Abstract
Scanning near-field optical microscopy (SNOM) circumvents the diffraction limit of conventional light microscopy and is able to achieve optical resolutions substantially below 100 nm. However, in the field of cell biology SNOM has been rarely applied, probably because previous techniques for sample-distance control are less sensitive in liquid than in air. Recently we developed a distance control based on a tuning fork in tapping mode, which is also well-suited for imaging in solution. Here we show that this approach can be used to visualize single membrane protein complexes kept in physiological media throughout. Nuclear envelopes were isolated from Xenopus laevis oocytes at conditions shown recently to conserve the transport functions of the nuclear pore complex (NPC). Isolated nuclear envelopes were fluorescently labeled by antibodies against specific proteins of the NPC (NUP153 and p62) and imaged at a resolution of ∼60 nm. The lateral distribution of epitopes within the supramolecular NPC could be inferred from an analysis of the intensity distribution of the fluorescence spots. The different number densities of p62- and NUP153-labeled NPCs are determined and discussed. Thus we show that SNOM opens up new possibilities for directly visualizing the transport of single particles through single NPCs and other transporters.
INTRODUCTION
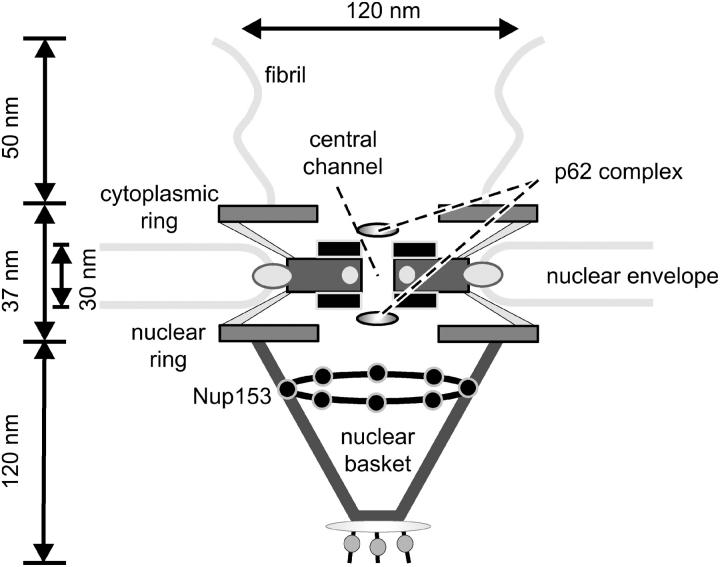
In eukaryotic cells, the genetic material is enclosed by an inner double membrane, the nuclear envelope (NE). A fundamental function of the NE is the protection of essential read and control processes of the genetic information by excluding the majority of molecules of the cytoplasm from the cell nucleus. The major gateway for an exchange of molecules between these two compartments is provided by a highly differentiated macromolecular assembly spanning the double membrane of the envelope. This so-called nuclear pore complex (NPC) consists of ∼30 different proteins (Rout et al., 2000), each occurring in multiple copies. The NPC has a main body of roughly cylindrical shape with a diameter of 120 nm and a length of 70 nm. On the cytoplasmic side the main body carries eight thin filaments radiating into the cytoplasm. On the nuclear surface also eight filaments exist, which are interconnected, however, at their ends to form a basket-like structure.
In the last decade, considerable insight has been gained into both the composition and structure of the NPC (Allen et al., 2000; Ryan and Wente, 2000; Adam, 2001; Fahrenkrog et al., 2001; Rout and Aitchison, 2001; Vasu and Forbes, 2001) and the mechanism of selective nuclear transport (Chook and Blobel, 2001; Conti and Izaurralde, 2001), which depends on specific signals and furthermore involves soluble transport receptors of the karyopherin family as well as the small GTPase Ran. However, the mechanism by which transport complexes are translocated through the NPC is not very well understood (Rout et al., 2000; Ben-Efraim and Gerace, 2001; Macara, 2001; Ribbeck and Görlich, 2001).
One promising approach (Kubitscheck et al., 2004) for obtaining more information about translocation through the NPC is based on the localization and tracking of single fluorescent molecules by far-field light microscopy (Goulian and Simon, 2000; Kues et al., 2001; Seisenberger et al., 2001). In general, however, conventional light microscopy is limited in resolution by diffraction to half the wavelength of light, so that, for example, single pores in the NE of Xenopus laevis oocytes, which have a mutual distance of ∼120 nm, cannot be resolved. Despite this fact, transport events through single NPCs were recently successfully measured with far-field optics by making use of a novel membrane patching method (OSTR, Optical Single Transporter Recording) (Keminer and Peters, 1999; Peters, 2003).
High-resolving methods from the family of scanning probe microscopy (SPM) have also been used for studying biomembranes and transporters. For example, by means of scanning force microscopy (SFM), an influence of Ca2+ on the topographic structure of the nuclear basket of the NPC (Stoffler et al., 1999a) or height changes of an NPC after addition of ATP to the surrounding medium have been measured recently (Rakowska et al., 1998). However, a purely force-microscopic approach seems to not be suited for investigating fast dynamic processes taking place below the immediate surface.
Nuclear transport could previously be studied in intact and permeabilized (Adam et al., 1990) cells only. It was therefore widely assumed that the NPC was a structure so delicate and intimately integrated into the cellular structure that it could not be removed from its native context in functional form. However, we have recently shown (Siebrasse et al., 2002) that the nuclear envelope of oocytes can be prepared such that the transport functions of the NPC are conserved and nuclear transport is fully reconstituted by recombinant transport factors. In particular, this method provides a suitable way to apply scanning probe techniques as a means for studying the nuclear envelope.
Here we demonstrate the optical investigation of single NPCs under physiological conditions by scanning near-field optical microscopy (SNOM). SNOM circumvents Abbe's principal diffraction limit by raster scanning a probe with a submicroscopic light source at a distance of only a few nanometers (Pohl et al., 1984; Betzig et al., 1991; for reviews, see Dunn, 1999; Hecht et al., 2000). The attainable optical resolution no longer depends on the wavelength of light, but scales with the size of the optical source and its distance to the sample. The most widely used light source in SNOM is a 50–100-nm large aperture at the tip of a light-guiding metal-coated tapered optical fiber. Apart from its high lateral resolution, SNOM furthermore features an even better optical resolution in axial direction, which is due to the exponential decrease of light intensity with increasing distance to the aperture.
To keep the near-field optical probe in closest proximity to the surface, a distance control on the basis of shear-forces acting between probe and sample is normally used (Betzig et al., 1992). Though excellently suited for applications under ambient conditions, the shear-force method appeared to be less sensitive in liquid environments (Moyer and Kämmer, 1996; Brunner et al., 1997; Gheber et al., 1998; Hwang et al., 1998; Lambelet et al., 1998; Rensen et al., 2000). The lack of an appropriate distance control resulted in a significant obstacle for the application of SNOM to biological issues requiring a physiological environment. Only recently shear-force imaging on a soft surface could be successfully applied under liquid using a novel method for liquid level control (Koopman et al., 2003). Furthermore, several alternative techniques for distance control have been proposed for imaging in liquid (Muramatsu et al., 1995; Keller et al., 1998; Tsai and Lu, 1998). We have recently shown that our own approach based on a tapping mode-like distance control (Höppener et al., 2003; see also Materials and Methods) works with equally high sensitivity in air and in liquid and is especially well-suited for the investigation of soft biological material in aqueous solutions.
A major aim of this article is to demonstrate that SNOM, combined with an appropriate distance control, is well-suited to investigate biomembranes under physiological conditions with an optical resolution on the nanoscale. Single nuclear pores of X. laevis oocytes kept in physiological media throughout are resolved for the first time by optical means. Our study shows that SNOM is able to give detailed insight into the structure of an NPC and thus provides a new possibility for nuclear transport investigations.
MATERIALS AND METHODS
Sample preparation
Microscope coverslips as substrates for the sample preparation were cleaned by sonication in a hot detergent (70°C, 1.0% Hellmanex II, Hellma GmbH, Müllheim, Germany) for 15 min, washed extensively in doubly distilled water, and finally immersed for 2 h in a 0.1 M solution of poly-L-lysine (molecular mass >70 kDa, Serva, Heidelberg, Germany). Tweezers and fine needles for handling the nucleus were immersed overnight in a 1% solution of BSA (Serva).
Stage VI oocytes were prepared from X. laevis according to the method described by Smith et al. (1991) and kept for further use in a buffer solution containing 88 mM NaCl, 1 mM KCl, 0.8 mM MgSO4, 1.4 mM CaCl2, 5 mM HEPES (pH 7.4), as well as 100 U/ml penicillin and 0.1 g/liter streptomycin. Just before isolation of the nuclei according to the method of Evans and Kay (1991), the oocytes were transferred into Mock3 buffer solution (90 mM KCl, 10 mM NaCl, 2 mM MgCl2, 0.1 mM CaCl2, 1.0 mM N-(2-hydroxyethyl)ethylene-diaminetriacetic acid (HEDTA), 10 mM HEPES, pH 7.3. All following incubation and washing steps for fluorescence labeling have been performed in Mock3-BSA (Mock3 containing additionally 15 g/l BSA) buffer solution.
For the investigation of the cytoplasmic face of the NE, nuclei were incubated 45 min in buffer containing monoclonal anti-p62 antibodies (mAb414, Babco, Berkeley Antibody Company, Richmond, CA), washed three times, incubated for 45 min in a buffer solution containing secondary antibodies (goat anti-mouse IgG labeled with Alexa 488, Molecular Probes, Leiden, The Netherlands), and washed again. Subsequently the nuclei were carefully opened with needles to remove the chromatin. Finally the membrane was spread out on a cover slip such that the cytoplasmic face was accessible.
For the investigation of the nuclear face of the NE, the nuclei were firmly attached to the cover slip and opened to remove the chromatin. After spreading out the membrane on the glass substrate the nuclear face of the NE was exposed. The following fluorescence labeling was identical to the labeling of the cytoplasmic face using monoclonal anti-p62 antibodies or alternatively antibodies against NUP153 (Walther et al., 2001) combined with a different secondary antibody (goat anti-rabbit IgG labeled with FITC or Alexa488).
SNOM imaging
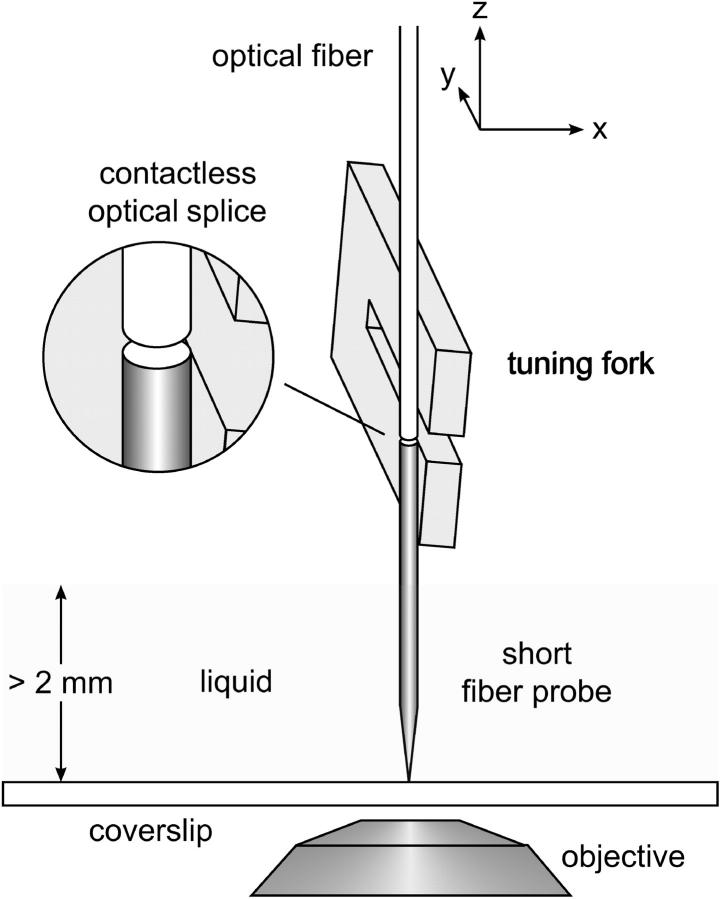
The basic stage of our home-made SNOM is mounted on an inverse optical microscope (Nikon, Düsseldorf, Germany, DIAPHOT 300) and comprises mainly a piezoelectric scanner with capacitive feedback control (Physik Instrumente, P-731.20, Karlsruhe, Germany) for positioning of the sample, and a SNOM head holding near-field probe and force sensor. The fluorescence light from the sample is collected by an oil immersion objective (Nikon, 100×, N.A. 1.25), passes a filter combination for suppressing the excitation light (Nikon, B-2H) and is detected by an avalanche photodiode (SPCM-AQR-16, PerkinElmer, Fremont, CA). For fluorescence excitation, the light (λ = 488 nm) of an argon-ion laser (Spectra-Physics, Darmstadt, Germany, FL 2020-03) is launched into a single mode optical fiber (Newport, F-SA) and is then guided through a fiber polarization control (Thorlabs (Newton, NJ), FPC560) to the SNOM head. There, the laser light is coupled into a short aperture fiber probe which is attached to a piezoelectric quartz tuning fork (resonance frequency ∼32 kHz). The tuning fork serves as a force sensor for distance control (Karraï and Grober, 1995) and is aligned in such a way that the prongs are vibrating in a direction normal to the sample surface (Fig. 1) (Edwards et al., 1997; Tsai and Lu, 1998). To maintain the force sensitivity of the force sensor only a short fiber probe (length 3–4 mm) with a corresponding low mass is glued to the end of the lower prong (Naber, 1999). The tip of the probe may protrude ∼2–3 mm from the prong, so that it is feasible to investigate a sample covered with up to 2 mm liquid without immersing the force sensor. In this way, the high quality factor of the resonance oscillation, and thus the force sensitivity of the tuning fork, is largely preserved (Naber et al., 1999). On the basis of forces acting normal to the surface, a defined and stable dynamic probe-sample interaction is established (tapping-mode-like force feedback). During imaging in liquid the decrease of the liquid level due to evaporation causes a gradual change of the resonance frequency of the tuning fork. Therefore an electronic self-excitation circuit has been implemented, which automatically tunes the frequency of the driving voltage to its momentary resonance frequency (Höppener et al, 2003). The high stability of our method was demonstrated, for example, by imaging double-stranded DNA molecules on a mica surface in a liquid environment.
FIGURE 1.
Tapping-mode-like distance control for SNOM measurements in liquid. The force sensor, a piezoelectric quartz tuning fork, is aligned parallel to the surface so that the prongs are oscillating normal to the sample. A short SNOM probe (length ∼3 mm) is attached to the lower prong. Though the SNOM probe is immersed ∼2 mm in the solution, the force sensor does not get in contact with the liquid and thus can preserve its high sensitivity. An optical fiber is used to couple light into the SNOM probe by bringing it close to the entrance face of the short fiber.
To couple light into the short fiber probe, the cleaved end face of the light-guiding optical fiber is positioned only a few micrometers in front of the entrance face of the short fiber probe. This coupling method is very efficient and produces only minor background signal. The precise adjustment can be done conveniently for the lateral position by observation through the inverse microscope (fourfold magnification) and for the axial position by means of the tuning fork as contact sensor.
The images in this article consist of 256 × 256 measured points and have been recorded with an integration time of 8 ms/pixel. Most images have been slightly low-pass filtered.
Characterization of SNOM probes
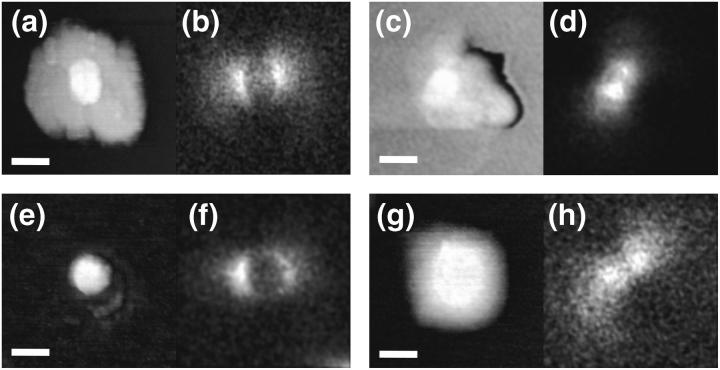
Before their use in an intended SNOM measurement, we characterized our near-field probes with high precision by means of a recently introduced method (Höppener et al., 2002). The basis of the characterization method is the near-field optical imaging of a single ∼20 nm sized fluorescent nanosphere (Molecular Probes). The nanospheres have been prepared in low concentration on a smooth glass substrate. The SNOM measurement (topography and optical image) of such a small fluorescing particle displays mainly the properties of the near-field probe, which is in analogy to the determination of the point-spread-function of a conventional optical microscope by imaging a point-like light source. From such a “fingerprint”, the optical resolution capability, brightness, and topographic properties of the probe can be easily determined. Fig. 2 shows the electric field distributions and topographies of the probes used for the measurements described in this article. The strong brightness at the metallic rim of the apertures results from a well-known field enhancement, which appears only in case of a fluorescing object getting very close to a sharp-edged aperture. The high definition of the used probes is the result of a mechanical modification, a controlled squeezing of the tip against a smooth substrate. A good optical quality can be obtained only at the expense of a flattened end face. Generally, such a tip is less well suited for topographic imaging, though the glass tip sticking out 10–20 nm from the flat end face is capable of detecting small topographic features.
FIGURE 2.
Topographic (left) and near-field optical (right) characterization of all SNOM probes used in this study. Characterizations have been done using a single ∼16–24-nm-sized fluorescent nanosphere as a sample (see text for details). The probes have been optimized by controlled squeezing against a smooth glass surface. The depicted SNOM probes were applied for the measurements: (a and b) Fig. 3; (c and d) Fig. 4; (e and f) Fig. 5, a, b, d, and e; (g and h) Fig. 5, g and h. Gray scales, 20–30 nm; scale bar, 80 nm.
RESULTS AND DISCUSSION
p62 labeling on the cytoplasmic face of the NE
For comparison, we carried out measurements of the cytoplasmic face of the NE on the same sample first under ambient conditions and then in water. For this purpose, we dehydrated a NE prepared on a glass substrate, performed a SNOM measurement, and then covered the sample again with water for the subsequent measurement in liquid. Before dehydration, the NPCs were optically marked by means of an antibody against the nucleoporin p62. In Fig. 3, a and b, a topographic and a simultaneously acquired near-field fluorescence image of the cytoplasmic face are shown. Though the distance control worked fine, the NPCs in the NE are not resolved topographically, probably because of the flat end face of the used near-field probe (see Fig. 2 a). The optical image on the other hand exhibits a large number of distinct fluorescence spots, most of which are separated clearly from each other. The minimum distance of adjacent spots is ∼100 nm which strongly suggests that a single spot indeed represents a single nuclear pore.
FIGURE 3.
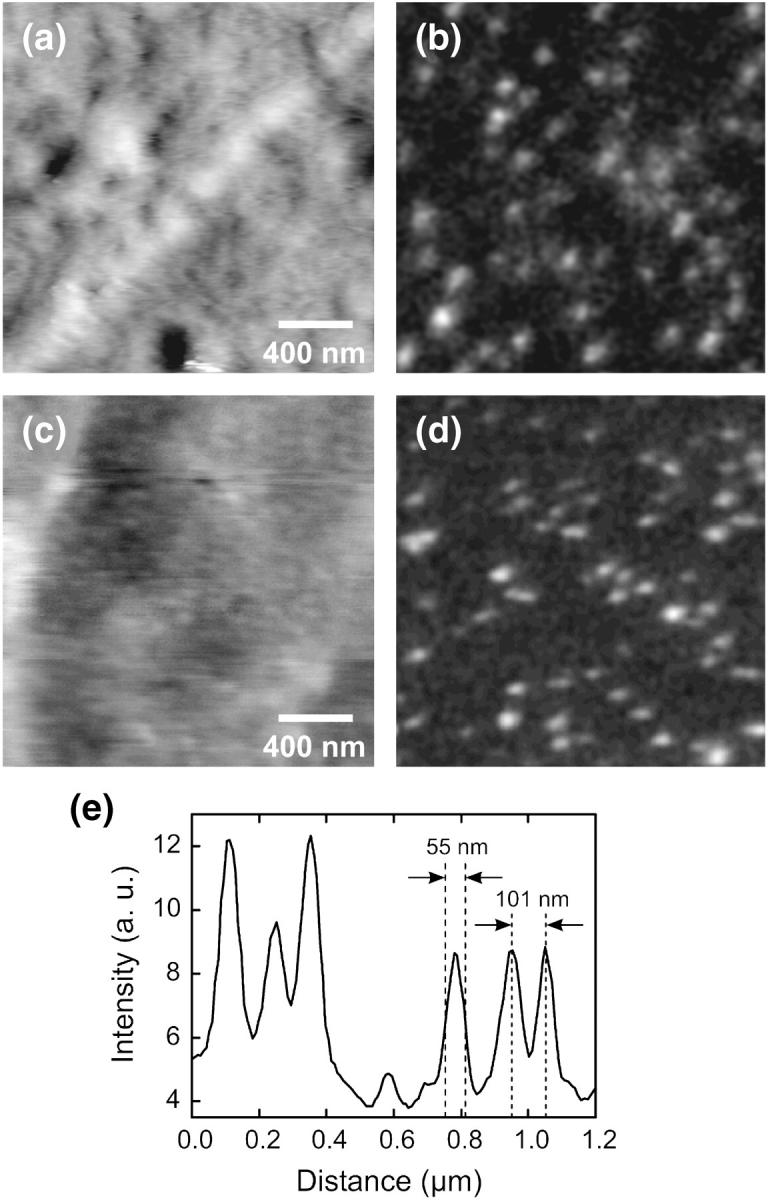
Topography (left) and simultaneously taken near-field fluorescence images (right) of the cytoplasmic face of a Xenopus laevis nuclear envelope. The membrane was fluorescently labeled by means of antibodies against the p62-complex of the nuclear pore complex. (a and b) Dehydrated membrane measured under ambient conditions (gray scale, 15 nm; 0.03–0.13 arbitrary units). (c and d) Subsequently taken SNOM images of the same sample in water (gray scale, 13 nm; 0.04–0.17 arbitrary units). (e) Cross section through several fluorescence spots in d (dotted line).
Subsequent measurements on the same sample under water are depicted in Fig. 3, c and d. Though the NE became rather soft by absorption of water, the force image is only slightly diminished in quality as compared to the corresponding image under ambient conditions (Fig. 3 a). The quality of the fluorescence image has not changed thus confirming that the good imaging conditions are preserved even in liquid environment. This leads us to the conclusion, that the soft membrane was not affected by the force exerted by the distance control. Compared to the measurements performed in air, the signal-to-noise ratio of the optical signal is even improved. A section through several fluorescence spots (Fig. 3 e) shows that the size of the spots, and thus the optical resolution, is below 60 nm (full width at half-maximum, FWHM). Even two closely neighbored NPCs can be easily distinguished in the optical image.
p62 labeling on the nucleoplasmic face of the NE
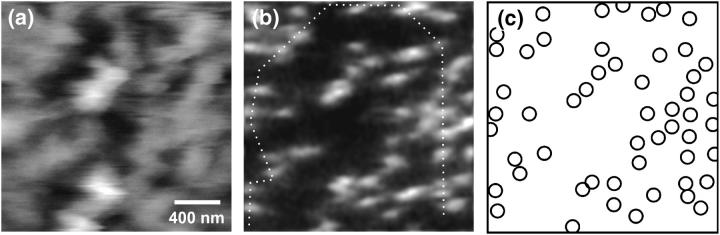
For all following measurements, the membrane patches have been prepared without an intermediate drying step and spread on the glass surface such that the nuclear face is exposed to the probing tip. For the SNOM image depicted in Fig. 4 a nuclear membrane was again labeled with antibodies against p62 but now the membrane was kept in Mock3 buffer solution throughout preparation and measurement. According to the flat end face of the used SNOM probe (see Fig. 2 c) single nuclear pore complexes could not be identified topographically (Fig. 4 a); however, the fluorescence image (Fig. 4 b) shows a distribution of clearly separated bright spots. To the best of our knowledge this image represents the first high-resolution SNOM measurement of a native biomembrane under physiological conditions. In Fig. 4 c, the positions of the nuclear pores as detected in the optical image are schematically represented by open circles with a diameter corresponding to the size of a pore. The average density of p62-tagged NPCs was determined to be ∼14 NPCs/μm2 for several equally prepared NE. This result agrees well with the average density of p62-labeled NPCs in Fig. 3, where the cytoplasmic face of the NE was imaged.
FIGURE 4.
Topographic (a) and near-field fluorescence image (b) of the nucleoplasmic side of a nuclear envelope kept in Mock3 buffer solution throughout. The membrane has been labeled using p62 antibodies (gray scale, 24 nm; 0.2–0.9 arb units). (c) Schematic distribution of fluorescence spots in b.
NUP153 labeling on the nucleoplasmic face of the NE
Fig. 5 shows several SNOM images of NEs labeled with antibodies against the nucleoporin α-NUP153. The measurements have been performed in water (Fig. 5, a–e) and in a Mock3 buffer solution (Fig. 5, g and h), respectively. The targeted epitope of NUP153 is located at the ring moiety of the nuclear basket (Walther et al., 2001).
FIGURE 5.
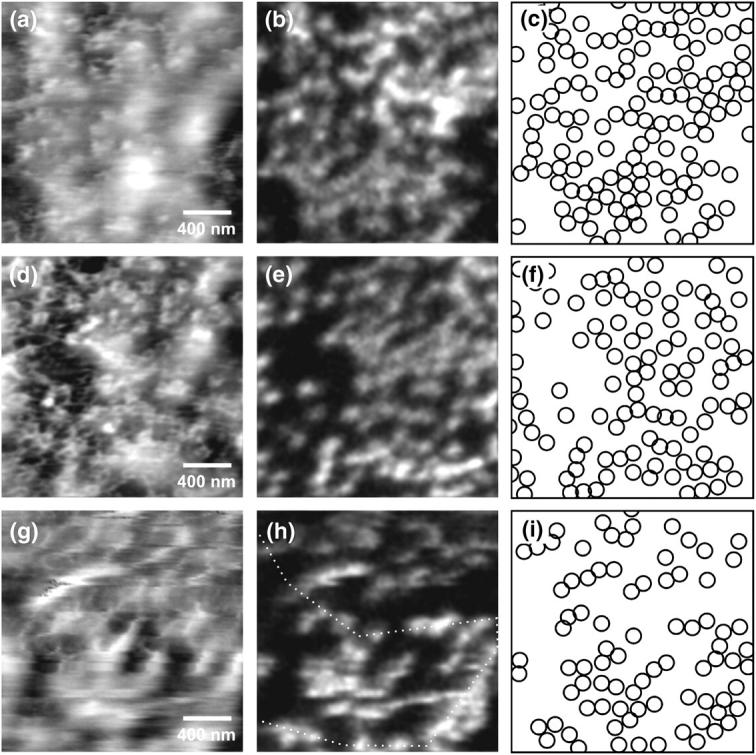
Topographic and near-field fluorescence images of the nucleoplasmic side of a nuclear envelope. The nuclear pore complexes have been labeled by antibodies against the nucleoporin NUP153. (a and b) Measurements performed in water (gray scale, 17 nm; 0.1–0.5 arb units). (d and e) Images of the same sample using an increased force as compared to a and b (gray scale, 27 nm; 0.1–0.3 arb units). (g and h) Measurements of a nuclear envelope in Mock3 buffer solution (gray scale, 26 nm; 0.3–1.3 arb units). (c, f, and i) Schematic distribution of fluorescence spots in respective near-field optical images.
By using a SNOM probe with a protruding glass tip at the aperture (Fig. 2, e and f) we were able to resolve simultaneously single NPCs topographically and optically, though the optical resolution of such a tip is slightly decreased. The two upper measurements, Fig. 5, a–f, have been performed on the same sample at different locations and with different exerted forces. In the force image of the first measurement, Fig. 5 a, only few details of the NE can be recognized because a thin, hazy film seemingly covered the membrane. To get closer to the membrane surface the exerted force of the distance control was gradually increased resulting in a much clearer topographic image with features which obviously represent single NPCs (Fig. 5 d). Both optical images (Fig. 5, b and e) show densely packed fluorescence spots of comparable brightness which can be just distinguished from each other. The weaker contrast in comparison to Figs. 3 d or 4 b is presumably due to the relatively low optical resolution capability of the used near-field probe, which exhibited an aperture diameter of ∼100 nm. In agreement with the lateral dimensions of a NPC, the distance of closely neighbored spots is ∼120 nm. Despite the dense distribution of fluorescent markers, the majority of tagged NPCs were optically resolved. A comparison of the simultaneously acquired topographical (Fig. 5 d) and optical image (Fig. 5 e) reveals that almost every topographic feature can be assigned to a single fluorescence spot. This result confirms that a single fluorescence spot indeed represents a single NPC. The average density of fluorescently tagged NPCs was found to be ∼30 NPCs/μm2, which is in good agreement to the density of NPCs found in our own AFM measurements (not shown) and AFM studies of other groups (Stoffler et al., 1999b). Thus we conclude that the staining protocol for NUP153 nucleoporins described above (see Materials and Methods) produces almost a complete labeling of available NPCs.
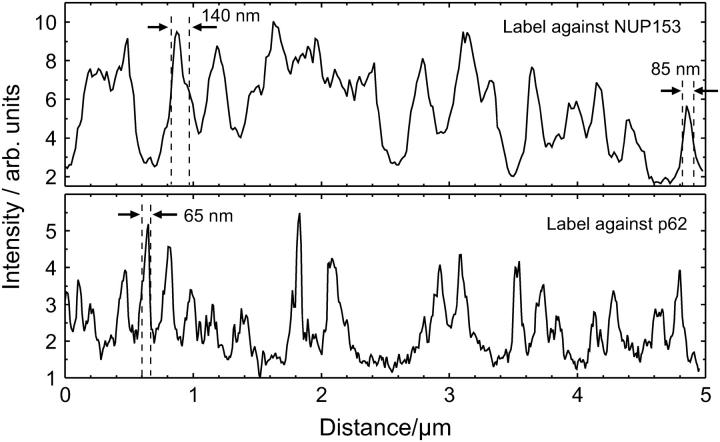
Fig. 5, g and h, depict a further SNOM measurement of a membrane in physiological buffer solution. In contrast to the previous measurements we selected a near-field probe, which did not exhibit a pronounced force tip but provided an improved optical resolution (see Fig. 2, g and h). Therefore, the optical image shows well-separated fluorescence spots though the topographic structure of the NE could not be resolved. However, despite the enhanced optical resolution capability of the SNOM probe, which is quite similar to the one used for the measurement of p62-labeled NE (Fig. 4), it is difficult to distinguish single NPCs within the densely packed regions of the membrane. To illustrate this in more detail, intensity profiles through several fluorescence spots of Figs. 4 b and 5 g (dotted lines) are compared in Fig. 6. From this comparison, it becomes obvious that the FWHM of the fluorescence spots of NUP153-labeled NPCs is much larger than the respective spots of p62-tagged NPCs, though both measurements have been prepared and performed in the same way. The marked differences cannot be attributed to the individual properties of the used SNOM probes as can be inferred from the characterization measurements shown in Fig. 2, c, d, g, and h.
FIGURE 6.
Comparison of cross sections through several fluorescence spots in Fig. 4 b (p62-labeled NE) and Fig. 5 h (NUP153-labeled NE) (location of section is indicated by a dotted line in both figures). The spot size (FWHM) of p62-tagged NPCs is ∼60 nm, whereas the spot size of NUP153-tagged NPCs varies between 80 nm and 140 nm. The different size of the fluorescence spots is likely due to the different distributions of epitopes of the targeted proteins within the NPC (cf. Fig. 7).
We conclude that the discrepancies between the attained FWHM and the determined optical resolution are caused by the different dye distributions resulting from the different binding sites of the used antibodies. The epitope of the used NUP153 antibody is known to be located at the ring moiety of the nuclear basket (see sketch in Fig. 7). Due to the octagonal symmetry of the NPCs eight binding sites for the NUP153-antibody are available. These binding sites are equally distributed over a ring with a diameter of ∼95 nm, so that the labeled sites are situated entirely at the periphery of the NPC. The distance of directly neighbored binding sites is ∼40 nm and thus below the achieved resolving power. It is likely that many pores are only partly labeled, that means, in some cases only two or three antibodies are expected to bind to the possible eight binding sites. Such a statistical fluctuation of label efficiency may explain that the FWHM of well-separated fluorescence spots largely varies between 80 nm and 140 nm as can be seen in Fig. 5 and as well in Fig. 6 b. Mathematically, the fluorescence spot of a pore fully occupied with NUP153 and imaged with an optical resolution of 80 nm would exhibit a FWHM of ∼130 nm (convolution of two Gaussians) which is in fair agreement to the upper value of the experimentally measured spot widths.
FIGURE 7.
Schematic diagram of the epitope-binding sites of some nucleoporins within the NPC (Stoffler et al., 1999b).
In contrast to NUP153, the p62-complex is located close to the aqueous channel and its lateral extension is largely restricted to the center of the pore (see Fig. 7). Consequently, the FWHM of the resulting fluorescence spots should mainly be governed by the optical resolution capability of the used near-field probe. This is exactly, what we have found. The FWHM of ∼60 nm for the fluorescence spots of the p62-labeled NPCs is in good agreement with the optical resolution capability as determined with our characterization method.
Number density of NPCs
We have observed a surprising discrepancy in the fluorescence spot density for membranes labeled with antibodies against the nucleoporins NUP153 and p62. For comparison, in some of the so far presented figures the positions of fluorescence spot are additionally displayed in an extra image and marked with open circles with a size adjusted to the NPC diameter (Fig. 4 c and Fig. 5, c, f, and i). This comparison clearly shows that the fluorescence spot density for p62-labeled NEs is significantly smaller (∼50%) than for NUP153- labeled NEs. As described above the labeling yield using NUP153 antibodies corresponded to 100% and the NPC density could be determined to ∼30 NPCs/μm2.
We can envision three possible reasons for this discrepancy. As described before, the labeling has been done after spreading the membrane on the glass substrate, so that only the nuclear face was exposed to the staining solution. Since the p62-complex is commonly assumed to be located at either end of the transport channel, each NPC should offer several binding sites to the antibodies in the solution. We want to point out here that even if the cytoplasmic face of the NPC were also stained, a fluorophore at the cytoplasmic side could hardly be excited by the near-field probe because the evanescent light intensity exponentially decays on a length scale of only several 10 nm (depending on the aperture size).
The first possible explanation for the measured relatively low label density is that the labeling yield for the monoclonal p62-antibody may be lower than for the polyclonal NUP153-antibody. This might be caused either by a lower binding affinity of the p62 antibody or a lower accessibility of the antibody to the epitope due to a steric hindrance, e.g. caused by the nuclear basket. However, a partial labeling of the NPCs using p62-antibodies has not been reported so far. In previous fluorescence measurements with a confocal scanning light microscope, for example, the minimum center-to-center distance of NPCs in p62-antibody labeled 3T3 cells have been determined, and the resulting distance was in good agreement to a value measured by electron microscopy or AFM (Kubitscheck et al., 1996). Thus we conclude that variations in the labeling yield are presumably not responsible for the observed low density of fluorescence spots.
Also, the different labeling densities may be interpreted as an indication for different states of activity of the p62-complex. Since soluble transport factors are known to interact with p62, it has been suggested that the p62 complex may have an important functional role in the nucleocytoplasmic transport process (Hu et al., 1996; Stoffler et al., 1999b). Hence, the specific p62-domain at the nucleoplasmic face of the NPC, which is targeted by the employed monoclonal antibody mAb414, might temporarily be inaccessible due to a conformational reorganization or a binding of the domain to bulky transport substrates such as RNPs in transit.
A further possible cause for a varying accessibility of the antibody to its epitope has interesting functional implications. A mobile domain of the p62 complex capable of “shuttling” through the transport channel between the nucleoplasmic and cytoplasmic face of the NPC could readily explain a lower labeling density, because the bulky labeling complex comprising primary and secondary antibodies cannot pass the channel. A nice feature of this speculative model, which has already been discussed by Stoffler et al. (1999b) and Fahrenkrog et al. (1998), is that the expected labeling density of ∼50% fits well to the measured density. Such a domain shuttling of a NPC component has already been shown for NUP153: different locations of various NUP153 domains within the NPC could be attributed to only a single NUP153 with a highly mobile C-terminal domain (Fahrenkrog et al., 2002; Fahrenkrog and Aebi, 2003). In our measurements related to NUP153, we used an antibody directed against the stationary domain; therefore, we did not expect, and actually did not observe, any effect of this mobility.
In summary, we think it is likely that the partial labeling of the NPCs using αp62-antibodies is caused by a specific biological activity of the p62-complex. Nonetheless further investigations are necessary to clarify this in more detail.
CONCLUSIONS
Up to now near-field optical studies of biological specimens under physiological conditions have been rare, because common methods for a probe-sample distance control are less-suited for use in a liquid than in air. We have shown here that, in combination with a tapping mode-like force feedback on the basis of a tuning fork, SNOM can be readily applied even to a soft biological membrane in liquid without affecting its integrity. By means of SNOM imaging in buffer solution, single pore complexes of X. laevis oocytes have been optically resolved for the first time under physiological conditions.
The fluorescence labeling of the pores has been done by means of antibodies against two different proteins of the NPC, NUP153 and p62. The different lateral distributions of the labels reflect the different sites of the antibody epitopes with respect to the center of the nuclear pores. Therefore, we could not only resolve single NPCs by SNOM, but also actually make visible structural details of this supramolecular complex. The surprisingly low density of p62-marked NPCs is discussed and tentatively interpreted as indication of a biological activity of the p62 complex.
We consider these first high-resolution SNOM measurements on a native membrane in physiological media as a substantial step toward a live observation of transport events through a single NPC. Next, we want to place a near-field optical probe close above a single NPC to study the time-resolved fluorescence signal of a labeled particle passing through the transport channel. The direction of translocation coincides in this case with the axial direction of the SNOM probe for which the optical resolution is even higher than in lateral direction. Thus, based on its ultra-high resolution and its single molecule sensitivity (Betzig and Chichester, 1993), SNOM is possibly able to provide detailed information of a biological transport process not easily obtained by other means.
Acknowledgments
We thank H. Oberleithner and his group for substantial help regarding the preparation of the nuclear envelope for scanning probe microscopy measurements. The polyclonal antibody against Nup153 was a kind gift of I. Mattaj (EMBL, Heidelberg, Germany). A.N. and C.H. are deeply indebted to H. Fuchs for his continuous support.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through a generous grant to A.N. (NA 382/1 and NA 382/2-1) and by the Center of Functional Nanostructures (CFN, Karlsruhe, Germany). All measurements have been carried out in the department of H. Fuchs at the Physics Institute, University of Münster, Germany. U.K. and R.P. gratefully acknowledge support through DFG grant KU 1507/1-1.
References
- Adam, S. A., R. Stern-Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, S. A. 2001. The nuclear pore complex. Genome Biol. 2:reviews0007.1–0007.6. [DOI] [PMC free article] [PubMed]
- Allen, T. D., J. M. Cronshaw, S. Bagley, E. Kiseleva, and M. W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651–1659. [DOI] [PubMed] [Google Scholar]
- Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin b for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig, E., P. L. Finn, and J. S. Weiner. 1992. Combined shear force and near field scanning optical microscopy. Appl. Phys. Lett. 60:2484–2486. [Google Scholar]
- Betzig, E., J. K. Trautman, T. D. Harris, J. S. Weiner, and R. L. Kostelak. 1991. Breaking the diffraction barrier: optical microscopy on a nanometric scale. Science. 251:1468–1470. [DOI] [PubMed] [Google Scholar]
- Betzig, E., and R. J. Chichester. 1993. Single molecules observed by near-field scanning optical microscopy. Science. 262:1422–1425. [DOI] [PubMed] [Google Scholar]
- Brunner, R., O. Hering, O. Marti, and O. Hollricher. 1997. Piezoelectrical shear-force control on soft biological samples in aqueous solution. Appl. Phys. Lett. 71:3628–3630. [Google Scholar]
- Chook, Y., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703–715. [DOI] [PubMed] [Google Scholar]
- Conti, E., and E. Izaurralde. 2001. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13:310–319. [DOI] [PubMed] [Google Scholar]
- Dunn, R. C. 1999. Near-field optical microscopy. Chem. Rev. 99:2891–2927. [DOI] [PubMed] [Google Scholar]
- Edwards, H., L. Taylor, W. Duncan, and A. J. Melmed. 1997. Fast, high-resolution atomic force microscopy using a quartz tuning fork as actuator and sensor. J. Appl. Phys. 82:980–984. [Google Scholar]
- Evans, J. P., and B. K. Kay. 1991. Biochemical fractionation of oocytes. Methods Cell Biol. 36:133–148. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., E. C. Hurt, U. Aebi, and N. Panté. 1998. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1 subcomplexes. J. Cell Biol. 143:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., D. Stoffler, and U. Aebi. 2001. Nuclear pore complex architecture and functional dynamics. Curr. Top. Microbiol. Immunol. 259:95–117. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., B. Marco, A. M. Fager, J. Köser, U. Sauder, K. S. Ullman, and U. Aebi. 2002. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140:254–267. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., and U. Aebi. 2003. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 4:757–766. [DOI] [PubMed] [Google Scholar]
- Goulian, M., and S. M. Simon. 2000. Tracking single proteins within cells. Biophys. J. 79:2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber, L. A., J. Hwang, and M. Edidin. 1998. Design and optimization of a near-field scanning optical microscope for imaging biological samples in liquid. Appl. Opt. 37:3574–3581. [DOI] [PubMed] [Google Scholar]
- Hecht, B., B. Sick, U. P. Wild, V. Deckert, R. Zenobi, O. J. F. Martin, and D. W. Pohl. 2000. Scanning near-field optical microscopy with aperture probes: fundamentals and applications. J. Chem. Phys. 112:7761–7774. [Google Scholar]
- Höppener, C., D. Molenda, H. Fuchs, and A. Naber. 2002. Simultaneous topographical and optical characterization of near-field optical aperture probes by way of imaging fluorescent nanospheres. Appl. Phys. Lett. 80:1331–1333. [Google Scholar]
- Höppener, C., D. Molenda, H. Fuchs, and A. Naber. 2003. Scanning near-field optical microscopy of a cell membrane in liquid. J. Microsc. 210:288–293. [DOI] [PubMed] [Google Scholar]
- Hu, T., T. Guan, and L. Gerace. 1996. Molecular and functional characterization of the p62Complex, an assembly of nuclear pore glycoproteins. J. Cell Biol. 134:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, J., L. A. Gheber, L. Margolis, and M. Edidin. 1998. Domains in cell plasma membranes investigated by near-field scanning optical microscopy. Biophys. J. 74:2184–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karraï, K., and R. D. Grober. 1995. Piezoelectric tip-sample distance control for near-field optical microscopes. Appl. Phys. Lett. 66:1842–1844. [Google Scholar]
- Keller, T. H., T. Rayment, and D. Klenerman. 1998. Optical chemical imaging of tobacco mosaic virus in solution at 60-nm resolution. Biophys. J. 74:2076–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keminer, O., and R. Peters. 1999. Permeability of single nuclear pores. Biophys. J. 77:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman, M., B. I. de Bakker, M. F. Garcia-Parajo, and N. F. van Hulst. 2003. Shear force imaging of soft samples in liquid using a diving bell concept. Appl. Phys. Lett. 83:5083–5085. [Google Scholar]
- Kubitscheck, U., P. Wedekind, O. Zeidler, M. Grote, and R. Peters. 1996. Single nuclear pores visualized by confocal microscopy and Image Processing. Biophys. J. 70:2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kues, T., A. Dickmanns, R. Lührmann, R. Peters, and U. Kubitscheck. 2001. High intranuclear mobility and dynamic clustering of the splicing factor U1 snRNP observed by single particle tracking. Proc. Natl. Acad. Sci. USA. 98:12021–12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambelet, P., M. Pfeiffer, A. Sayah, and F. Marquis-Weible. 1998. Reduction of tip–sample interaction forces for scanning near-field optical microscopy in a liquid environment. Ultramicroscopy. 71:117–121. [Google Scholar]
- Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, P. J., and S. B. Kämmer. 1996. High-resolution imaging using near-field scanning optical microscopy and shear force feedback in water. Appl. Phys. Lett. 68:3380–3382. [Google Scholar]
- Muramatsu, H., N. Chiba, K. Homma, K. Nakajima, T. Ataka, S. Ohta, A. Kusumi, and M. Fujihira. 1995. Near-field optical microscopy in liquids. Appl. Phys. Lett. 66:3245–3247. [Google Scholar]
- Naber, A. 1999. The tuning fork as sensor for dynamic force distance control in scanning near-field optical microscopy. J. Microsc. 194:307–310. [DOI] [PubMed] [Google Scholar]
- Naber, A., H.-J. Maas, K. Razavi, and U. C. Fischer. 1999. Dynamic force distance control suited to various probes for scanning near-field optical microscopy. Rev. Sci. Instrum. 70:3955–3961. [Google Scholar]
- Peters, R. 2003. Optical single transporter recording: Transport kinetics in microarrays of membrane patches. Annu. Rev. Biophys. Biomol. Struct. 32:47–67. [DOI] [PubMed] [Google Scholar]
- Pohl, D. W., W. Denk, and M. Lanz. 1984. Optical stethoscopy: image recording with resolution λ/20. Appl. Phys. Lett. 44:651–653. [Google Scholar]
- Rakowska, A., T. Danker, S. W. Schneider, and H. J. Oberleithner. 1998. ATP-induced shape change of nuclear pores visualized with the atomic force microscope. J. Membr. Biol. 15:129–136. [DOI] [PubMed] [Google Scholar]
- Rensen, W. H. J., N. F. van Hulst, and S. B. Kämmer. 2000. Imaging soft samples in liquid with tuning fork based shear force microscopy. Appl. Phys. Lett. 77:1557–1559. [Google Scholar]
- Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. Chait. 2000. The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593–16596. [DOI] [PubMed] [Google Scholar]
- Ryan, K. J., and S. R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361–371. [DOI] [PubMed] [Google Scholar]
- Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Bräuchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 294:1929–1932. [DOI] [PubMed] [Google Scholar]
- Siebrasse, J., E. Coutavas, and R. Peters. 2002. Reconstitution of nuclear protein export in isolated nuclear envelopes. J. Cell Biol. 158:849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, I. D., X. Weilong, and R. L. Varnold. 1991. Oogenesis and oocyte isolation. Methods Cell Biol. 36:45–60. [DOI] [PubMed] [Google Scholar]
- Stoffler, D., B. Fahrenkrog, and U. Aebi. 1999b. The nuclear pore complex: from architecture to functional dynamics. Curr. Opin. Cell Biol. 11:391–401. [DOI] [PubMed] [Google Scholar]
- Stoffler, D., K. N. Goldie, B. Feja, and U. Aebi. 1999a. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J. Mol. Biol. 287:741–752. [DOI] [PubMed] [Google Scholar]
- Tsai, D. P., and Y. Y. Lu. 1998. Tapping-mode tuning fork force sensing for near-field scanning optical microscopy. Appl. Phys. Lett. 73:2724–2726. [Google Scholar]
- Vasu, S. K., and D. J. Forbes. 2001. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13:363–375. [DOI] [PubMed] [Google Scholar]
- Walther, T. C., M. Fornerod, H. Pickersgill, M. Goldberg, T. D. Allen, and I. W. Mattaj. 2001. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20:5703–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]