Abstract
Endosymbionts, which are widely observed in nature, have undergone reductive genome evolution because of their long-term intracellular lifestyle. Here we compared the complete genome sequences of two different endosymbionts, Buchnera and a protist mitochondrion, with their close relatives to study the evolutionary rates of functional genes in endosymbionts. The results indicate that the rate of amino acid substitution is two times higher in symbionts than in their relatives. This rate increase was observed uniformly among different functional classes of genes, although strong purifying selection may have counterbalanced the rate increase in a few cases. Our data suggest that, contrary to current views, neither the Muller's ratchet effect nor the slightly deleterious mutation theory sufficiently accounts for the elevated evolutionary rate. Rather, the elevated evolutionary rate appears to be mainly due to enhanced mutation rate, although the possibility of relaxation of purifying selection cannot be ruled out.
Eubacteria that live within eukaryotic cells—endocellular symbionts—obtain metabolic precursors synthesized by their hosts and have lost many genes for biosynthetic pathways relative to their free-living cousins (1, 2). Such genome reduction is manifested in Buchnera sp. APS (3, 4), a maternally transmitted obligate γ-proteobacterial endosymbiont of aphids. This endosymbiont produces essential amino acids for its insect host, which in turn provides nonessential amino acids and other metabolites for the bacterium (1). More extreme genome reduction is observed in mitochondria (2), ATP-producing eukaryotic organelles that stem from endosymbiotic eubacteria and that have relinquished their genome entirely in their anaerobic forms, hydrogenosomes (5).
In addition, the rate of amino acid substitution of functional genes is often higher in endosymbionts than in their sister species or bacteria that are considered to be closest to their ancestral species (6–10). This accelerated rate of evolution in endosymbionts is generally believed to occur by Muller's ratchet effect (see below) or by fixation of slightly deleterious mutations in small endosymbiont populations (6–10). However, the acceleration of evolutionary rate may also be caused by a high mutation rate or relaxation of purifying selection in endosymbionts. To distinguish between these two types of hypotheses, we conducted genomic comparison of the endosymbionts Buchnera sp. APS and Reclinomonas americana mitochondrion (mt) with their closest relatives Escherichia coli and Rickettsia prowazekii, respectively, and estimated the relative rates of amino acid substitution for different groups of proteins.
Materials and Methods
Identification of Orthologs.
To compare rates of amino acid substitution in endocellular symbionts and relatives, we identified orthologous genes, that is, the genes that were neither duplicated before the divergence of species under study nor horizontally transferred subsequently. We first compared the genome of Buchnera sp. APS with that of its free-living γ proteobacterial relative E. coli K-12 (11) (Fig. 1). After completion of this article, the genomic sequence of another Buchnera species, Buchnera aphidicola Sg, was published (15). In this study, however, we did not use this genomic sequence, because the Buchnera genomes are highly conserved (15), and the new sequence data are expected to give essentially the same results. For the comparison of the genes from the R. americana mt (16), an extremely reduced α proteobacterial symbiont, we used homologs from the α proteobacterial parasite R. prowazekii (17) (Fig. 1). Buchnera is believed to have diverged from the E. coli lineage about 200 million years ago (18), whereas the mt apparently became a symbiont of eukaryotic cells before eukaryotic kingdoms diverged, more than 1.5 billion years ago (19). Ortholog candidates between two species were defined primarily by reciprocal top scores of Gapped blast with manual corrections (20, 21). The Buchnera genome encodes only 564 proteins (1) and is thus highly reduced in comparison with the E. coli genome, which contains 4,289 genes (11). Almost all genes present in Buchnera (560; 99%) are also present in E. coli (Table 1 and Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). Moreover, 57 (85%) of the 67 proteins encoded in the Reclinomonas mt genome are present in Buchnera (Table 1), and 56 of these 57 counterparts are shared by all four genomes studied here.
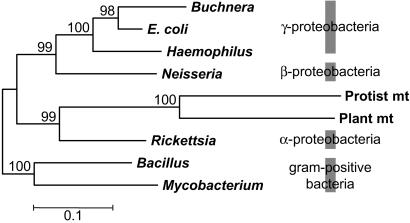
Figure 1.
Phylogenetic tree of 16S rRNA genes from eubacteria and mitochondria: Buchnera (Buchnera sp.); E. coli, Haemophilus (Haemophilus influenzae), Neisseria (Neisseria meningitidis), protist mt (R. americana mt), plant mt (Arabidopsis thaliana mt), Rickettsia (Rickettsia prowazekii), Bacillus (Bacillus subtilis), and Mycobacterium (Mycobacterium tuberculosis). We used 1,358 nucleotides, excluding gap sites. The proteobacterial portion of the tree was rooted by using Gram-positive bacteria. This tree was reconstructed by the neighbor-joining method with the Tamura–Nei-γ distance (12, 13). A γ parameter was estimated to be 0.665 by Gu and Zhang's method (14). Numbers besides branches indicate percent bootstrap values with 1,000 replications.
Table 1.
Numbers of shared protein-coding genes among eubacterial and mitochondrial genomes
| No. of protein-coding genes | No. of shared genes with*
|
|||
|---|---|---|---|---|
| Buchnera | Rickettsia | E. coli† | ||
| Reclinomonasmt | 67 | 57 (85.1%) | 64 (95.5%) | 63 (94.0%) |
| Buchnera | 564 | — | 331 (58.7%) | 560 (99.3%) |
| Rickettsia | 834 | — | — | 562 (67.3%) |
Percentages of shared genes found in the species of the left column are in parentheses.
The number of protein-coding genes in E. coli is 4,289.
In the estimation of the ratio of the evolutionary rates of Buchnera and E. coli, we discarded duplicate genes from the analysis, because they may have experienced higher evolutionary rates. We used Haemophilus influenzae (22) as the outgroup for the Buchnera/E. coli comparison (Fig. 1). In this case, we used the H. influenzae proteins that showed reciprocal top scores to both Buchnera and E. coli ortholog candidates. However, because horizontal gene transfer is known to occur in bacteria (23, 24), these proteins are not necessarily orthologs, and E. coli and H. influenzae proteins may form sister groups in some cases (see Figs. 3 and 4, which are published as supporting information on the PNAS web site). To identify orthologs that can be used for comparing amino acid substitution rates, an additional proteobacterium, Neisseria meningitidis serogroup B (25), was included so that we can examine whether proteins of Buchnera and E. coli form a sister group among four taxa. In total, 340 proteins were found to be present in all four genomes. We reconstructed 340 phylogenetic trees by the neighbor-joining method (12) with p distance and Grishin distance [Poisson-correction (PC) γ distance with a γ parameter (a) of 0.65] (26). A phylogenetic tree of 16S rRNA shows the clear relationship of these proteobacteria, implying that E. coli and Buchnera form a sister group (Fig. 1). However, less than one-third (96) of the 340 genes supported the sister group relationship between Buchnera and E. coli with significant bootstrap values (95% or more) by both p and PC γ distances, whereas 85 supported a sister group of E. coli and H. influenzae, and six supported a Buchnera and H. influenzae cluster at the same bootstrap threshold. This result indicates that a considerable amount of horizontal gene transfer may have occurred among the proteobacteria examined, and that phylogenetic analysis gives much more stringent criteria (27) than reciprocal blast search (21, 28) for identifying orthologous genes. To compare evolutionary rates between Buchnera and E. coli with H. influenzae as the outgroup, the topologies of 96 protein trees supporting the Buchnera/E. coli sister group were further examined; we collected other homologous sequences found in swiss-prot 39 and reconstructed neighbor-joining trees to confirm the 96 topologies. As a result, only 89 (16%) of the 560 genes were found to be shared by Buchnera and E. coli when the H. influenzae proteins were used as the outgroup. Likewise, we identified 60 ortholog data sets of the Reclinomonas mt and R. prowazekii proteins (16, 17) and compared their rates by using E. coli homologs as the outgroup.
Estimation of Relative Substitution Rates.
Protein sequences were aligned by using clustal w (29). We concatenated sequences excluding all gap sites (30) and estimated the number of amino acid substitutions per site by using the Dayhoff distance (Poisson correction γ distance with a = 2.25) (26). We obtained essentially the same results when we used the γ parameters obtained by Gu and Zhang's method (14). To conduct a relative rate test, we constructed a three-species tree including the outgroup. The branch lengths of the tree were estimated by the least-squares method (26), and the variance of the ratio of branch lengths was computed by the standard delta method.
Results and Discussion
Accelerated Evolutionary Rates in Endosymbionts.
The relative evolutionary rates of Buchnera and E. coli genes can be computed by comparing the branchpoint (R)-to-tip distances (amino acid substitutions per 100 sites) for Buchnera (a) and E. coli (b) when H. influenzae (c) was used as the outgroup (see Fig. 1). The distances for R to a, R to b, and R to c were 27.7 ± 0.4, 13.8 ± 0.2, and 30.6 ± 0.4, respectively, when all 89 orthologous proteins were used (Table 2). This finding implies that Buchnera proteins evolved on average two times (2.01 ± 0.04) faster than E. coli proteins. This increased rate in Buchnera was observed in all six different functional groups of proteins examined, consistent with previous results observed for a few proteins (6, 10, 31), although the latter results depend on the assumption that the divergence time of the Buchnera species used is nearly half that between E. coli and its related species Salmonella. Similarly, when 60 proteins were used with E. coli as the outgroup, the branchpoint-to-tip distances for Reclinomonas mt, Rickettsia, and the outgroup were 69.4 ± 1.2, 32.7 ± 0.9, and 67.2 ± 1.2, respectively. Thus, Reclinomonas mt proteins also evolved about two times (2.13 ± 0.07) faster than Rickettsia homologues (Table 2). Because the parasite Rickettsia showed a longer branchpoint-to-tip distance than the free-living bacterium E. coli when Bacillus subtilis was used as the outgroup (data not shown), the increase of the evolutionary rate found in Reclinomonas mt appears much greater than that in Buchnera.
Table 2.
Root-to-tip amino acid substitutions per 100 sites for endosymbionts and their close relatives
| Function* | No. of proteins | Branch length†
|
Ratio | No. of proteins | Branch length‡
|
Ratio | ||
|---|---|---|---|---|---|---|---|---|
| Buchnera | E. coli | Protist mt | Rickettsia | |||||
| Small-molecule metabolism | 34 | 30 ± 1 | 16 ± 0 | 1.9 | 23§ | 38 ± 1 | 38 ± 1 | 1.0 |
| (amino acid synthesis)¶ | (10) | 30 ± 1 | 18 ± 1 | 1.7 | NA‖ | |||
| Broad regulatory functions | 3 | 17 ± 1 | 8 ± 1 | 2.1 | NA‖ | |||
| Macromolecule metabolism | 32 | 26 ± 1 | 13 ± 0 | 2.1 | 32** | 89 ± 2 | 32 ± 1 | 2.8 |
| Cellular processes | 9 | 25 ± 1 | 13 ± 1 | 2.0 | NA‖ | |||
| Others | 11 | 38 ± 2 | 16 ± 1 | 2.3 | 5 | 98 ± 5 | 56 ± 4 | 1.7 |
| All sequences | 89 | 28 ± 0‡‡ | 14 ± 0‡‡ | 2.0 | 60 | 69 ± 1‡‡ | 33 ± 1‡‡ | 2.1 |
NA, not applicable.
For the definition of functional categories, see the gene list at http://buchnera.gsc.riken.go.jp/.
Outgroup is H. influenzae.
Outgroup is E. coli.
Respiration and ATP synthesis.
Amino acid biosynthesis proteins separated from small molecule metabolism proteins.
Mitochondrial proteins were not available.
Translation and transcription.
Differences of the branch lengths were significant (P < 0.0001) for endosymbionts and their close relatives by relative rate tests (26).
Enhanced Mutation Rate and Relaxation of Selection as Causes of the Increased Evolutionary Rate.
Although previous authors proposed Muller's ratchet or the slightly deleterious mutation theory to explain the enhanced evolutionary rate in endosymbionts, as mentioned earlier, a simpler hypothesis in the framework of neutral evolution is to assume that the mutation rate has been enhanced in endosymbionts or that the extent of purifying selection has been relaxed because of changes in functional requirements of individual genes (32). For simplicity, let us express the rate of amino acid substitution per year (α) for a given protein by α = vTf, where vT is the total mutation rate per year, and f is the fraction of neutral or nearly neutral mutations that are determined by functional constraints of the protein (32). If f remains the same for the entire evolutionary time, α is expected to increase as vT increases. Different loci may have different f values, but the proportional increases of α would be the same for all proteins as long as f remains the same for each protein. By contrast, if the increase in α is caused by an increase in f, the proportional increase of α is expected to vary from protein to protein because the extent of relaxation of purifying selection is unlikely to be the same for all proteins (33).
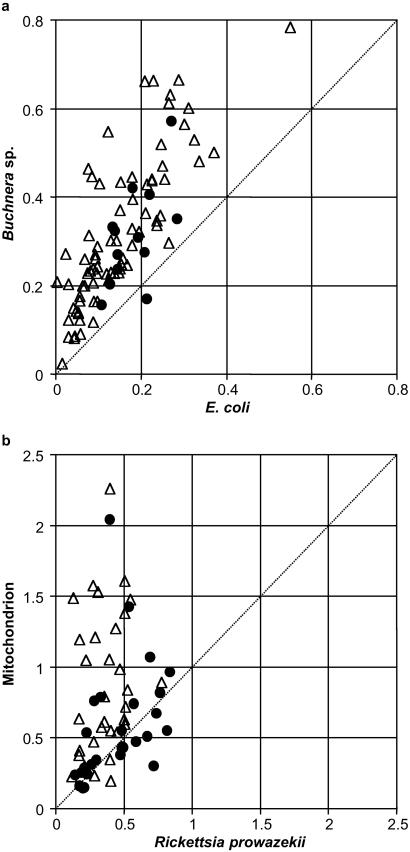
In the case of the Buchnera/E. coli comparison, the evolutionary rate increased for 88 of the 89 orthologs identified (Fig. 2). For 67 of the 88 proteins, the increase was significant at P < 0.05 (see Table 4, which is published as supporting information on the PNAS web site), suggesting that elevated mutation rate rather than relaxed selection is responsible for the uniformly enhanced rate of protein evolution in Buchnera. This is consistent with the observations that Buchnera lacks several important genes involved in DNA repair (1, 15) and that the synonymous substitution rate is apparently two times higher in Buchnera species than in the E. coli/Salmonella comparison (10, 31). It might be worth noting that elevation of evolutionary rates has been reported in other parasitic or symbiotic species in which DNA repair-related genes were absent (34). These observations suggest that the elevated rates of protein evolution are primarily caused by higher mutation rates due to loss of DNA repair genes (see below for relaxation of selection).
Figure 2.
Branch lengths of orthologous proteins between (a) E. coli and Buchnera sp. and (b) R. prowazekii and the mt of R. americana. Closed circles indicate the proteins involved in amino acid biosynthesis in Buchnera and those involved in respiration and ATP synthesis in the mt, respectively. Triangles are proteins in all other functional categories. The abscissa and the ordinate represent the number of amino acid replacements per site.
Elevated evolutionary rate differs among functional categories in the Rickettsia/Reclinomonas mt comparison. Only 12 of the 23 orthologs involved in respiration and ATP synthesis showed an increased rate in Reclinomonas mt (Fig. 2), and none of these increases was statistically significant (see Table 5, which is published as supporting information on the PNAS web site). Of the 37 remaining orthologs, 33 (89%) showed an increased rate in Reclinomonas mt (Fig. 2 and Table 2), and the increase was significant at P < 0.05 in 19 of them (see Table 5, which is published as supporting information on the PNAS web site). Findings from the latter class of proteins suggest that mutation rate has increased in Reclinomonas mt relative to Rickettsia. In the Caenorhabditis elegans mt, which encodes fewer proteins than Reclinomonas mt DNA, an increase in mutation rate has also been reported (35).
That no acceleration of substitution rate was observed in proteins of respiration and ATP synthesis suggests that purifying selection can counterbalance the effect of increased mutation rate in this important functional class, which is the sole known source of net ATP synthesis in both the Reclinomonas mt (16) and the obligate aerobe Rickettsia (17). Similarly, the relatively small increase of substitution rate observed in proteins involved in amino acid biosynthesis in Buchnera (Table 2) also suggests that stronger purifying selection is operating in these proteins on which the host aphids heavily depend (1).
Although we have argued that the major factor for the enhanced rate of protein evolution is an increased mutation rate, we cannot exclude the possibility that relaxation of purifying selection is also partially responsible. One way to distinguish between the two hypotheses is to compute the synonymous (dS) and nonsynonymous (dN) nucleotide substitutions. If increased mutation rate is the main factor, dS would be two times higher in endosymbionts than in free-living bacteria, and the dN/dS ratio would be nearly the same for both endosymbionts and free-living bacteria. By contrast, if the relaxation-of-selection hypothesis is correct, dS would remain nearly the same, whereas dN/dS is expected to be higher in endosymbionts than in free-living bacteria. Unfortunately, synonymous substitutions were saturated for the Buchnera/E. coli and for the Reclinomonas/Rickettsia comparison, so that we could not use this approach. However, previous comparisons of two species of Buchnera and of E. coli with Salmonella (10, 31) have suggested that dS is about two times higher in Buchnera than in the E. coli/Salmonella comparison after adjustment of the divergence times. This result is consistent with the high mutation theory.
Previous studies (10, 31) have also suggested that dN/dS is about four times higher in Buchnera than in the E. coli/Salmonella comparison. If this is true, we would expect that the rate of amino acid substitution should be about eight times higher in Buchnera than in E. coli and Salmonella, because dS is two times higher in the former than in the latter. However, our observation indicates that the rate of protein evolution is only two times higher in the former than in the latter. The reason for this discrepancy is unclear at present, but it is possible that the previous estimates are biased because the number of genes examined was small (10, 31), and plasmid-derived genes as well as chromosomal genes were included (10), suggesting that dN/dS may have been overestimated. It is also possible that nonorthologous genes, which we found to predominate in similar taxon samples (see above), were compared. Consistent with our interpretation are the observations that the dN/dS ratios for several other free-living bacteria are on the same order of magnitude as that of Buchnera, and the dN/dS for the E. coli/Salmonella comparison is exceptionally high (31). If this interpretation is correct, currently available data would generally support the high mutation hypothesis. Nevertheless, we cannot completely exclude the possibility of relaxation of selection at present, because some of the data we used have been obtained indirectly.
Advantageous Mutations.
Theoretically, advantageous mutations are expected to enhance the rate of amino acid substitution. Fares et al. (36) reported that certain amino acid substitutions in the heat-shock protein GroEL of Buchnera are apparently caused by positive Darwinian selection. However, the gene for this protein shows a low dN/dS ratio like other genes (33). Therefore, the effect of this positive selection on the overall rate of amino acid substitution appears to be small.
Muller's Ratchet and Slightly Deleterious Mutations.
Several authors (6–10) have suggested that the enhanced evolutionary rate observed in genes of Buchnera and organelles can be explained by Muller's ratchet (37, 38), which refers to the event that deleterious mutations accumulate in asexual genomes because they cannot be eliminated through recombination. Indeed, theoretical studies have shown that slightly deleterious mutations accumulate at a faster rate in asexual genomes than in sexual genomes (38, 39). However, if deleterious mutations accumulate continuously in a gene, the gene will eventually deteriorate and lose its original function (39–41). Note that here we are considering only the genes that still maintain the original function after 200 million or 1.5 billion years of endosymbiosis. Furthermore, Muller's ratchet cannot work in the presence of back mutation, which surely occurs at the amino acid level (42). Therefore, the applicability of the ratchet hypothesis to the present case is questionable.
Another hypothesis that is often used to explain enhanced substitution rate is the slightly deleterious mutation theory (9, 43–45). According to this hypothesis, slightly deleterious mutations can be fixed in small populations as though they were neutral, and therefore the substitution rate would increase in endosymbiotic organisms, because the effective population size of endosymbionts such as Buchnera is likely to be small. However, if deleterious mutations accumulate continuously for hundreds of millions of years, as in the present case (31), the gene would eventually become nonfunctional, as mentioned above. To maintain the same protein function, fixation of slightly deleterious mutations must be followed by fixation of slightly advantageous mutations, and the frequencies of fixation of the two types of mutations must be more or less the same. If this happens, the rate of amino acid substitution may be enhanced compared with that in organisms of large population size. However, this evolutionary scheme is essentially the same as neutral evolution, because the product of effective population size and selection coefficient must be very small (32). If the proportion of slightly deleterious mutations is greater than that of slightly advantageous mutations, as is often assumed (45), the gene would again eventually deteriorate in long-term evolution. One hypothesis to prevent this situation is to assume that a large-effect advantageous mutation occasionally occurs and rescues the gradual deterioration of gene function. However, this hypothesis is unrealistic, because it is unclear how a single mutation can nullify the effects of many deleterious mutations at the molecular level.
In addition, definitely deleterious mutations are likely to occur quite often in any gene. Therefore, unless all deleterious mutations are eliminated by purifying selection, most genes would become nonfunctional. Once the occurrence of deleterious mutation is balanced by purifying selection, amino acid substitutions are expected to occur following the neutral model of evolution irrespective of population size. Definitely advantageous mutations, which may occur occasionally, would increase the rate of protein evolution, but as long as protein function remains the same, their contribution is expected to be small.
It has been suggested that compensatory mutations, which interact in the three-dimensional protein structure, as in the case of the stem regions of ribosomal RNA, may enhance the substitution rate (6, 45). Here the first mutation is assumed to be deleterious, but the second compensatory mutation is supposed to restore the original function. However, involvement of deleterious mutations in the evolutionary process is expected to reduce the substitution rate compared with the neutral rate (46). In fact, mathematical studies have shown that this is indeed the case (47, 48).
Note that here we are not considering the strict neutrality of alleles, which would never exist in nature, but we are defining amino acid substitutions as neutral or nearly neutral as long as the gene function remains essentially the same. Under this definition, which was often used in the early days of study of molecular evolution (49–52), the compensatory mutations as formulated by Hartl and Taubes (53) belong to the realm of neutral mutations, because they do not change the gene function in long-term evolution (no adaptation). If we consider short-term evolution and intraspecific polymorphism, there may be plenty of slightly deleterious mutations, but they are not our concern in this paper. For the above reasons, we believe the higher rate of amino acid substitution is caused primarily by a higher mutation rate. It would be interesting to test this hypothesis by experiments.
Supplementary Material
Acknowledgments
We thank Shuji Shigenobu for valuable comments on the Buchnera genome annotation and Hiroshi Akashi, Xun Gu, Dan Hartl, Fumio Tajima, Tom Whittam, and Jianzhi Zhang for comments on the manuscript. This work was supported in part by grants from the National Institutes of Health (GM 20293) and the National Aeronautics and Space Administration (NCC2-1057) (to M.N.).
Abbreviation
- mt
mitochondrion
References
- 1.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Nature (London) 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 2.Gray M W, Burger G, Lang B F. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 3.Ochman H, Moran N A. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 4.Silva F J, Latorre A, Moya A. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- 5.Martin W, Müller M. Nature (London) 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 6.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch M. Mol Biol Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- 8.Lynch M. Mol Biol Evol. 1997;14:914–925. doi: 10.1093/oxfordjournals.molbev.a025834. [DOI] [PubMed] [Google Scholar]
- 9.Brynnel E U, Kurland C G, Moran N A, Andersson S G. Mol Biol Evol. 1998;15:574–582. doi: 10.1093/oxfordjournals.molbev.a025958. [DOI] [PubMed] [Google Scholar]
- 10.Clark M A, Moran N A, Baumann P. Mol Biol Evol. 1999;16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- 11.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 14.Gu X, Zhang J. Mol Biol Evol. 1997;14:1106–1113. doi: 10.1093/oxfordjournals.molbev.a025720. [DOI] [PubMed] [Google Scholar]
- 15.Tamas I, Klasson L, Canback B, Naslund A K, Eriksson A S, Wernegreen J J, Sandstrom J P, Moran N A, Andersson S G. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 16.Lang B F, Burger G, O'Kelly C J, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Gray M W. Nature (London) 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 17.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. Nature (London) 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 18.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc London Ser B. 1993;253:167–171. [Google Scholar]
- 19.Javaux E J, Knoll A H, Walter M R. Nature (London) 2001;412:66–69. doi: 10.1038/35083562. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh T, Takemoto K, Mori H, Gojobori T. Mol Biol Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- 22.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 23.Doolittle W F. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 24.Ochman H, Lawrence J G, Groisman E S. Nature (London) 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 25.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, et al. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 26.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ. Press; 2000. [Google Scholar]
- 27.Eisen J A. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 28.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nei M, Xu P, Glazko G. Proc Natl Acad Sci USA. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochman H, Elwyn S, Moran N A. Proc Natl Acad Sci USA. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura M. The Neutral Theory of Molecular Evolution. New York: Cambridge Univ. Press; 1983. [Google Scholar]
- 33.Wernegreen J J, Moran N A. Mol Biol Evol. 1999;16:83–97. doi: 10.1093/oxfordjournals.molbev.a026040. [DOI] [PubMed] [Google Scholar]
- 34.Koonin E V, Mushegian A R, Rudd K E. Curr Biol. 1996;6:404–416. doi: 10.1016/s0960-9822(02)00508-0. [DOI] [PubMed] [Google Scholar]
- 35.Denver D R, Morris K, Lynch M, Vassilieva L L, Thomas W K. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- 36.Fares M A, Barrio E, Sabater-Munoz B, Moya A. Mol Biol Evol. 2001;19:1162–1170. doi: 10.1093/oxfordjournals.molbev.a004174. [DOI] [PubMed] [Google Scholar]
- 37.Muller H J. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pamilo P, Nei M, Li W-H. Genet Res. 1987;49:135–146. doi: 10.1017/s0016672300026938. [DOI] [PubMed] [Google Scholar]
- 40.Kondrashov A S. J Theor Biol. 1995;175:583–594. doi: 10.1006/jtbi.1995.0167. [DOI] [PubMed] [Google Scholar]
- 41.Rispe C, Moran N A. Am Nat. 2000;156:425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- 42.Dayhoff M O. Atlas of Protein Sequence and Structure. Silver Spring, MD: Biomedical Research Foundation; 1972. [Google Scholar]
- 43.Ohta T. Nature (London) 1974;252:351–354. doi: 10.1038/252351a0. [DOI] [PubMed] [Google Scholar]
- 44.Lambert J D, Moran N A. Proc Natl Acad Sci USA. 1998;95:4458–4462. doi: 10.1073/pnas.95.8.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohta T. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 46.Li W-H, Nei M. Genetics. 1977;86:901–914. doi: 10.1093/genetics/86.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura M. J Genet. 1985;64:7–19. [Google Scholar]
- 48.Innan H, Stephan W. Genetics. 2001;159:389–399. doi: 10.1093/genetics/159.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freese E, Yoshida A. In: Evolving Genes and Proteins. Bryson V, Vogel H J, editors. New York: Academic; 1965. pp. 341–355. [DOI] [PubMed] [Google Scholar]
- 50.Margoliash E, Fitch W M, Dickerson R E. Brookhaven Symp Biol. 1968;21:259–305. [PubMed] [Google Scholar]
- 51.King J L, Jukes T H. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 52.Nei M. Molecular Population Genetics and Evolution. Amsterdam: North–Holland; 1975. [PubMed] [Google Scholar]
- 53.Hartl D L, Taubes C H. J Theor Biol. 1996;182:303–309. doi: 10.1006/jtbi.1996.0168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.