Abstract
The transcription factor signal transducer and activator of transcription 1 (STAT1) requires phosphorylation at both Tyr-701 and Ser-727 for full activation. IFN-γ induces phosphorylation of both residues, whereas stress signals like UV or lipopolysaccharide stimulate phosphorylation of Ser-727 only. Using p38α mitogen-activated protein kinase (MAPK)-deficient cells, we show that the stress-induced phosphorylation of Ser-727 requires p38α MAPK activity, whereas IFN-γ-stimulated Ser-727 phosphorylation occurs independently of the p38α pathway. Consistently, IFN-γ stimulated expression of the STAT1 target gene IRF1 to a similar extent in both wild-type and p38α-deficient cells. However, stress-induced activation of the p38 MAPK pathway considerably enhanced the IFN-γ-induced expression of both the endogenous IRF1 gene and a reporter driven by the IFN-γ-activated sequence element of the IRF1 promoter. This enhancement occurred independently of increased phosphorylation of Ser-727 by the p38 pathway. Taken together, these results demonstrate an interaction between IFN-γ signaling and the p38 pathway that leads to increased transcriptional activation by STAT1 independently of phosphorylation at Ser-727.
The signal transducer and activator of transcription 1 (STAT1) is an essential transcription factor for the expression of the majority of IFN-induced genes (1, 2). IFNs bind to their cognate receptors and activate the receptor-associated tyrosine kinases janus kinase (JAK)1 and tyrosine kinase (TYK)2 in case of type I IFN (IFN-α and -β), and JAK1 and -2 in case of type II IFN (IFN-γ). Activated JAKs phosphorylate the receptor chains, thus creating docking sites for STATs that are in turn tyrosine phosphorylated by JAKs (reviewed in refs. 3–5). In response to IFN-γ, STAT1 is tyrosine phosphorylated at Tyr-701, forms homodimers, translocates to the nucleus, and binds to the γ-activated sequence (GAS) elements of the target promoters (reviewed in ref. 6). IFN-α and -β induce tyrosine phosphorylation of STAT1 and -2. Consequently, STAT1/2 heterodimers and, to lesser extent, STAT1/1 homodimers are formed. STAT1/2 heterodimers translocate to the nucleus, where they associate with the p48 (IRF9) protein to form the interferon-stimulated gene factor (ISGF)3 complex that binds to interferon-stimulated response element (ISRE) elements within the promoters of type I IFN-regulated genes.
In addition to Tyr-701 phosphorylation, both types of IFNs induce phosphorylation of STAT1 at Ser-727 (7–10). The Ser-727 phosphorylation causes STAT1 to acquire its full transactivation potential that is reduced by ≈80% after mutation of the Ser-727 to alanine (7). STAT1-deficient U3A cells reconstituted with STAT1-S727A mutant did not display antiproliferative and antiviral responses to treatment with IFN (11, 12).
The signaling cascade leading to the IFN-induced phosphorylation of STAT1 at Ser-727 is not well understood. Although the residue Ser-727 lies within a good mitogen-activated protein kinase (MAPK) consensus sequence (PMS727P), most experimental data do not support an involvement of MAPKs in the IFN-stimulated Ser-727 phosphorylation of STAT1 (10, 13–15). Recently, Ca(2+)/calmodulin-dependent kinase (CaMK) II and protein kinase C (PKC)δ were reported to phosphorylate STAT1 at Ser-727 on treatment with IFN-γ and -α, respectively (16, 17), acting possibly via the PI3K/Akt pathway (15). In addition, the IFN-induced phosphorylation of STAT1 at Tyr-701 is needed for subsequent Ser-727 phosphorylation (14).
STAT1 can be phosphorylated at Ser-727 independently of IFNs (and phosphorylation at Tyr-701) as well. The IFN-independent induction of Ser-727 phosphorylation correlates with activation of the p38 MAPK (8, 13, 18). p38 is also able to phosphorylate STAT1 at Ser-727 in vitro (13). Combined treatment with IFN-γ and the p38 activator lipopolysaccharide (LPS) caused stronger phosphorylation at Ser-727 as compared with treatment with IFN-γ alone, a finding that correlated with enhanced expression of an IFN-γ-dependent reporter gene (8). Ser-727 rather than Tyr-701 was needed for expression of Fas/Fas-ligand and certain caspase genes, suggesting that serine phosphorylation alone causes STAT1 to be transcriptionally active (19, 20). Thus, p38 impinges on the activity of STAT1 and/or IFN-induced gene expression in several different ways. Whether p38 might contribute to IFN-γ-stimulated STAT1-dependent gene expression independent of Ser-727 phosphorylation has not been previously examined.
By using well defined knockout cells [p38α(−/−)] rather than biochemical, pharmacological, or overexpression experiments, we show that in mouse fibroblasts, p38 MAPK is neither involved in the IFN-induced phosphorylation of STAT1 at Ser-727 nor required for the IFN-γ-stimulated expression of the IRF1 gene. Instead, p38 activity is needed for the IFN-independent Ser-727 phosphorylation. Together, the p38 MAPK pathway and IFN-γ interact to cause an increased transcription of STAT1 target genes. Surprisingly, the p38-dependent enhancement of IFN-γ-induced expression of the IRF1 gene occurs independently of phosphorylation at Ser-727.
Materials and Methods
Cells, Cytokines, Drugs, and Treatments.
3T3 fibroblasts from STAT1-deficient mice (1) and the derivative cell lines reconstituted with STAT1-wild type (WT) and STAT1-S727A have been previously described (14). Human T98G glioblastoma cells stably transfected with dominant-negative IκB (GL-IκB-DN) and vector control (GL-Neo) were obtained from G. Stark's laboratory. To obtain p38α(−/−) and p38α(+/+) fibroblasts, embryonic day 10.5 mouse embryo fibroblasts were immortalized by infection with a simian virus 40 large T-containing murine retrovirus followed by G-418 selection, as previously described (21). All cells were maintained in DMEM containing 10% FCS. Murine recombinant IFN-γ and human recombinant tumor necrosis factor (TNF) α were used, respectively, at a concentration of 5 and 20 ng/ml for the periods indicated in the figure legends. UV irradiation was with UVC (254 nm, 40 J/m2) followed by incubation for 30 min. Anisomycin and SB203580 were purchased from Calbiochem and used, respectively, at a concentration of 100 ng/ml and 5 μM for times indicated in the figure legends. Sodium arsenite was purchased from Sigma and used at a concentration of 50 μM for times indicated in the figure legends.
Transient Transfections and Luciferase Assays.
p38α(−/−) and p38α(+/+) mouse embryo fibroblasts (MEFs) were transfected with reporter plasmid IRF1-GAS-Luc containing the luciferase gene under the control of four GAS elements (22). To control for transfection efficiency, the ecdysone inducible system (Invitrogen) was used. For this, cells were cotransfected with the plasmids pIND/LacZ and pVgRXR (in addition to IRF-GAS-Luc plasmid), and expression of β-galactosidase was induced by addition of ponasterone. Transfections were performed by using the Polyfect reagent (Qiagen, Hilden, Germany). Sixteen hours after transfection, cells were treated for 6 h with ponasterone (10 μM) alone or together with arsenite, IFN-γ, or both. Luciferase activity was assayed in duplicate according to standard protocols (23). Two equal aliquots of cell extracts were used for the luciferase assay and one for measurement of β-galactosidase activity. β-Galactosidase activity was assayed by adding 50 μl of extract to 500 μl of buffer BG (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgCl2/50 mM β-mercaptoethanol/0.4 mg/ml of ortho-nitro-phenyl-galactoside) followed by 60-min incubation at 37°C. After stopping the reaction by addition of 250 μl of 1 M Na2CO3, the β-galactosidase activity was determined by measuring the extinction at 420 nm. The luciferase cpm light emission of each sample was normalized to its β-galactosidase value. Luciferase induction was calculated by dividing normalized cpm light emission of stimulated cells by that of unstimulated cells.
Antibodies.
Antisera to the STAT1 C terminus and to phosphoSer727-STAT1 have recently been described (8). Rabbit antiserum to Tyr-701-phosphorylated STAT1 was purchased from NEB (Beverly, MA). A monoclonal antibody to the STAT1 N terminus was purchased from Transduction Laboratories (Lexington, KY). Phosphospecific antibodies to p38 MAPK and jun N-terminal kinase (JNK) were bought from NEB. Antibodies to p38-MAPK and IκB were purchased from NEB and those to JNK from Santa Cruz Biotechnology. Monoclonal antibodies to extracellular signal-regulated kinase (ERKs) (pan-ERK) were purchased from Transduction Laboratories.
Immunoprecipitation, Western Blot, and Electrophoretic Mobility-Shift Assay (EMSA).
After treatment, cell extracts were prepared as described (15). Cell extracts were incubated overnight at 4°C with antiserum to the C terminus of STAT1 (at a 1:250 dilution) together with Protein A-Sepharose beads. Immunoprecipitates were washed four times with lysis buffer, and the immunocomplexes were eluted by boiling in Laemmli sample buffer. A protocol for Western blotting and EMSA has recently been described (8).
Quantitation of Gene Expression by Using Real-Time PCR.
Two sets of real-time PCR experiments were run on the Light Cycler (Roche Diagnostics): (i) amplification of the hypoxanthine phosphoribosyltransferase (HPRT) housekeeping gene chosen as an endogenous control for normalization of the RNA load, and (ii) amplification of the specific gene of interest, IRF1. The isolation of total RNA, reverse transcription (RT), and real-time PCR and its quantitation were carried out as described recently (14).
Results
IFN-γ-Stimulated Phosphorylation of STAT1 at Ser-727 Does Not Require p38 MAPK, Whereas Stress-Induced Ser-727 Phosphorylation Is Strictly p38 MAPK-Dependent.
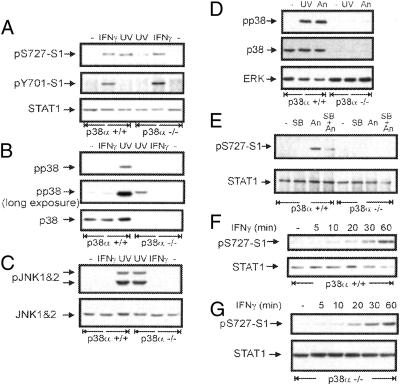
p38 MAPK was implicated in the stress-induced Ser-727 phosphorylation of STAT1, whereas the IFN pathway was in most cases shown not to depend on p38 (13–15, 18). To confirm the differential requirement for the p38 MAPK in phosphorylation at Ser-727, we took advantage of cells with targeted disruption in the gene coding for p38α MAPK, the most abundant p38 isoform (24). p38α(+/+) and p38α(−/−) immortalized MEFs were treated with IFN-γ or UV light, and phosphorylation of STAT1 at Ser-727 was analyzed by Western blotting with phosphoSer727-specific antibodies. IFN-γ-induced phosphorylation at Ser-727 (Fig. 1A Top) as well as Tyr-701 (Fig. 1A Middle) was increased to similar levels in both p38α(+/+) and p38α(−/−). In contrast, the UV-induced phosphorylation at Ser-727 was significantly stronger in p38α(+/+) than in p38α(−/−) cells (Fig. 1A). The weak induction of phosphorylation at Ser-727 in UV-treated p38α(−/−) cells is most likely caused by other p38 isoforms that are still expressed and weakly activated by UV light in p38α(−/−) cells (Fig. 1B, long exposure). IFN-γ did not lead to detectable activation of p38 (Fig. 1B). Activation by UV light of the JNK members of the stress-activated MAPKs was similar in both p38α(+/+) and p38α(−/−) cells (Fig. 1C). This result is in agreement with our previous finding that JNK is not involved in Ser-727 phosphorylation (14).
Figure 1.
Stimulation of phosphorylation of STAT1 at Ser-727 does not require p38 MAPK on IFN-γ treatment, whereas it is p38-dependent upon stress treatment. (A) Western blots of extracts from IFN-γ, UV-irradiated, or untreated p38α(+/+) and -(−/−) MEFs were stained with antiserum to phosphorylated Ser-727 (Top), reprobed with antibodies to phosphorylated Tyr-701 (Middle), and antibody to STAT1 N terminus to control for loading (Bottom). (B) The same extracts as in A were analyzed by Western blotting by using antibodies to phosphorylated p38 MAPK (Top). Longer exposure of the blot shows weak activation of nondisrupted p38 isoforms in the p38α(−/−) cells (Middle). The blot was reprobed by using antibodies to p38 (Bottom). (C) The same extracts as in A were Western-blotted and analyzed by using antibodies to phosphorylated JNK (Upper), the blot was reprobed for JNK1 and -2 to control for equal loading. Note similar activation of JNK in both p38α(+/+) and -(−/−) cells. (D) Whole-cell extracts from UV-irradiated, anisomycin-treated (30 min), or untreated p38α(+/+) and -(−/−) cells were Western-blotted by using antibodies to phosphorylated p38 (Top). The blot was reprobed by using antibodies to p38 (Middle) and ERK (Bottom) to control for loading. (E) STAT1 was immunoprecipitated from p38α(+/+) and -(−/−) cells treated with SB203580 for 1 h (SB) or anisomycin for 30 min (An) or pretreated with SB203580 for 30 min followed by 30-min treatment with anisomycin (SB + An). Western blotting revealed p38-dependent induction of STAT1 Ser-727 phosphorylation by anisomycin (Upper) and similar loading (Lower). (F) Kinetics (for time indicated) of IFN-γ-stimulated Ser-727 phosphorylation (labeled pS727-S1) in p38α(+/+) and (G) p38α(−/−) cells, and the appropriate loading control (labeled as STAT1).
To prove that stress signals other than UV cause the same Ser-727 phosphorylation, we used anisomycin, which is routinely used to activate p38 (25). Anisomycin treatment caused strong activation of p38 in p38α(+/+) cells, whereas there was almost no induction of p38 activity in p38α(−/−) cells (Fig. 1D Top). Anisomycin activated phosphorylation of STAT1 at Ser-727 in p38α(+/+) cells, whereas the Ser-727 phosphorylation in p38α(−/−) cells was barely detectable (Fig. 1E). Consistently, the anisomycin-induced Ser-727 phosphorylation of STAT1 was reduced by 30-min pretreatment with the p38 inhibitor SB203580. These results prove that anisomycin causes phosphorylation of STAT1 at Ser-727 through the p38 pathway.
Defects in signaling pathways often cause aberrant kinetics of activation events rather than different peak levels of activation. To find out whether a lesion in the p38 pathway influences the stress or IFN-γ-induced activation profile of STAT1, we conducted kinetic studies. These experiments showed that the kinetics of IFN-γ-stimulated phosphorylation at Ser-727 was similar in both p38α(+/+) (Fig. 1F) and p38α(−/−) (Fig. 1G) cells. The activation profile of the IFN-γ-induced phosphorylation of STAT1 at Tyr-701 was also not affected by the p38 deficiency (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). In contrast, kinetic studies demonstrated that the stress-induced Ser-727 phosphorylation remained low in p38α(−/−) cells during the entire period that was relevant for our experiments (Fig. 8, which is published as supporting information on the PNAS web site). These data clearly demonstrate that IFN-γ signaling toward STAT1 is independent of the p38 pathway, whereas the stress-induced phosphorylation of STAT1 at Ser-727 requires p38 activity.
IFN-γ-Stimulated Expression of the STAT1 Target Gene IRF1 Does Not Require the Activity of p38 MAPK.
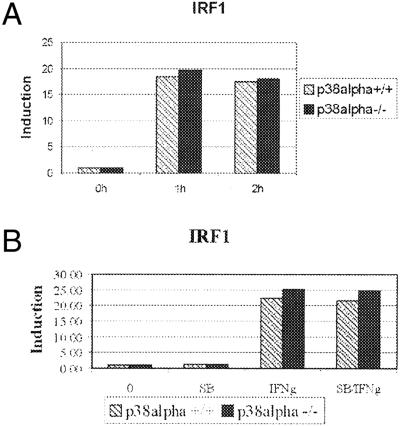
Because the two key IFN-γ-induced STAT1-activating modifications, phosphorylation at Tyr-701 and Ser-727, were not affected by the p38 deficiency, we assumed that the expression of IFN-γ-stimulated genes occurs independently of p38 as well. We used real-time RT-PCR analysis to quantitatively compare the expression levels of the IRF1 gene in IFN-γ-treated p38α(+/+) and p38α(−/−) cells. The promoter of the IRF1 gene contains a GAS element, and the IFN-γ-induced expression from this promoter is strictly STAT1-dependent (1, 2). As shown in Fig. 2A, the IFN-γ-induced expression of IRF1 was similar in p38α(+/+) and p38α(−/−) cells. This result was further confirmed by carrying out the expression analyses in the presence or absence of the p38 kinase inhibitor SB203580. As shown in Fig. 2B, SB203580 did not affect the IFN-γ-induced expression in either cell line. All real-time RT-PCR analyses were done in at least three independent experiments carried out in duplicate. The expression levels of IRF1 were normalized to the housekeeping gene hypoxanthine phosphoribosyltransferase (HPRT).
Figure 2.
IFN-γ-stimulated expression of the IRF1 gene does not require p38. p38α(+/+) and -(−/−) cells were treated with IFN-γ for 1 or 2 h (A) or treated with SB203508 for 30 min before stimulation with IFN-γ for 60 min (B), and total RNA was isolated. After RT, the amounts of IRF1 cDNA were assayed by using real-time PCR in duplicate in three independent experiments. The values of IRF1 expression after treatment with IFN-γ alone (IFNg) or with both SB203580 and IFN-γ (SB/IFNg) were normalized to those of untreated cells.
These results demonstrate that p38 activity is dispensable for the IFN-γ-induced gene expression of IRF1.
p38 MAPK Enhances IFN-γ-Induced Gene Expression.
It has been reported that pretreatment with the stress stimulus LPS enhanced the IFN-γ-induced expression of a STAT1-driven reporter gene (13). This enhancement correlated with activation of the p38 kinase. On the basis of these data, we asked whether p38 was indeed capable of enhancing the IFN-γ response. We used anisomycin as stress stimulus for MEFs instead of LPS, because the expression of some essential components of the LPS signaling pathway is restricted to hematopoietic cells (26). We also ruled out UV light as inducer, because it has been shown that this stimulus down-regulates IFN-γ-induced Tyr-701 phosphorylation of STAT1 by unknown mechanisms (27).
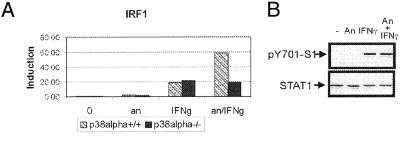
To investigate the effect of activated p38 on the IFN-γ-induced expression of STAT1 target genes, p38α(+/+) and -(−/−) cells were treated for 20 min with anisomycin followed by 1-h treatment with IFN-γ. Expression of IRF1 was determined in three independent experiments by using real-time RT-PCR performed in duplicate. Fig. 3A shows a representative result. As expected, there was no difference between p38α(+/+) and p38α(−/−) cells with respect to the expression of IRF1 induced by IFN-γ alone. Strikingly, pretreatment of cells with anisomycin caused a 3-fold enhancement of IFN-γ-induced expression of the IRF1 gene in p38α(+/+) cells but not in p38α(−/−) cells. Anisomycin alone did not cause significant changes in expression of IRF1. The levels of IFN-γ-induced tyrosine-phosphorylated STAT1 were not influenced by anisomycin treatment (Fig. 3B).
Figure 3.
Anisomycin treatment enhances IFN-γ-induced transcription in a p38-dependent way. (A) p38α(+/+) and -(−/−) cells were treated with IFN-γ for 60 min with (an/IFNg) or without (IFNg) 20-min pretreatment with anisomycin or for 80 min with anisomycin alone (an), or left untreated (−). Total RNA was isolated, and the expression of IRF1 was determined in three independent experiments by using real-time RT-PCR carried out in duplicate (a representative result is shown). (B) p38α(+/+) cells were treated with anisomycin for 60 min (An) or with IFN-γ for 30 min (IFNγ) or pretreated with anisomycin (100 ng/ml) for 30 min followed by treatment with IFN-γ (30 min) (An + IFNγ). Tyr-701 phosphorylation (Upper) of STAT1 and equal loading (Lower) were assayed in whole-cell extracts by Western blotting by using pTyr701-S1 and STAT1 N-terminal antibodies, respectively.
We conclude that activated p38 kinase strongly enhances the IFN-γ-stimulated expression of the STAT1-driven gene IRF1.
p38 MAPK-Mediated Enhancement of IFN-γ-Stimulated Gene Expression Does Not Require Phosphorylation of STAT1 at Ser-727 but Is STAT1-Dependent.
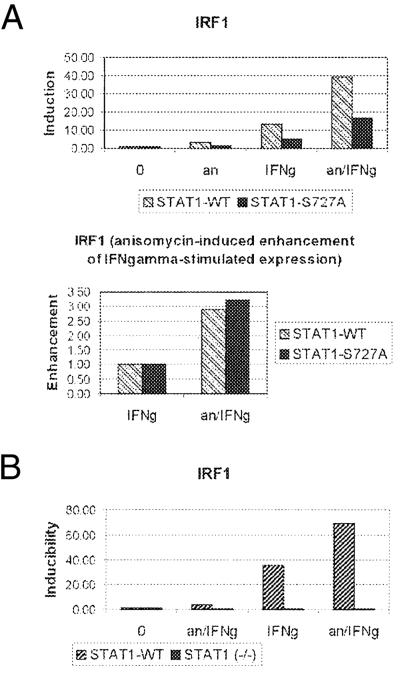
The effect of activated p38 on the IFN-γ-induced expression would be consistent with increased phosphorylation of STAT1 at Ser-727 by combined treatment with IFN-γ and anisomycin. To investigate the role of Ser-727 phosphorylation in the p38-dependent enhancement, we performed expression studies in STAT1-deficient cells stably reconstituted with either WT STAT1 (STAT1-WT cells) or the S727A mutant of STAT1 (STAT1-S727A cells) that were described recently (14). As expected, IFN-γ-induced expression of IRF1 was reduced to 40% by the S727A mutation (Fig. 4A). Surprisingly, activation of p38 by pretreatment with anisomycin enhanced the IFN-γ-stimulated gene expression of IRF1 in both STAT1-WT and -S727A cells. The enhancement was ≈3-fold in both cell lines (Fig. 4A Lower). Similar expression data using real-time PCR were obtained in five independent experiments performed in duplicate or triplicate. These results confirm that phosphorylation of STAT1 at Ser-727 is needed for efficient IFN-γ-stimulated IRF1 expression. However, Ser-727 phosphorylation is not required for the p38-dependent enhancement of IFN-γ-induced expression of IRF1. Recently, IFN-γ has been shown to regulate gene expression also independently of STAT1 (28, 29). To address the issue of requirement for STAT1 in our system, the expression of IRF1 in STAT1-WT and -(−/−) MEFs (the parental cells of STAT1-WT cells) was analyzed. As shown in Fig. 4B, neither treatment with IFN-γ or anisomycin alone nor the combined treatment caused a significant induction of IRF1 expression in cells lacking STAT1. This result proved that both IFN-γ-stimulated IRF1 expression as well as the transcriptional enhancement caused by p38 required STAT1.
Figure 4.
p38-mediated enhancement of IFN-γ-induced gene expression is STAT1-dependent and does not require phosphorylation of STAT1 at Ser-727. STAT1(−/−) MEFs reconstituted with STAT1-WT or -S727A mutant (A) or the parental STAT1(−/−) and the STAT1-WT reconstituted cells (B) were treated with anisomycin for 80 min (an) or with IFN-γ for 60 min (IFNg) or pretreated with anisomycin for 20 min followed by treatment with IFN-γ (60 min) (an/IFNg). Total RNA was isolated, and expression of IRF1 was determined in five (A) or two (B) independent experiments by using real-time RT-PCR carried out in triplicate or duplicate (a representative result is shown). The IRF1 value of each treatment was normalized to that of untreated cells (A Upper and B). Anisomycin-dependent enhancement is ≈3-fold in both STAT1-WT and -S727A cells, as shown by normalization of the values for the combined treatment with anisomycin (an) and IFN-γ (an/IFNg) to those of IFN-γ alone (IFNg) (A Lower). Note no significant induction of IRF1 in STAT1(−/−) cells by either treatment.
On IFN-α treatment, the activity of STAT1 is increased by arginine methylation that causes tyrosine-phosphorylated STAT1 to associate less efficiently with its inhibitor PIAS1 (30). Consequently, more STAT1 is available to bind DNA. Thus, p38 MAPK might enhance the DNA binding of STAT1 by increasing arginine methylation. We used electrophoretic mobility-shift assay by using a β-casein GAS probe to determine the amount of STAT1 capable of binding to DNA. No difference of the DNA-binding activity in extracts from cells treated with the combination of IFN-γ and anisomycin or IFN-γ alone was observed (Fig. 9, which is published as supporting information on the PNAS web site), indicating that arginine methylation is not likely to play a role in the p38-mediated enhancement of the STAT1-driven transcription.
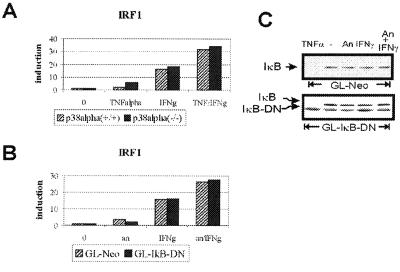
Simultaneous activation of STAT1 by IFNs and the transcription factor NF-κB by TNFα causes a synergistic increase of the IRF1 transcription (31). In our experimental setting, effects of treatment with anisomycin might be explained by the activation of NF-κB. To address this issue, p38α(+/+) and -(−/−) cells were treated with IFN-γ, TNFα, or both, and the expression of IRF1 was measured. Combined treatment with both cytokines led to synergistic increase of the IRF1 expression (Fig. 5A) that was, however, essentially the same in both p38α(+/+) and -(−/−) cells. Thus, the NF-κB-mediated synergistic stimulation of IFN-γ-induced IRF1 expression does not require p38 and thereby differs from the anisomycin-mediated enhancement. This finding was further confirmed by using cells expressing the dominant-negative variant of the NF-κB inhibitor IκB (IκB-DN) that lacks the N-terminal 36 amino acids and was shown to inhibit activation of NF-κB (32). T98G glioblastoma cells stably transfected with the IκB-DN construct (GL-IκB-DN) or vector control (GL-Neo) were stimulated with IFN-γ for 1 h with or without 20-min pretreatment with anisomycin, and the expression of IRF1 was analyzed as described above. Pretreatment with anisomycin caused an increase of the IFN-γ-induced IRF1 expression in both GL-Neo and GL-IκB-DN cell lines, confirming that the enhancement was independent of NF-κB activation (Fig. 5B). Western blot analyses revealed that IκB was not degraded in cells stimulated by anisomycin, IFN-γ, or both (Fig. 5C). Control treatment with TNFα proved that the degradation of endogenous IκB proceeded normally, whereas the IκB-DN mutant remained resistant to treatment.
Figure 5.
NF-κB is not required for p38-mediated transcriptional enhancement. (A) p38α(+/+) and -(−/−) cells were treated for 60 min with TNFα, IFN-γ, or both, and the expression of IRF1 was determined. (B) Glioblastoma cells stably transfected with dominant negative IκB (GL-IκB-DN) or vector control (GL-Neo) were treated with IFN-γ with or without 20-min pretreatment with anisomycin. RNA was isolated, and the expression of IRF1 was analyzed. In both A and B, the expression was determined in two independent experiments by using real-time RT-PCR carried out in duplicate (representative results are shown). (C) GL-IκB-DN and GL-Neo cells were treated with IFN-γ with or without 20-min pretreatment with anisomycin, TNFα (25 min), or left untreated. Whole-cell extracts were prepared and analyzed by Western blotting for degradation of IκB by using IκB antibodies. The endogenous IκB and mutant IκB-DN bands are indicated.
We conclude that activated p38 enhances expression of IFN-γ-stimulated genes by a mechanism that does not require Ser-727 of STAT1 and is independent of NF-κB.
The IRF1-GAS Element Is Sufficient to Mediate p38-Dependent Enhancement of IFN-γ-Stimulated Expression of IRF1.
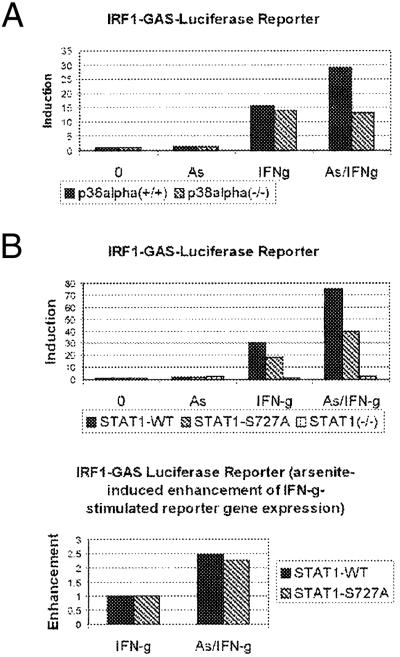
We have ruled out the involvement of NF-κB in the p38-mediated transcriptional enhancement of IRF1 expression. To exclude involvement of other transcription factors downstream of p38, we performed reporter gene assays by using a construct with four GAS elements (of the IRF1 gene) followed by the reporter luciferase. Although the IRF1-GAS was reported to bind STAT1 strongly and NF-κB weakly, the binding of the two transcription factors to the GAS is mutually exclusive (31), and an involvement of NF-κB in the p38-dependent transcriptional enhancement of IRF1 expression was excluded (see Fig. 5). We assayed luciferase activity of the IRF1-GAS reporter in p38α(+/+) and -(−/−) as well as in STAT1-WT, -S727A, and -(−/−) cells treated or untreated with IFN-γ for 6 h with or without 20-min pretreatment with arsenite. To control for transfection efficiency, the inducible β-galactosidase expression system was used. The expression of β-galactosidase was induced by addition of ponasterone at the time of treatment with IFN-γ, arsenite, or both. Arsenite was used as p38 agonist instead of anisomycin, because the latter drug inhibits translation and therefore interferes negatively with enzymatic reporter assays. In control experiments, we confirmed that arsenite caused p38-dependent enhancement of IFN-γ-stimulated expression of the endogenous IRF1 gene, similar to anisomycin (data not shown). As shown in Fig. 6A, IFN-γ-stimulated expression of the GAS-driven luciferase reporter was strongly enhanced by arsenite in p38α(+/+) but not in p38α(−/−) cells. For IFN-γ induction of the GAS-driven reporter, STAT1 was required; for full induction, Ser-727 was also needed (Fig. 6B Upper). Consistent with the data in Fig. 4, p38-mediated transcriptional enhancement was independent of Ser-727 (Fig. 6B Lower). This finding suggests that the GAS element in the IRF1 promoter is sufficient for the p38-mediated transcriptional enhancement of IRF1 expression.
Figure 6.
Reporter driven by GAS element displays p38-mediated Ser-727-independent enhancement of IFN-γ-stimulated expression. p38α(+/+) and -(−/−) cells (A) or STAT1(−/−), -WT, and -S727A (B) cells were transiently transfected with IRF1-GAS-luciferase reporter together with pIND/LacZ and pVgRXR plasmids to control for transfection efficiency by using inducible β-galactosidase expression system. Cells were treated with IFN-γ for 6 h with or without 20-min pretreatment with arsenite (As) (50 μM in A, 10 μM in B). In addition, all samples were treated with ponasterone to induce expression of β-galactosidase for normalization of transfection. The induction of luciferase activity was determined in three independent experiments (a representative result is shown). The p38-mediated enhancement of the IFN-γ response in STAT1-WT and -S727A cells was calculated (B Lower).
Discussion
p38 Is Required for Stress-Induced but Not IFN-Induced Phosphorylation of STAT1 at Ser-727.
On the basis of recent studies, a scenario for the phosphorylation of STAT1 at Ser-727 is emerging: (i) in the IFN pathway, Tyr-701 phosphorylation of STAT1 is necessary for subsequent Ser-727 phosphorylation to occur; and (ii) in the IFN-independent pathway, only Ser-727 and no Tyr-701 phosphorylation is induced (14, 15). Recently, PKCδ and CaMKII have been described as IFN-α- and -γ-activated, respectively, STAT1 Ser-727 kinases (16, 17). The IFN-independent (or stress-induced) serine kinase is most likely the p38 MAPK (10, 13, 15, 18). In HeLa S3 cells, p38 appears to be required for the IFN-induced signaling pathway (9). Because the requirement for p38 in Ser-727 phosphorylation was not completely clear, and the findings were based predominantly on the use of the p38 inhibitor SB203580, we addressed the issue by using cells that lack the most abundant p38 isoform, p38α. We also made the assumption that the other remaining p38 isoforms (p38β, -γ, and -δ) would not significantly influence the outcome of the experiments, because the activation of p38 by stress stimuli was barely detectable in p38α(−/−) cells. Thus, our results clearly establish that, (i) stress-induced Ser-727 phosphorylation is p38 dependent, and (ii) neither active nor inactive p38 is required for the IFN-γ signaling toward Ser-727 of STAT1. In the p38α(−/−) cells, a minor role of the remaining p38 isoforms in the IFN-γ signaling cannot be excluded.
IFN-γ-Stimulated Transcriptional Activity of STAT1 Does Not Require p38 MAPK.
IFN-induced phosphorylation of STAT1 at Ser-727 is required for its full transcriptional activity. Our findings provide evidence that p38 MAPK is not required for IFN-induced Ser-727 phosphorylation. To find out whether the p38 nevertheless impinges on IFN-γ-stimulated gene expression, we investigated the transcriptional activity of STAT1 in p38-deficient cells. We performed real-time RT-PCR analysis of the IFN-γ-induced expression of the STAT1 target gene IRF1 that contains a GAS element in its promoter conferring strong inducibility by IFNs (22, 33). Our results show that in response to IFN-γ, STAT1 does not need the activity or physical presence of p38 to achieve its full transcriptional potential. Interestingly, IFN-α-induced transcription of a STAT1 dimer-driven reporter gene was shown to be reduced by inhibition of p38 activity (10), indicating that IFN-α and -γ might differ regarding their requirement for p38 in the expression of STAT1 dimer target genes. IFN-α-activated p38 MAPK may enhance gene expression in a way similar to the activation of p38 by anisomycin before stimulation with IFN-γ (see below).
IFN-γ-Stimulated Gene Expression Can Be Enhanced by Activation of the p38 MAPK.
Stress stimuli like UV, LPS, TNFα, and osmotic stress cause phosphorylation of STAT1 at Ser-727 without phosphorylation at Tyr-701 (10, 13, 18). According to the paradigm of IFN-γ signaling, STAT1 needs phosphorylation at Tyr-701 and the phosphorylation at Ser-727 for its transcriptional activity. Therefore, one possible function of IFN-independent Ser-727 phosphorylation may be an enhancement of the STAT1 transactivation function by increasing the number of serine-phosphorylated STAT1 molecules. In fact, short pretreatment of macrophages with LPS (before stimulation with IFN-γ) increased Ser-727 phosphorylation and enhanced considerably the IFN-γ-induced expression of a GAS-driven reporter gene. This enhancement offers a link between stress or inflammatory signals and IFN-γ in the physiological activation of macrophages (34).
Here we provide evidence that p38 can enhance IFN-γ-induced gene expression of a STAT1 target gene by a mechanism that does not require phosphorylation of STAT1 at Ser-727. First, we show that the stress-induced 3-fold increase of IFN-γ-stimulated expression of IRF1 is indeed strictly dependent on p38. Second, in both STAT1-WT and -S727A cells, activation of p38 caused a 2.5-fold increase of the IFN-γ-induced expression of IRF1. Overall induction of IRF1 was reduced by the S727A mutation (consistent with published data to ≈30%), but the enhancement factor was not affected by the mutation. This finding, together with our results in STAT1-deficient fibroblasts, suggests that the main input of p38 MAPK on promoters with GAS sequences depends on the presence of the STAT1 dimer but not through an increase of Ser-727 phosphorylation over the levels achieved by IFN-γ alone.
In the context of the endogenous IRF1 promoter, the transcription factor NF-κB and arginine methylation of STAT1 were excluded as potential mediators of the p38-dependent transcriptional enhancement. Moreover, a synthetic promoter with only a GAS element (i.e., STAT1-binding site) transmitted the p38-dependent enhancing signal. These findings suggest that no other transcription factor is required for the p38-mediated transcriptional enhancement. Instead, p38 might activate the general transcription machinery by phosphorylation. In fact, proline-directed kinases like cyclin-dependent kinases and ERKs have been implicated in activation of transcription by regulating phosphorylation of the C-terminal domain of RNA polymerase II or by becoming direct components of the transcription machinery (35, 36). Recently, p38 was found to indirectly activate phosphorylation and acetylation of histone H3 at specific promoters, thereby marking them for enhanced transcription (37). Thus, the p38-mediated enhancement of IFN-γ-induced expression of IRF1 could involve direct recruitment of p38 by STAT1 to the IRF1 promoter. In yeast, the p38 homologue Hog1 is recruited to promoters on osmotic stress conditions and becomes an integral part of the transcription activation complexes (38). By direct recruitment, the regulatory function of p38 would be limited only to promoters that possess a p38-recruiting factor. The advantages of such a mechanism are increased specificity and efficiency of the signaling processes.
In this work, we conclusively show that IFN-γ-induced gene expression via STAT1 is in principle independent of the p38 MAPK. Nevertheless, p38 feeds in by enhancing the IFN-γ-induced expression. The molecular basis of the enhancement has yet to be elucidated, although, at least in case of the IRF1 gene, it does not require phosphorylation of STAT1 at Ser-727.
Supplementary Material
Acknowledgments
We are grateful to David Livingston (Dana–Farber Cancer Institute) for providing the simian virus 40 large T-containing retrovirus, and to Hannah Nguyen (Cleveland Clinic Foundation) and George Stark (Cleveland Clinic Foundation) for providing the glioblastoma cell lines. The technical help of R. Glinitzer, S. Urschitz, and D. Grote is greatly appreciated. We also thank Rodger Novak for critical reading of the manuscript. This work was supported by the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung through Grant P14945 (P.K.) and by the Grant Agency of the Czech Republic through Grant 301/00/0564.
Abbreviations
- TNFα
tumor necrosis factor α
- MAPK
mitogen-activated protein kinase
- JNK
c-jun N-terminal kinase
- JAK
janus kinase
- ERK
extracellular signal-regulated kinase
- MEFs
mouse embryo fibroblasts
- RT
reverse transcription
- STAT1
signal transducer and activator of transcription 1
- GAS
γ-activated sequence
- LPS
lipopolysaccharide
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 2.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, et al. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 3.Bach E A, Aguet M, Schreiber R D. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 4.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 5.Schindler C, Strehlow I. Adv Pharmacol. 2000;47:113–174. doi: 10.1016/s1054-3589(08)60111-8. [DOI] [PubMed] [Google Scholar]
- 6.Decker T, Kovarik P, Meinke A. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 7.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 8.Kovarik P, Stoiber D, Novy M, Decker T. EMBO J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh K C, Haque S J, Williams B R. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uddin, S., Lekmine, F., Sharma, N., Majchrzak, B., Mayer, I., Young, P. R., Bokoch, G. M., Fish, E. N. & Platanias, L. C. (2000) J. Biol. Chem., 27634–27640. [DOI] [PubMed]
- 11.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath C M, Darnell J E., Jr J Virol. 1996;70:647–650. doi: 10.1128/jvi.70.1.647-650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovarik P, Stoiber D, Eyers P A, Menghini R, Neininger A, Gaestel M, Cohen P, Decker T. Proc Natl Acad Sci USA. 1999;96:13956–13961. doi: 10.1073/pnas.96.24.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovarik P, Mangold M, Ramsauer K, Heidari H, Steinborn R, Zotter A, Levy D E, Muller M, Decker T. EMBO J. 2001;20:91–100. doi: 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen H, Ramana C V, Bayes J, Stark G R. J Biol Chem. 2001;3:33361–33368. doi: 10.1074/jbc.M105070200. [DOI] [PubMed] [Google Scholar]
- 16.Nair J S, DaFonseca C J, Tjernberg A, Sun W, Darnell J E, Jr, Chait B T, Zhang J J. Proc Natl Acad Sci USA. 2002;23:5971–5976. doi: 10.1073/pnas.052159099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uddin S, Sassano A, Deb D K, Verma A, Majchrzak B, Rahman A, Malik A B, Fish E N, Platanias L C. J Biol Chem. 2002;11:14408–14416. doi: 10.1074/jbc.M109671200. [DOI] [PubMed] [Google Scholar]
- 18.Gollob J A, Schnipper C P, Murphy E A, Ritz J, Frank D A. J Immunol. 1999;162:4472–4481. [PubMed] [Google Scholar]
- 19.Kumar A, Commane M, Flickinger T W, Horvath C M, Stark G R. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 20.Stephanou A, Scarabelli T M, Brar B K, Nakanishi Y, Matsumura M, Knight R A, Latchman D S. J Biol Chem. 2001;276:28340–28347. doi: 10.1074/jbc.M101177200. [DOI] [PubMed] [Google Scholar]
- 21.Brown M, McCormack M, Zinn K G, Farrell M P, Bikel I, Livingston D M. J Virol. 1986;60:290–293. doi: 10.1128/jvi.60.1.290-293.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pine R, Canova A, Schindler C. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Adams R H, Porras A, Alonso G, Jones M, Vintersten K, Panelli S, Valladares A, Perez L, Klein R, Nebreda A R. Mol Cell. 2000;6:109–116. [PubMed] [Google Scholar]
- 25.Fukunaga R, Hunter T. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haziot A, Ferrero E, Kontgen F, Hijiya N, Yamamoto S, Silver J, Stewart C L, Goyert S M. Immunity. 1996;4:407–414. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- 27.Aragane Y, Kulms D, Luger T A, Schwarz T. Proc Natl Acad Sci USA. 1997;94:11490–11495. doi: 10.1073/pnas.94.21.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gil M P, Bohn E, O'Guin A K, Ramana C V, Levine B, Stark G R, Virgin H W, Schreiber R D. Proc Natl Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramana C V, Gil M P, Han Y, Ransohoff R M, Schreiber R D, Stark G R. Proc Natl Acad Sci USA. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mowen K A, Tang J, Zhu W, Schurter B T, Shuai K, Herschman H R, David M. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 31.Pine R. Nucleic Acids Res. 1997;25:4346–4354. doi: 10.1093/nar/25.21.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sims S H, Cha Y, Romine M F, Gao P Q, Gottlieb K, Deisseroth A B. Mol Cell Biol. 1993;13:690–702. doi: 10.1128/mcb.13.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nacy C A, Meltzer M S. Curr Opin Immunol. 1991;3:330–335. doi: 10.1016/0952-7915(91)90033-w. [DOI] [PubMed] [Google Scholar]
- 35.Hengartner C J, Myer V E, Liao S M, Wilson C J, Koh S S, Young R A. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 36.Lis J T, Mason P, Peng J, Price D H, Werner J. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 37.Saccani S, Pantano S, Natoli G. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 38.Alepuz P M, Jovanovic A, Reiser V, Ammerer G. Mol Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.