tempering (PT) simulations (Swendsen and Wang, 1986; Marinari and Parisi, 1992; Sugita and Okamoto, 1999). The PT method, also known as replica exchange, combines multiple molecular dynamics simulations with a temperature-exchange Monte Carlo process (Frenkel and Smit, 2002). The method has been proved useful in folding/unfolding studies on peptides, including α-helices (Nymeyer and García, 2003), a β-hairpin (Zhou, 2004; García and Sanbonmatsu, 2001), protein A (García and Onuchic, 2003), and Trp-cage (Zhou, 2004; Yang et al., 2004). In this work we employ the parallel tempering technique to overcome the free energy barriers for the formation of pB and study the conformational differences between the receptor and signaling states of PYP.
METHODS
During signal transduction, the photoactive yellow protein (PYP) undergoes conformational transitions toward the signaling state at a microsecond-and-millisecond timescale. In this work we investigate the formation of this signaling state by performing five independent parallel tempering (PT) simulations, each based on a different starting structure. The crystal structures of PYP in the dark and the bleached state served as two of the starting configurations, PDB codes 2PHY (Borgstahl et al., 1995) and 2PYP (Genick et al., 1997), respectively. Coordinates based on NMR constraints (Dux et al., 1998) served as input for two more PT simulations. Conformation 11 in the ensemble of solution structures (PDB code 3PHY) has a hydrogen bond between the chromophore and Glu46, and hence was selected as a starting structure. As a starting point for the formation of pB, we replaced the original chromophore coordinates in the NMR structure by coordinates from a crystal structure of a cryotrapped photocycle intermediate (Kort et al., 2004) in cis configuration. This replacement did not result in unfavorable atomic interactions. The ground state pG differs from the signaling state pB not only in conformation (including the trans or cis configuration of the chromophore), but also in the protonation of the chromophore and Glu46. In pG, the phenolic oxygen of the chromophore is deprotonated, and Glu46 is protonated. Conversely, the chromophore is protonated and Glu46 is deprotonated in the pB state (Fig. 2). The altered NMR structure with the cis chromophore is not yet the pB signaling state. Instead, this configuration represents the pB′ state in the photocycle of PYP. The last of the five PT simulations was initiated from a selected on-way conformation in the NMR pB′ run. This last run was included to speed up the slow equilibration toward the pB state.
Polar and aromatic hydrogen atoms were added to all four starting structures, taking into account the correct protonation state of Glu46 and the chromophore. Aliphatic groups were included as heavy carbon atoms (united-atom model). Subsequently, the protein configurations were placed in a periodic dodecahedral box, and immersed in SPC water (Berendsen et al., 1981). The box size included the protein and a radius of 1.5 nm around it. Water molecules that overlapped with the protein or resided in internal hydrophobic cavities were removed. Six water molecules at the most electronegative positions were replaced by sodium ions to neutralize the charge of −6 on the protein. Next, the four systems were energy-minimized using 200 steps of the conjugate gradient method (Lindahl et al., 2001), and equilibrated to dissipate excess energy and relax the box volume. Positions of the water molecules and the hydrogens were relaxed for 10 ps, followed by 100 ps of equilibration of the whole system in the NpT ensemble. The GROMOS96 force field was used to describe the bonded interactions between the atoms (van Gunsteren and Berendsen, 1987; van Buuren et al., 1993; Mark et al., 1994). Van der Waals interactions were treated with a cutoff (Lindahl et al., 2001) of 1.4 nm, and particle-mesh Ewald handled the long-range electrostatics (Lindahl et al., 2001). Using constraints, LINCS for interactions between protein atoms (Hess et al., 1997) and SETTLE for water interactions (Miyamoto and Kollman, 1997), allowed a time step of 2 fs. Parameters for the chromophore were taken from Groenhof et al. (2002a). Prepared as such, the systems were used as input for the PT simulations.
The GROMACS software package was used for equilibration and parallel tempering, in combination with a PERL script that performed the temperature swaps. Every 1 ps, attempts to exchange temperatures between systems were made. The Berendsen thermostat (Berendsen et al., 1984), with a coupling constant of 0.1 ps, allowed fast adaptation of the systems to temperatures ranging from 280 K to 640 K. Although the Nosé-Hoover algorithm is, in principle, the correct thermostat in constant temperature simulations, we found it adjusted too slowly to equilibrate within 1 ps. However, it not likely that changing the thermostat will alter the qualitative results in this work.
Coordinates before every temperature-swapping attempt were written out and used for subsequent analysis. The simulations were performed on a homebuilt Beowulf cluster, using 32 AMD processors, each running two replicas simultaneously. The temperatures for the xtal simulations ranged from 283 K to 630 K. In the NMR simulations the temperatures were set between 300 K and 560 K for both the pG and pB′ simulations. The temperatures in the pB parallel tempering run varied between 282 K and 645 K. The temperature gap was initially estimated by a linear dependence on the inverse temperature, and turned out afterward to give rise to a reasonably uniform acceptance ratio of ∼30%, around the entire temperature domain. After a 2 ns equilibration period, the five independent parallel tempering runs were continued for on average 8–10ns, amounting to a total simulation time of 64 × 10 × 5 ≈ 3200 ns.
Various analysis tools included in the GROMACS molecular dynamics package were used here to calculate fluctuations and several order parameters: the distance between the centers of mass between two groups, the number of hydrogen bonds, and the radius of gyration (Lindahl et al., 2001). To analyze the extent of solvation in a protein region we subtracted the number of solvent-protein hydrogen bonds (Nprotein–solvent) from (two times) the intraprotein hydrogen bonds (Nprotein–protein) to obtain the hydrogen-bond difference parameter:
 |
(1) |
This parameter is positive when the protein is not solvated and becomes negative when more water enters the protein region included in the analysis, replacing the intraprotein hydrogen bonds. A donor-acceptor distance <0.35 nm and a donor-hydrogen-acceptor angle of <60° defined a hydrogen bond. The number of water molecules surrounding a residue and visual inspection in VMD (Humphrey et al., 1996) were also part of the analysis. Free energy landscapes or profiles as a function of (a combination of) the above order parameters can give insight into the (meta)stability of PYP. They can be computed by taking the negative natural logarithm of one- or two-dimensional probability histograms, which result from the sampling of the order parameters at a fixed temperature during the course of a PT simulation.
RESULTS
Using the PT technique, we have investigated the functional conformational transitions during the photocycle of PYP. To probe the differences between simulations starting from crystal structures and conformations based on NMR constraints, both the crystal structure 2PHY (Borgstahl et al., 1995) and a conformation from the NMR solution structure 3PHY (Dux et al., 1998) served as starting points for simulations of the receptor state pG. To extend this comparison to the signaling state, the crystal structure of a photocycle intermediate, 2PYP (Genick et al., 1997) and a manually altered NMR conformation from 3PHY (Dux et al., 1998), initiated simulations of the pB state. The label xtal or crystal denotes simulations starting from crystal structure coordinates, and NMR indicates simulations that started with a conformation from a solution structure (see Methods for a more detailed description).
Fig. 3 shows the root mean square fluctuation in the atomic displacements as a function of the residue number. The values are averaged over all atoms in each residue. The error bars indicate the variance of the fluctuations using simulation blocks of 1 ns. The peaks in the graphs correspond to loops in the protein structure, whereas the stable parts (below 0.2 nm) correspond to strands of the central β-sheet. With the increase of temperature, the fluctuations in the flexible loops increase also, whereas the β-strands fluctuate at a value of ∼0.2 nm. Additional fluctuation peaks arise around residues in the chromophore binding pocket (CBP), in particular for residues 42–52, 68–72, and 96–100. The first two stretches are part of a helical structure, and the last stretch is a loop connecting two β-strands. Higher temperatures cause larger fluctuations in these parts of the protein. The N-terminal domain shows large fluctuations of 0.5–1.0 nm for the first two residues, independent of temperature and starting configuration. Regarding the secondary structure in the N-terminal domain, as assigned on basis of the structural elements in the ground-state crystal structure (Borgstahl et al., 1995), the first helix (residues 11–15) is more stable than the second (residues 19–23). In the NMR simulations of pG and pB the difference in fluctuation between the first and the second helices is ∼0.2 nm, and at higher temperatures this difference is more pronounced. Unfortunately, the fluctuation graphs do not provide detailed atomistic information on the rearrangements in either the chromophore-binding pocket or the N-terminal domain.
FIGURE 3.
Fluctuations in the protein. For pG (left panel) and pB (right panel), deviations in atomic displacement are averaged in nm for each residue in PYP at 300 K, 417 K, and 510 K. Using fluctuations over 1-ns time intervals, the error bars show the drift during sampling. The labels xtal and NMR indicate the starting structure for the PT. At the bottom, the secondary structure in the protein is indicated by thick, shaded bars for α-helices and smaller, solid bars for β-strands.
The convergence of parallel tempering results starting from entirely different conformations, but with identical chemical structure (i.e., simulation topology), would serve as a good test for the quality of the simulation method. Although conventional MD simulations are not able to reach global equilibrium for PYP when starting from different conformations, the fluctuations of the crystal PT simulations overlap remarkably well with those of the NMR simulations. This overlap was only achieved after the highest temperature in the crystal PT runs was set to 640 K, whereas the NMR simulations had a maximum temperature of 560 K only. Differences in the interaction between protein and solvent might explain this behavior. Indeed, the potential energy of this nonbonded interaction was ∼200 kJ/mol higher for the NMR-based simulations than for the crystal-based simulations.
Focusing on the rearrangements taking place in the chromophore binding pocket, Fig. 4 shows the time evolution of the two-dimensional free energy diagrams of the NMR-based pB′ simulation as a function of the distance dHC4–Glu46 between the centers of mass of the phenol(ate) ring of the chromophore and the side chain of Glu46, and the hydrogen-bond difference ΔCBP in the CBP (for a definition of Δ, see Methods). Included as part of the hydrogen-binding pocket are Tyr42, Glu46, Thr50, Cys69, Phe96, Met100, and the chromophore itself. The values of ΔCBP range from +14 in the crystal structure to −24 in the completely solvated pB conformation. The first frame in Fig. 4 shows the profile for a chromophore that is buried in the protein and participates in a hydrogen-bonding network formed by Tyr42, Glu46, and Thr52. Here, the free energy minimum lies at a Glu46-chromophore distance of 0.63 nm and a hydrogen-bond difference of ΔCBP = 5. The subsequent time frames show a consistent shift of the sampling toward an increasingly negative value for ΔCBP, combined with a larger distance between Glu46 and the chromophore dHC4–Glu46. A new minimum appears at dHC4–Glu46 = 1.64 nm and ΔCBP = −2, in a region that has fewer intra-CBP-hydrogen bonds and a larger distance between the chromophore and Glu46. Visual inspection shows that the negative charge on Glu46 destabilizes the hydrogen-bonded connections and causes the intrusion of water molecules in the protein interior, as indicated by the increasingly negative value for ΔCBP. Ultimately, Glu46 breaks loose from the hydrogen-bonding network and becomes exposed to solvent. During the solvent exposure of the chromophore a hydrogen bond forms between the backbone amide of Cys69 and the carbonyl oxygen on the chromophore, stabilizing the solvent-oriented conformation. This bond is absent while the chromophore is still buried in the protein.
FIGURE 4.
Time evolution of a strained chromophore in the confinement of the protein environment. The free energy diagrams for the chromophore binding pocket are plotted for each nanosecond of the parallel tempering simulation starting from a protonated cis-chromophore (Kort et al., 2004) in an equilibrated NMR configuration of PYP. The free energy is plotted as a function of the distance between the chromophore and Glu46, and the hydrogen-bond difference ΔCBP (Eq. 1) in the chromophore-binding pocket. The contour lines indicate the kBT levels, decreasing with darker shading. Open areas have not been sampled.
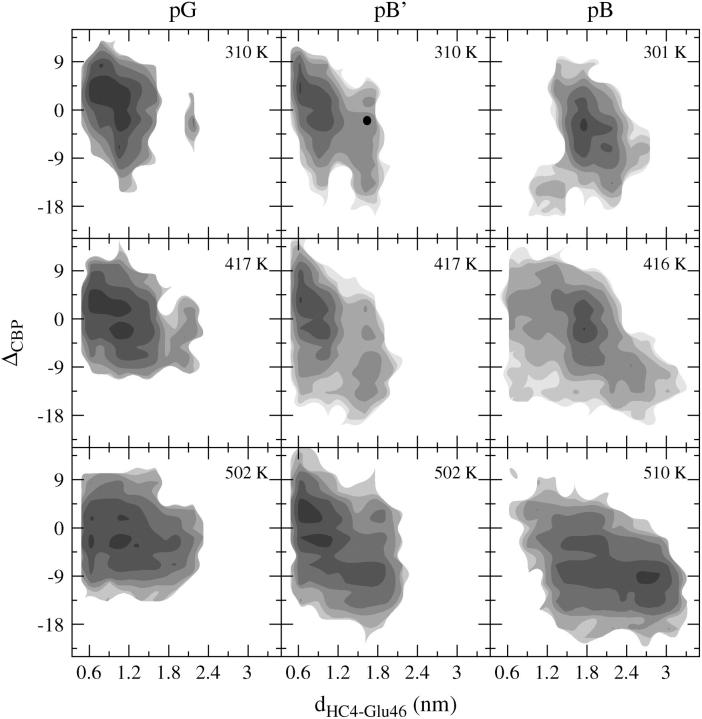
In Fig. 5 the PT free energy of the CBP at three different temperatures is shown for the NMR simulations of the receptor state pG, the initial signaling state pB′, and a final signaling state pB. The complete sampling of the entire configuration space between pB′ and pB is too slow, even for the PT simulations. To speed up the sampling, a protein configuration from the 310 K run of the pB′ PT simulation that has both the chromophore and Glu46 solvent-exposed was selected as the starting point for a new set of PT replicas. The solid circle in the 310 K frame of the pB′ simulation indicates this chosen configuration and the label pB indicates the new PT simulation run.
FIGURE 5.
Free energy diagrams at three different temperatures as a function of the distance between the chromophore and Glu46, and the hydrogen-bond difference ΔCBP (Eq. 1) in the chromophore-binding pocket for PT NMR simulations of the pG, pB′, and pB states, indicated by the labels. The contour lines indicate the kBT levels, decreasing with darker shading. Open areas have not been sampled. A protein conformation within the solid area in the 310-frame of the pB′ simulation is selected as a starting point for a new PT run.
In all simulations, the potential energy contributions become less relevant at higher temperatures, which leads to broader minima and shallower free energy profiles, as the free energy profiles at ∼500 K clearly show. The regions sampled in the pG simulation and in the pB′ simulation are similar and show minima at dHC4–Glu46 = 0.63 nm and ΔCBP = 5. In both the pG and the pB′ states the hydrogen-bonding network fluctuates between a tightly connected and a more loose structure. The free energy profile of pG at 310 K shows a high barrier that separates a state where the chromophore-Glu46 distance is 1.63 nm at the most, from a state where this distance has a value of 2.19 nm, thus stabilizing the ground state. This free energy barrier disappears at higher temperatures. No barriers larger than a few kBT are present in the free energy profiles of pB′ at 310 K, and of pB at 301 K, suggesting these states are much less stable. The pB state at 301 K has a different profile in comparison to the pB′ simulation with a second minimum at dHC4–Glu46 = 1.78 nm and ΔCBP = 3. At 416 K the free energy profile of pB extends into two directions; one is a return to the region also sampled by the pB′ simulation, with a small dHC4–Glu46 value and ΔCBP larger than zero, the other samples chromophore-Glu46 distances of above 3 nm and values for ΔCBP that indicate that solvent molecules entered into almost all CBP protein-protein hydrogen bonds.
Fig. 6 displays the free energy profiles for the pG and pB simulations initiated from a crystal structure. The profile for the pG state at 302 K shows similar states to those in the profile of the pG NMR simulation at 310 K. The barrier separating the states is less high, and the free energy profile is shallower. At 505 K the sampled region extends to values >3.4 nm. Extension of the PT simulation temperature range to higher temperatures, which may be the cause for these differences with regard to the NMR simulation of pG. The pB xtal simulation seems to be in a state that lies between the states sampled in the pB′ and pB NMR simulations. At a temperature of 302 K two states occur, at dHC4–Glu46 = 1.15 nm and ΔCBP = 2 and at dHC4–Glu46 = 2.20 nm and ΔCBP = −15. The former is more similar to the pB′ NMR simulation and contains a buried chromophore and Glu46, whereas the latter is closer to the NMR-based pB results, with the chromophore and Glu46 exposed to solvent. At higher temperatures both the free energy profiles resemble those sampled for pB in the NMR simulation.
FIGURE 6.
Free energy diagrams at three different temperatures as a function of the distance between the chromophore and Glu46, and the hydrogen-bond difference ΔCBP (Eq. 1) in the chromophore-binding pocket for PT xtal simulations of the pG and pB states, indicated by the labels. The contour lines indicate the kBT levels, decreasing with darker shading. Open areas have not been sampled.
Upon triggering of the photocycle, not only does the chromophore-containing part in PYP undergo a conformational transition, but the N-terminal cap, comprising the first 29 amino acids of PYP, also partially unfolds. Its role in the conformational transitions and the degree of unfolding is still unclear, largely because the crystal structure shows more prominent α-helices than the NMR experimental solution structure predicts. One measure for unfolding is the radius of gyration Rgyr of the hydrophobic core. Three phenylalanine residues at positions 6, 28, and 112 make up the hydrophobic core in the N-terminal domain. A second order parameter for unfolding is the hydrogen-bond difference ΔN–term in the helical residues in the N-terminal cap, measuring the solvent exposure. Since some helical residues are always solvent-exposed, ΔN–term has more negative values in comparison to ΔCBP, ranging from ΔN–term = 11 for the crystal structure to ΔN–term = −40 for complete solvation. Fig. 7 top shows the free energy profiles for the N-terminal domain for the NMR-based pG and pB PT simulations, whereas Fig. 7 bottom shows those for the crystal-based simulations. The conformations sampled for the N-terminal cap at ∼300 K in the pG simulations have a minimum at (Rgyr = 0.50 nm, ΔN–term = −16) for the NMR simulation and at (Rgyr = 0.45 nm, ΔN–term = −5) for the crystal simulation. The values for ΔN–term differ significantly, relating to the fluctuating N-terminal helices in the NMR simulation and the well-defined helical structures in the xtal simulation. The free energy profile of the pG crystal simulation shows a second minimum at Rgyr = 0. 75 nm and ΔN–term = −25, closely resembling a similar state in the NMR-pG simulation. The large values for the radius of gyration in this state indicate that expansion of the hydrophobic core is closely linked to loss of α-helical structure. At higher temperatures, the free energy profiles of both pG simulations are similar.
FIGURE 7.

Free energy profiles of the N-terminal cap in the receptor state pG and the signaling state pB as a function of the radius of gyration of the hydrophobic core and the hydrogen-bond difference ΔN–term (Eq. 1) in the helical residues, with (top) NMR simulations and (bottom) xtal simulations. The contour lines indicate the kBT levels, decreasing with darker shading. Open areas have not been sampled.
The free energy profiles for the NMR and crystal simulations exhibit large differences (see Fig. 7). In the xtal simulation of the pB state the hydrophobic core is very compact, since Rgyr expands beyond 0.6 nm only at a temperature of 505 K. ΔN–term varies widely, from 5 to −30, unrelated to the core compactness. In contrast, the pB-NMR simulation free energy profile has an minimum at Rgyr = 0.60 nm with few intraprotein hydrogen bonds, since ΔN–term varies between −18 and −27. Separated by a barrier, a second minimum at Rgyr = 0.80 nm and less negative values for ΔN–term occur, and this also appears in the pG simulations. A third state, at (Rgyr = 1.00 nm, ΔN–term = −20), represents a widely expanded hydrophobic core with few α-helical hydrogen bonds. At higher temperatures the barrier separating these states decreases to a few kBT and eventually disappears at 510 K.
DISCUSSION
The main interest in this work is the elucidation of the conformational transitions during the photocycle of the PYP, especially those linked to the formation of the signaling state. Several experiments hinted at the extent and details of the structural changes that occur during pB formation, including molecular simulation studies (Shiozawa et al., 2001; Groenhof et al., 2002a; Itoh and Sasai, 2004), although it is inefficient to use MD simulations to access the long timescales necessary for escaping a local minimum. This has prevented most studies from sampling the signaling state. Parallel tempering (PT) combines conventional MD replicas at different temperatures with a Monte Carlo scheme for exchanging temperatures. In this way PT overcomes free energy barriers, and hence enabled us to expand the exploration of the configurational space of PYP. The implementation of the PT algorithm in the case of PYP involved the choice of what starting point would be most relevant. Complete and well-defined crystal structures are most accurate and thus serve best to initiate molecular simulation procedures, whereas solution structures suffer from the fact that less well-defined protein regions introduce additional flexibility (Fan and Mark, 2004). On the other hand, Rajagopal and co-workers state that the crystalline lattice restrains PYP (and the N-terminal domain in particular) from losing helical structure during the signaling process (Rajagopal et al., 2005).
In our PT simulations, different initial starting structures do not result in different time-averaged fluctuation profiles, as illustrated in Fig. 3. The average fluctuation per residue does not differ significantly for simulations based on coordinates from x-ray diffraction experiments or NMR spectroscopy. Looking in more detail at the chromophore binding pocket (Figs. 5 and 6), a similar picture emerges: loss of structure, exposure of protein interior, and the intrusion of water molecules in the protein core occur at a similar level in simulations that were initiated from different starting structures. However, there is an exception, related to a topic of debate in the literature: the role and extent of unfolding of the N-terminal cap. In the crystal simulations the N-terminal domain is more compact, with clearly defined helices, whereas in the NMR simulations it shows a looser conformation in which water molecules can enter more easily. The latter conformation agrees with the observation that the interaction energy between water molecules and the protein is higher in the NMR simulations than for the xtal simulations. At higher temperatures, the crystal simulations also visit the looser conformation. The structure of the N-terminal domain in crystal structures may represent a conformation, induced by crystal contacts, which is packed too tightly to represent the protein in an aqueous environment, in agreement with the crystallographic work on a mutant, E46Q, of PYP (Rajagopal et al., 2005; Anderson et al., 2004).
The fluctuations shown in Fig. 3 indicate that the second N-terminal helix is less well defined in comparison to the first. Residues 11–15 have in each PT simulation lower average fluctuations than residues 19–23. Imamoto et al. (2002a) find that the removal of the second helix does not affect the change in radius of gyration of the protein when exposed to light, whereas the removal of the first helix induces an increase in volume during the photocycle (Imamoto et al., 2002b). Moreover, removing the first helix also affects this structural change (Harigai et al., 2003). These observations agree well with the explanation that the second α-helix in the N-terminal domain fluctuates between two configurations—a well-structured, helical form and a disordered, looplike form. Both occur in our simulations of the receptor and the signaling state, although in the latter only at higher temperatures for the crystal simulations. The first α-helix in the N-terminal cap is ordered in the pG state, but loses structure, and suffers water intrusion upon solvent exposure of the chromophore.
Figs. 5 and 6 lead to the conclusion that a different chemical composition (pG versus pB′ or pB) of the chromophore-binding pocket leads to a different free energy profile. If the temperature is sufficiently high, water molecules enter the binding pocket and disrupt its integrity regardless of its state. The location of the negative charge determines the mechanism of CBP-disruption. In the receptor state pG, the chromophore contains a negative charge, delocalized over its whole length. A strong interaction exists between the chromophore and the positively charged Arg52, the latter being in contact with solvent molecules. A fluctuation causing Arg52 to move more toward the solvent leaves the chromophore prone to solvent exposure. Balancing the favorable interaction with solvent are surrounding residues that contribute to a hydrogen-bonding network that further stabilizes the negative charge on the chromophore. When these connections inside the CBP are broken at high temperature, the chromophore shifts toward the solvent and disrupts the chromophore-binding pocket.
In the case of the signaling state pB, the negative charge is located at Glu46 and localized over a smaller set of atoms in comparison to the chromophore. This has two consequences with regard to the CBP-disruption mechanism: first, the negatively charged Glu46 destabilizes the hydrogen-bonding network inside the protein and allows water molecules to access the protein interior and second, the interaction between Arg52 and the chromophore has become less favorable. Consequently, the sequence of events has reversed in the signaling state with respect to the receptor state; first Glu46 becomes solvent-exposed, followed by emergence of the chromophore into the solvent. The final situation in both the pG and pB states is the same, as depicted in Fig. 8: a huge disruption of the chromophore-binding pocket, resulting in loss of α-helical content, in agreement with literature. Of course, the most important difference is that at room temperature the CBP disruption in receptor state pG is much more unlikely than in the pB state, as is found in experiments.
FIGURE 8.

Ribbon representation of PYP in (top) the crystal structure of the receptor state pG and (bottom) a typical conformation of the signaling state pB at ambient conditions, taken from the PT run and clearly exhibiting pB features. Red indicates the N-terminal cap, with the Phe6, Phe28, and Phe121 in space-filling model representation to show the hydrophobic core. The yellow stick models represent the chromophore and Glu46.
The conformational rearrangements in the CBP relate to those in the N-terminal cap. When Glu46 becomes fully solvent-exposed, it interacts with the backbone of the N-terminal hydrophobic core, thus disrupting it. This observation leads to the conclusion that our simulations have predicted the conformation of the signaling state pB of PYP. This state has the following characteristics: The hydrogen-bonding network in the CBP has disappeared, and both the chromophore and Glu46 are fully solvent-exposed. These rearrangements cause the loss of α-helical structure in the first and last helices in the PAS-core. Glu46 interacts with the N-terminal cap, causing conformational instabilities in the N-terminal hydrophobic core and α-helices. Fig. 8 summarizes these observations and shows the crystal structure of the receptor state next to a typical room temperature conformation of the signaling state. Recent results from NMR experiments on a truncated form of PYP, where removal of the N-terminal cap has led to a extended pB lifetime, agree very well with our prediction (C. Bernard, K. Houben, N. Derix, D. Marks, M. van der Horst, K. Hellingwerf, R. Boelens, R. Kaptein, and N. van Nuland, unpublished results).
In this work, chromophore protonation occurred through removal of the proton at Glu46 to place it at the chromophore, neglecting energetic considerations, such as whether the protein environment had assumed a configuration favorable for proton transfer. Although a mechanism involving a direct proton transfer mechanism from Glu46 to the chromophore is certainly possible, this manual transfer is probably too crude to describe the change of protonation states in the chromophore-binding pocket. Our PT results indicate that the proton transfer in PYP during its photocycle might actually be more complicated, involving solvent intermediates. The reverse reaction, relevant for the recovery of the receptor state, may also include multiple pathways.
We should stress that the parallel tempering simulations, although expanding the exploration of the PYP conformation space, are still not completely converged. The complete equilibration of all states requires that each replica makes many trips from the lowest to the highest temperature, which might take a multiple amount of the simulation time yet invested. The GROMACS force field might also have deficiencies and underestimate the stability of partially unfolded protein structures. An exhaustive comparison between different force fields is beyond the scope of this work. However, despite all this, we believe that the qualitative results obtained in this work are reproducible, and that the main conclusions are warranted.
CONCLUSION
In this work we have used parallel tempering simulations to study the conformational transitions associated with the photocycle of the PYP. The main goal of this work is the prediction of the mechanism of formation and the structure of the signaling state pB (see Fig. 8). Comparing several independent PT simulation series we found the following mechanism for pB formation. After the initial isomerization and proton transfer from Glu46 to the chromophore, the negative charge is located at Glu46 and localized over a smaller set of atoms in comparison to the previous location of the negative charge at the chromophore. This negative charge has two effects on the chromophore binding pocket: first, the negatively charged Glu46 destabilizes the hydrogen-bonding network inside the protein and allows water molecules to access the protein interior; and second, the interaction between Arg52 and the chromophore has become less favorable. The next step is the solvent exposure of Glu46, followed by the solvent exposure of the chromophore. The solvent exposure of Glu46 also has an effect on the stability of the hydrophobic core in the N-terminal domain, resulting in the partial unfolding seen in several experiments. In summary, we have predicted the structure and the formation mechanism of the signaling state in the photocycle of PYP. Our results are qualitative, but compare well to very recent NMR experiments (C. Bernard, K. Houben, N. Derix, D. Marks, M. van der Horst, K. Hellingwerf, R. Boelens, R. Kaptein, and N. van Nuland, unpublished results).
Future simulation work will focus on the recovery reaction from the signaling state to the ground state, thus completing a theoretical model description of the photocycle.
The PT has proved very powerful in sampling rugged energy landscapes such as occur in protein conformational transitions. However, the technique cannot give detailed information of the kinetics of the conformation transitions. In the near future we will employ other advanced simulation techniques such as transition path sampling and related techniques to access the relevant kinetic information in PYP (Bolhuis, 2005).
References
- Anderson, S., V. Srajer, R. Pahl, S. Rajagopal, F. Schotte, P. Anfinrud, M. Wulff, and K. Moffat. 2004. Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein. Structure. 12:1039–1045. [DOI] [PubMed] [Google Scholar]
- Antes, I., W. Thiel, and W. F. van Gunsteren. 2002. Molecular dynamics simulations of photoactive yellow protein (PYP) in three states of its photocycle: a comparison with x-ray and NMR data and analysis of the effects of Glu46 deprotonation and mutation. Eur. Biophys. J. 31:504–520. [DOI] [PubMed] [Google Scholar]
- Berendsen, H., J. Postma, W. van Gunsteren, A. DiNola, and J. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans, editors. 1981. Pullman intermolecular forces. In Interaction Models for Water in Relation to Protein Hydration. D. Reidel Publishing, Dordrecht, The Netherlands. 331–342.
- Bolhuis, P. G. 2005. Kinetic pathways of β-hairpin (un)folding in explicit solvent. Biophys. J. 88:50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgstahl, G. E. O., D. R. Williams, and E. D. Getzoff. 1995. 1.4 Ångstrom structure of photoactive yellow protein, a cytosolic photoreceptor—unusual fold, active-site, and chromophore. Biochemistry. 34:6278–6287. [DOI] [PubMed] [Google Scholar]
- Chen, E. F., T. Gensch, A. B. Gross, J. Hendriks, K. J. Hellingwerf, and D. S. Kliger. 2003. Dynamics of protein and chromophore structural changes in the photocycle of photoactive yellow protein monitored by time-resolved optical rotatory dispersion. Biochemistry. 42:2062–2071. [DOI] [PubMed] [Google Scholar]
- Craven, C. J., N. M. Derix, J. Hendriks, R. Boelens, K. J. Hellingwerf, and R. Kaptein. 2000. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in isolation by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry. 39:14392–14399. [DOI] [PubMed] [Google Scholar]
- Derix, N. M., R. W. Wechselberger, M. A. van der Horst, K. J. Hellingwerf, R. Boelens, R. Kaptein, and N. A. J. van Nuland. 2003. Lack of negative charge in the E46Q mutant of photoactive yellow protein prevents partial unfolding of the blue-shifted intermediate. Biochemistry. 42:14501–14506. [DOI] [PubMed] [Google Scholar]
- Dux, P., G. Rubinstenn, G. W. Vuister, R. Boelens, F. A. A. Mulder, K. Hard, W. D. Hoff, A. R. Kroon, W. Crielaard, K. J. Hellingwerf, and R. Kaptein. 1998. Solution structure and backbone dynamics of the photoactive yellow protein. Biochemistry. 37:12689–12699. [DOI] [PubMed] [Google Scholar]
- Fan, H., and A. E. Mark. 2004. Relative stability of protein structures determined by x-ray crystallography or NMR spectroscopy: a molecular dynamics simulation study. Proteins. 53:111–120. [DOI] [PubMed] [Google Scholar]
- Frenkel, D., and B. Smit. 2002. Understanding Molecular Simulation. From Algorithms to Applications, 2nd Ed. Academic Press, New York.
- García, A. E., and J. N. Onuchic. 2003. Folding a protein in a computer: an atomic description of the folding/unfolding of protein A. Proc. Natl. Acad. Sci. USA. 100:13898–13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, A. E., and K. Y. Sanbonmatsu. 2001. Exploring the energy landscape of a β-hairpin in explicit solvent. Proteins. 42:345–354. [DOI] [PubMed] [Google Scholar]
- Genick, U. K., G. E. O. Borgstahl, K. Ng, Z. Ren, C. Pradervand, P. M. Burke, V. Srajer, T. Y. Teng, W. Schildkamp, D. E. McCree, K. Moffat, and E. D. Getzoff. 1997. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science. 275:1471–1475. [DOI] [PubMed] [Google Scholar]
- Gensch, T., E. F. Chen, A. B. Gross, J. C. Hendriks, K. J. Hellingwerf, and D. S. Kliger. 2002. Dynamics of alteration of secondary structure in the photocycle of photoactive yellow protein (PYP) as monitored by time-resolved optical rotary dispersion (TRORD). Biophys. J. 82:314A. [Google Scholar]
- Gensch, T., J. Hendriks, and K. J. Hellingwerf. 2004. Tryptophan fluorescence monitors structural changes accompanying signaling state formation in the photocycle of photoactive yellow protein. Photochem. Photobiol. Sci. 3:531–536. [DOI] [PubMed] [Google Scholar]
- Groenhof, G., M. F. Lensink, H. J. C. Berendsen, and A. E. Mark. 2002a. Signal transduction in the photoactive yellow protein. II. Proton transfer initiates conformational changes. Proteins. 48:212–219. [DOI] [PubMed] [Google Scholar]
- Groenhof, G., M. F. Lensink, H. J. C. Berendsen, J. G. Snijders, and A. E. Mark. 2002b. Signal transduction in the photoactive yellow protein. I. Photon absorption and the isomerization of the chromophore. Proteins. 48:202–211. [DOI] [PubMed] [Google Scholar]
- Groenhof, G., M. Bouxin-Cademartory, B. Hess, S. P. de Visser, H. J. C. Berendsen, M. Olivucci, A. E. Mark, and M. A. Robb. 2004. Photoactivation of the photoactive yellow protein: why photon absorption triggers a trans-to-cis isomerization of the chromophore in the protein. J. Am. Chem. Soc. 126:4228–4233. [DOI] [PubMed] [Google Scholar]
- Harigai, M., Y. Imamoto, H. Kamikubo, Y. Yamazaki, and M. Kataoka. 2003. Role of an N-terminal loop in the secondary structural change of photoactive yellow protein. Biochemistry. 42:13893–13900. [DOI] [PubMed] [Google Scholar]
- Hendriks, J., W. D. Hoff, W. Crielaard, and K. J. Hellingwerf. 1999. Protonation deprotonation reactions triggered by photoactivation of photoactive yellow protein from Ectothiorhodospira halophila. J. Biol. Chem. 274:17655–17660. [DOI] [PubMed] [Google Scholar]
- Hendriks, J., T. Gensch, L. Hviid, M. A. van der Horst, K. J. Hellingwerf, and J. J. van Thor. 2002. Transient exposure of hydrophobic surface in the photoactive yellow protein monitored with Nile Red. Biophys. J. 82:1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks, J., I. H. M. van Stokkum, and K. J. Hellingwerf. 2003. Deuterium isotope effects in the photocycle transitions of the photoactive yellow protein. Biophys. J. 84:1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., B. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraints solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- Hoff, W. D., I. H. M. van Stokkum, H. J. van Ramesdonk, M. E. van Brederode, A. M. Brouwer, J. C. Fitch, T. E. Meyer, R. van Grondelle, and K. J. Hellingwerf. 1994. Measurement and global analysis of the absorbency changes in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila. Biophys. J. 67:1691–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff, W. D., A. Xie, I. H. M. van Stokkum, X. J. Tang, J. Gural, A. R. Kroon, and K. J. Hellingwerf. 1999. Global conformational changes upon receptor stimulation in photoactive yellow protein. Biochemistry. 38:1009–1017. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- Imamoto, Y., H. Kamikubo, M. Harigai, N. Shimizu, and M. Kataoka. 2002a. Light-induced global conformational change of photoactive yellow protein in solution. Biochemistry. 41:13595–13601. [DOI] [PubMed] [Google Scholar]
- Imamoto, Y., M. Kataoka, and R. S. H. Liu. 2002b. Mechanistic pathways for the photoisomerization reaction of the anchored, tethered chromophore of the photoactive yellow protein and its mutants. Photochem. Photobiol. 76:584–589. [DOI] [PubMed] [Google Scholar]
- Itoh, K., and M. Sasai. 2004. Dynamical transitions and proteinquake in photoactive yellow protein. Proc. Natl. Acad. Sci. USA. 101:14736–14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandori, H., T. Iwata, J. Hendriks, A. Maeda, and K. J. Hellingwerf. 2000. Water structural changes involved in the activation process of photoactive yellow protein. Biochemistry. 39:7902–7909. [DOI] [PubMed] [Google Scholar]
- Kort, R., H. Vonk, X. Xu, W. D. Hoff, W. Crielaard, and K. J. Hellingwerf. 1996. Evidence for trans-cis isomerization of the p-coumaric acid chromophore as the photochemical basis of the photocycle of photoactive yellow protein. FEBS Lett. 382:73–78. [DOI] [PubMed] [Google Scholar]
- Kort, R., K. J. Hellingwerf, and R. B. G. Ravelli. 2004. Initial events in the photocycle of photoactive yellow protein. J. Biol. Chem. 279:26417–26424. [DOI] [PubMed] [Google Scholar]
- Lee, B. C., P. A. Croonquist, T. R. Sosnick, and W. D. Hoff. 2001a. PAS domain receptor photoactive yellow protein is converted to a molten globule state upon activation. J. Biol. Chem. 276:20821–20823. [DOI] [PubMed] [Google Scholar]
- Lee, B. C., A. Pandit, P. A. Croonquist, and W. D. Hoff. 2001b. Folding and signaling share the same pathway in a photoreceptor. Proc. Natl. Acad. Sci. USA. 98:9062–9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Modeling. 7:306–317. [Google Scholar]
- Marinari, E., and G. Parisi. 1992. Simulated tempering—a new Monte Carlo scheme. Europhys. Lett. 19:451–458. [Google Scholar]
- Mark, A. E., S. P. van Helden, P. E. Smith, L. H. M. Janssen, and W. F. van Gunsteren. 1994. Convergence properties of free energy calculations: cyclodextrin complexes as a case study. J. Am. Chem. Soc. 116:6293–6302. [Google Scholar]
- Meyer, T. E., G. Tollin, J. H. Hazzard, and M. A. Cusanovich. 1989. Photoactive yellow protein from the purple phototrophic bacterium, Ectothiorhodospira halophila—quantum yield of photobleaching and effects of temperature, alcohols, glycerol, and sucrose on kinetics of photobleaching and recovery. Biophys. J. 56:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, T. E., M. A. Cusanovich, and G. Tollin. 1993. Transient proton uptake and release is associated with the photocycle of the photoactive yellow protein from the purple phototrophic bacterium Ectothiorhodospira halophila. Arch. Biochem. Biophys. 306:515–517. [DOI] [PubMed] [Google Scholar]
- Miyamoto, S., and P. A. Kollman. 1997. SETTLE: an analytical version of the SHAKE and the RATTLE algorithms for rigid water molecules. J. Comput. Chem. 13:952–962. [Google Scholar]
- Nymeyer, H., and A. E. García. 2003. Simulation of the folding equilibrium of α-helical peptides: a comparison of the generalized born approximation with explicit solvent. Proc. Natl. Acad. Sci. USA. 100:13934–13939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, D. H., A. Philip, W. D. Hoff, and R. A. Mathies. 2004. Time-resolved resonance Raman structural studies of the pB′ intermediate in the photocycle of photoactive yellow protein. Biophys. J. 86:2374–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellequer, J. L., K. A. Wager-Smith, S. A. Kay, and E. D. Getzoff. 1998. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. USA. 95:5884–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perman, B., V. Srajer, Z. Ren, T. Y. Teng, C. Pradervand, T. Ursby, D. Bourgeois, F. Schotte, M. Wulff, R. Kort, K. Hellingwerf, and K. Moffat. 1998. Energy transduction on the nanosecond time scale: early structural events in a xanthopsin photocycle. Science. 279:1946–1950. [DOI] [PubMed] [Google Scholar]
- Rajagopal, S., S. Anderson, V. Srajer, M. Schmidt, R. Pahl, and K. Moffat. 2005. A structural pathway for signaling in the E46Q mutant of photoactive yellow protein. Structure. 13:55–63. [DOI] [PubMed] [Google Scholar]
- Ren, Z., B. Perman, V. Srajer, T. Y. Teng, C. Pradervand, D. Bourgeois, F. Schotte, T. Ursby, R. Kort, M. Wulff, and K. Moffat. 2001. A molecular movie at 1.8 Ångstrom resolution displays the photocycle of photoactive yellow protein, a eubacterial blue-light receptor, from nanoseconds to seconds. Biochemistry. 40:13788–13801. [DOI] [PubMed] [Google Scholar]
- Salamon, Z., T. E. Meyer, and G. Tollin. 1995. Photobleaching of the photoactive yellow protein from Ectothiorhodospira halophila promotes binding to lipid bilayers—evidence from surface-plasmon resonance spectroscopy. Biophys. J. 68:648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, V., R. Pahl, V. Srajer, S. Anderson, Z. Ren, H. Ihee, S. Rajagopal, and K. Moffat. 2004. Protein kinetics: structures of intermediates and reaction mechanism from time-resolved x-ray data. Proc. Natl. Acad. Sci. USA. 101:4799–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa, M., M. Yoda, N. Kamiya, N. Asakawa, J. Higo, Y. Inoue, and M. Sakurai. 2001. Evidence for large structural fluctuations of the photobleached intermediate of photoactive yellow protein in solution. J. Am. Chem. Soc. 123:7445–7446. [DOI] [PubMed] [Google Scholar]
- Sprenger, W. W., W. D. Hoff, J. P. Armitage, and K. J. Hellingwerf. 1993. The eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorption-spectrum of the photoactive yellow protein. J. Bacteriol. 175:3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita, Y., and Y. Okamoto. 1999. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314:141–151. [Google Scholar]
- Swendsen, R., and J. Wang. 1986. Replica Monte Carlo simulation of spin-glasses. Phys. Rev. Lett. 57:2607–2609. [DOI] [PubMed] [Google Scholar]
- Takeshita, K., Y. Imamoto, M. Kataoka, F. Tokunaga, and M. Terazima. 2002. Thermodynamic and transport properties of intermediate states of the photocyclic reaction of photoactive yellow protein. Biochemistry. 41:3037–3048. [DOI] [PubMed] [Google Scholar]
- van Aalten, D. M. F., W. D. Hoff, J. B. C. Findlay, W. Crielaard, and K. J. Hellingwerf. 1998. Concerted motions in the photoactive yellow protein. Protein Eng. 11:873–879. [DOI] [PubMed] [Google Scholar]
- van Beeumen, J. J., B. V. Vreese, S. M. van Bun, W. D. Hoff, K. J. Hellingwerf, T. E. Meyer, D. E. McCree, and M. A. Cusanovich. 1993. Primary structure of a photoactive yellow protein from the phototrophic bacterium Ectothiorhodospira halophila, with evidence for the mass and the binding-site of the chromophore. Protein Sci. 2:1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brederode, M. E., W. D. Hoff, I. H. M. van Stokkum, M. L. Groot, and K. J. Hellingwerf. 1996. Protein folding thermodynamics applied to the photocycle of the photoactive yellow protein. Biophys. J. 71:365–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buuren, A. R., S. J. Marrink, and H. J. C. Berendsen. 1993. A molecular dynamics study of the decane/water interface. J. Phys. Chem. 97:9206–9212. [Google Scholar]
- van Gunsteren, W. F., and H. J. C. Berendsen. 1987. GROMOS-87 Manual. BIOMOS BV, Groningen, The Netherlands.
- van der Horst, M. A., I. H. van Stokkum, W. Crielaard, and K. J. Hellingwerf. 2001. The role of the N-terminal domain of photoactive yellow protein in the transient partial unfolding during signalling state formation. FEBS Lett. 497:26–30. [DOI] [PubMed] [Google Scholar]
- Yang, W. Y., J. W. Pitera, W. C. Swope, and M. Gruebele. 2004. Heterogeneous folding of the Trpzip hairpin: full atom simulation and experiment. J. Mol. Biol. 336:241–251. [DOI] [PubMed] [Google Scholar]
- Zhou, R. H. 2004. Exploring the protein folding free energy landscape: coupling replica exchange method with P3ME/RESPA algorithm. J. Mol. Graph. Modeling. 22:451–463. [DOI] [PubMed] [Google Scholar]