Abstract
Zinc finger transcription factors (TFsZF) were designed and applied to transgene and endogenous gene regulation in stably transformed plants. The target of the TFsZF is the Arabidopsis gene APETALA3 (AP3), which encodes a transcription factor that determines floral organ identity. A zinc finger protein (ZFP) was designed to specifically bind to a region upstream of AP3. AP3 transcription was induced by transformation of leaf protoplasts with a transformation vector that expressed a TFZF consisting of the ZFP fused to the tetrameric repeat of herpes simplex VP16's minimal activation domain. Histochemical staining of β-glucuronidase (GUS) activity in transgenic AP3∷GUS reporter plants expressing GUS under control of the AP3 promoter was increased dramatically in petals when the AP3-specific TFZF activator was cointroduced. TFZF-amplified GUS expression signals were also evident in sepal tissues of these double-transgenic plants. Floral phenotype changes indicative of endogenous AP3 factor coactivation were also observed. The same AP3-specific ZFPAP3 was also fused to a human transcriptional repression domain, the mSIN3 interaction domain, and introduced into either AP3∷GUS-expressing plants or wild-type Arabidopsis plants. Dramatic repression of endogenous AP3 expression in floral tissue resulted when a constitutive promoter was used to drive the expression of this TFZF. These plants were also sterile. When a floral tissue-specific promoter from APETALA1 (AP1) gene was used, floral phenotype changes were also observed, but in contrast the plants were fertile. Our results demonstrate that artificial transcriptional factors based on synthetic zinc finger proteins are capable of stable and specific regulation of endogenous genes through multiple generations in multicellular organisms.
Keywords: floral development‖APETALA3‖APETALA1
In nature, eukaryotic nuclear genes are tightly regulated at both the transcriptional and translational levels. Much of this control is achieved through DNA-binding transcription factors. The manipulation of plant traits in agricultural biotechnology would be greatly facilitated if preselected endogenous genes could be turned on or off in a controlled and selective manner. A conceptual approach to such manipulation is the engineered expression of specific native transcription factors that have evolved to control particular genes. Advances in whole-genome sequencing of Arabidopsis (1) and more recently rice (2), combined with informatics-based analysis have allowed the identification of numerous putative plant transcription factors (2, 3). However, the identification and characterization of the molecular targets of these transcription factors is still at a very early stage, and consequently it is not yet possible to use them broadly as gene-specific tools for controlled regulation of endogenous gene expression. Rational design of artificial transcription factors that target specific DNA sequences with non-native nucleotide binding domains fused to transcriptional activation or repression domains is therefore an attractive option. An especially promising approach of this kind utilizes synthetic DNA binding domains of the zinc finger protein (ZFP) class.
Numerous zinc finger DNA-binding domain motifs have been identified in genes originating from plants and other biological systems (4–7). Among these, the Cys-2–His-2 type of ZFP has been the subject of the most extensive structural, biochemical, and genetic studies (4–24). This highly modular zinc finger domain has been found to be particularly amenable to rational manipulation of target binding site specificity. Several design and selection strategies have been developed for construction of synthetic zinc finger-based DNA binding proteins that can be highly specific for given target sequences (8, 10–24). Recently, several studies have demonstrated targeting of endogenous genes in cultured mammalian cells using synthetic ZFP-based artificial transcription factors where the DNA binding domain of the ZFP has been fused to transcriptional activation or repression domains (18–24). Application of this technology to agriculture by means of designing plant-specific zinc finger transcription factors (TFsZF) would potentially enable a range of diverse applications. However, two critical issues remain to be addressed: the function and stability of TFsZF in a multicellular organism that has been regenerated from a transformed cell, and the ability of these genes to be stably inherited in subsequent generations.
In this study, we describe artificial transcription factors based on a designed polydactyl ZFP that specifically targets the AP3 floral development gene of Arabidopsis thaliana. Wild-type Arabidopsis flowers have four organ types (sepal, petal, stamen, carpel) arranged in concentric whorls (25). AP3, a member of the MADS box gene family (26, 27), is involved in specifying the organ identity of floral whorl 2 (petal) and whorl 3 (stamen) (28–30). Altered expression of AP3 results in homeotic mutations where whorl-specific organ identity is perturbed. Severe ap3 mutant alleles and mutations in the related gene pistillata cause organ identity changes and sterility (31, 32). Such readily observed phenotypes make the Arabidopsis flower a useful system in which to study manipulated gene regulation. A suitable target site for ZFP design using GNN repeat motifs (16–19) is located approximately 50 bp upstream of the AP3 TATA box. A six-finger ZFP with predicted specificity for this 18-bp site was designed and synthesized (19). A TFsZF for gene activation was prepared by fusion of the synthetic tetrameric repeat of herpes simplex VP16's minimal activation domain (VP64) to the designed ZFP. For gene-specific repression, a mammalian repression domain, mSin3 interaction domain (Sid), was fused to the ZFP (19, 21). Transformation of these TFsZF into Arabidopsis yielded transgenic phenotypes similar to known ap3 mutant alleles. Introduction of AP3-specific TFsZF into Arabidopsis plants expressing a β-glucuronidase (GUS) reporter gene under control of the AP3 promoter resulted in GUS expression changes reflective of the various promoters used to drive TFsZF expression. Our results demonstrate that artificial transcription factors based on engineered ZFPs can be used to manipulate endogenous transgene gene expression in multicellular organisms in both a transient and stable fashion.
Materials and Methods
Plant Materials and Promoters.
A 1.9-kb AP1 promoter sequence and transgenic plants expressing GUS under control of the AP3 promoter originally generated by Tom Jack's laboratory (25) was obtained from Detlef Weigel (The Salk Institute, La Jolla, CA).
Construction and Characterization of AP3-Specific ZFPAP3.
A ZFP was synthesized to bind to the complementary strand of the18-bp sequence 5′-TACTTCTTCAACTCCATC-3′ found at −112 to −95 relative to the start of translation of the APETALA3 genomic sequence (33). The gene was constructed and the protein expressed and purified as a fusion with maltose binding protein as described. ELISA specificity and electrophoretic mobility shift assays were performed as described.
Construction of Zinc Finger-Effector Domain Fusions and Transformation of Plants.
TFsZF bearing the VP64 activation domain and the Sid repression domain were prepared as described (19). For constitutive expression, the TFsZF were cloned into a dicot expression vector pNOV102 (34) downstream from the UBQ3 promoter (35, 36) and upstream of the nos transcriptional terminator. The resulting constructs (UBQ3∷ZFPAP3-VP64//nos and UBQ3∷Sid-ZFPAP3//nos) were transformed into A. thaliana plants (Columbia) using the agrobacteria-mediated transformation method (37). Protoplast transient transformation and assay were conducted as described (34). Putative transgenic plants were selected for hygromycin resistance as described (34). A nontarget activation construct UBQ3∷ZFPm4-VP64//nos was generated and transformed into Arabidopsis at the same time. ZFPm4 targets the sequence of maize myoinositol 1-phosphate synthase. ZFPm4-VP64 activates transcriptions of myoinositol 1-phosphate synthase in maize cells (34). A transformation control vector UBQ3∷GFP//nos was used for all transformations. For floral tissue-specific regulation, a 1.9-kb fragment containing the AP1 promoter was substituted for the UBQ3 promoter fragment. The resulting constructs (AP1∷ZFPAP3-VP64//nos and AP1∷Sid -ZFPAP3//nos) were transformed into Arabidopsis as described above.
RT-PCR Analysis of AP3 Expression Level.
Plant tissues (either protoplasts or floral tissue from stages 1 to approximately stage 15; ref. 38) were collected and frozen immediately in liquid nitrogen. Total RNA was prepared from these samples by using the Ambion RNAwiz kit (Ambion). The expression levels of the ZFP artificial transcription factors and endogenous AP3 were monitored by RT-PCR. RT-PCRs were carried out in 25-μl volumes by using 200 ng of RNA and the Qiagen 1-step RT-PCR kit. AP3-specific primers were Ap3-F, 5′-GGCGAGAGGGAAGATCCAG-3′ and Ap3–4R, 5′-CTCCTCTAATACGACTCACTATAGGGACACTCACCTAGCCTCTG-3′. The thermocycler settings were 50°C for 30 min, 95°C for 15 min (94°C for 30 s, 60°C for 30 s, 72°C for 1 min) × 30 cycles, and 72°C for 10 min.
Quantitative PCR Analysis of AP3 Expression Level.
Total RNA used for RT-PCR was assayed quantitatively on an ABI Prism 7900 Sequence Detector (Taqman) according to the manufacturer's instructions (Beckman Coulter). All probes and primers were designed with the program PRIMER EXPRESS with the default setting (Perkin–Elmer). For AP3 detection, the probe sequence 5′-CCATTTCATCCTCAAGACGACGCAGCT-3′ was used with primers 5′-TTTGGACGAGCTTGACATTCAG-3′ (forward) and 5′-CGCGAACGAGTTTGAAAGTG-3′ (reverse). Taqman PCRs were carried out by using 250 ng total RNA and Taqman one-step RT-PCR master mix reagent (Perkin–Elmer). Thermal cycling conditions were 48°C for 30 min, 95°C for 10 min for 1 cycle, then 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative expression was quantified by using the comparative Ct method with the thioredoxin gene TRX3 as an internal expression standard. The probe sequence used for TRX3 was 5′-AGACTTCACTGCAACATGGTGCCCAC-3′ with primers 5′-GTGTGGAAATGACACAGATTGTGA-3′ (forward) and 5′-AGACGGGTGCAATGAAACG-3′ (reverse).
GUS Histochemical Staining and Analysis of Expression Pattern.
GUS histochemical staining was conducted as described (39, 40). Freshly excised floral tissues were immersed immediately in GUS staining solution containing 0.25 mM 5-bromo-4-chloro-3-indolyl β-D-glucuronide cyclohexylammonium salt (X-glu, Rose Scientific, Edmonton, AB, Canada) in 50 mM sodium phosphate buffer (pH 7.3), and then incubated 20 h at 37°C. Stained tissues were dehydrated in an ethanol series and photographed before infiltration with Histo-Clear (International Diagnostics, Atlanta), and paraffin-cast specimen blocks (41) were sectioned on a microtome (Microm HM315, Mikron) and stained in situ with 0.1% aqueous Safranin-O for 5–60 s. Stained surfaces of specimen blocks were treated with immersion oil and visualized under a dissecting microscope (Olympus, New Hyde Park, NY).
Results
Synthesis and Characterization of ZFP.
The target sequence of the designed AP3-specific ZFPAP3 is an 18-bp sequence located in the AP3 promoter region, at −112 to −95 relative to the ATG codon (Fig. 1A). This six-domain ZFP was assembled from zinc finger domains of predefined specificity as described (16, 17, 19, 23). ZFPAP3 was expressed in Escherichia coli and purified. ZFPAP3 demonstrated good specificity in ELISA assays using a panel of oligonucleotide target sequences (data not shown). The affinity of the ZFPAP3 for its designed target was determined to be 2.3 nM by electrophoretic mobility assays. This affinity is in the range we have previously determined to be required for endogenous gene regulation (21, 23).
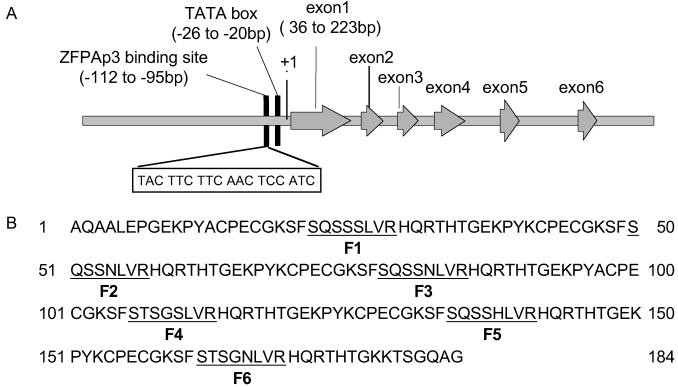
Figure 1.
(A) ZFPAP3 target sequence (boxed) and its position in the AP3 5′ UTR. Numbers indicate the distance from the ATG translation initiation codon. The arrowed boxes indicate the exon of AP3. (B) DNA recognition helix sequences of the ZFPAP3 protein. The underlined amino acids are the components of the new zinc fingers that provide specificity for the selected nucleotide sequences indicated in A. The recognition helices of fingers 1–6 (F1–F6) are underlined.
Transient Activation of Endogenous AP3 in Arabidopsis Leaf Cells.
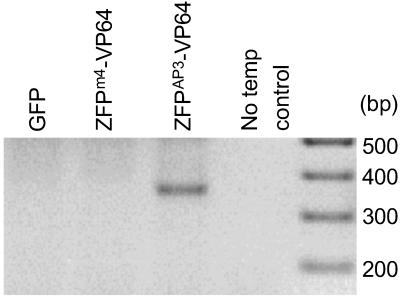
AP3 is normally expressed exclusively in developing flowers, and no expression of the gene has been reported in leaf mesophyll protoplasts. Such protoplasts were used to investigate transcriptional activation of AP3. The AP3-specific activation construct UBQ3∷ZFPAP3-VP64//nos and a nontarget activation construct UBQ3∷ZFPm4-VP64//nos (which targets the sequence of maize myoinositol 1-phosphate synthase) were transformed into Arabidopsis leaf protoplasts. UBQ3∷GFP//nos was used as a transformation control. All samples were analyzed by RT-PCR to determine the baseline levels of endogenous AP3 transcripts. The AP3-specific primers were designed to yield a 600-bp product if amplification is based on a genomic DNA template (indicating contamination) and 334 bp for transcribed AP3 messages. The originally silent AP3 gene was activated in cells that were transformed with the AP3-specific activation construct UBQ3∷ZFP AP3-VP64//nos but not with any other constructs (Fig. 2). These results indicate that the ZFPAP3 domain is able to bind AP3 DNA and direct the activation domain (VP64) to the specific and otherwise silenced endogenous target in vivo. Furthermore, TFsZF targeted to different genes (e.g., ZFPm4) did not activate AP3 in a nonspecific manner.
Figure 2.
Activation of the silenced endogenous AP3 gene in Arabidopsis leaf cells. RT-PCR was used to detect AP3 expression in Arabidopsis leaf protoplasts transformed with GFP (control), ZFPm4-VP64 (nonspecific activation control), and ZFPAP3-VP64 (AP3-specific activation). The last lane (−) is no template control amplification.
Phenotypic Consequences of in Planta Activation of AP3.
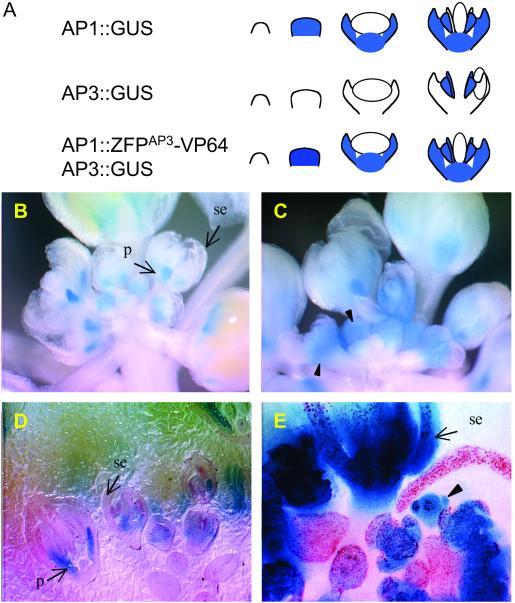
Attempts to stably transform Arabidopsis with the construct UBQ3∷ZFPAP3-VP64//nos to constitutively activate AP3 expression were inefficient. Few transformants could be obtained with this vector. Most had rearrangements that lacked the VP64 domain. None expressed ZFPAP3-VP64. To circumvent this problem, we restricted AP3 activation to floral tissue. For this, the APETALA1 (AP1) promoter was used to direct expression of ZFPAP3-VP64. To dissect AP1-specific regulation of AP3 from the morphological consequence of altered AP3 transcription factor activity, we examined the effects of AP1∷ZFPAP3- VP64//nos first in an AP3-driven GUS reporter background (AP3∷GUS). This reporter has been characterized (25) and established a characteristic AP3-specific pattern of GUS staining. Floral tissue-specific activation construct AP1∷ZFPAP3- VP64//nos and control construct AP1∷ZFPAP3//nos were transformed into homozygous Arabidopsis plants harboring construct AP3∷GUS. The expression of AP3 starts from stage 3 of flower development and is accumulated in petal and stamen primordia only. As a result of this late expression, AP3∷GUS plants accumulate a moderate level of the stable reporter in the petals and stamens, as illustrated in Fig. 3A. In contrast, AP1 is expressed at the very early stages (stages 1–3) of flower development (42, 43), when the floral organ primordia are just beginning to initiate. This results in an AP1 signal that is uniformly distributed throughout the young floral primordia and later in development is restricted to the sepal and petal primordia (Fig. 3A). Consequently, we predict that a plant double-transformed with constructs AP1∷ZFPAP3-VP64//nos and AP3∷GUS will initiate AP3-directed expression at the very early stage of floral primordia development (stages 1–3), coinciding with AP1 expression, and will also show the petal and stamen pattern at later stages of the characteristics of normal AP3 expression. To test this hypothesis, flowers from many double transgenic plants were stained for GUS and observed either with or without tissue mounting. The GUS staining patterns shown in Fig. 3 revealed strong activation of the AP3 promoter in an AP1-dependent manner as predicted: (i) the GUS signals expanded throughout the entire flower primordia, starting at the very earliest stages of development (Fig. 3 C and E); (ii) the GUS signal was detected in the sepals of mature flowers (Fig. 3 C and E); and (iii) the intensity of the GUS signal increased dramatically in the petals of mature flowers (Fig. 3 C and E). These patterns were distinct from the petal and stamen staining of the AP3∷GUS line. In our system, both the transgenic GUS gene and the native AP3 transcription factor gene should each be subject to AP1-dependent activation by the AP3-targeted TFsZF. Reflective of this, we found that several independent transgenic plants showed distinct phenotypic changes in floral development, suggesting an altered pattern of AP3 transcription factor activity. In these plants, some whorl 1 organs are replaced by whorl 2 organs, resulting in flowers having five or more petals (Fig. 4B) and reduced or absent sepals. The stamens appear normal and all plants are fertile. An interesting floral phenotype was observed in some of the floral-specific activation events obtained using construct AP1:ZFPAP3-VP64. These plants exhibited a markedly higher percentage of young flowers in the inflorescence tissue as compared with controls (data not shown). Activation of the AP3∷GUS transgene and the endogenous AP3 gene were both found to be stable genetically and were transmitted faithfully to progeny over two subsequent generations (T3).
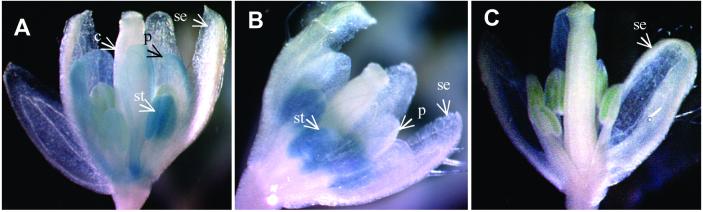
Figure 3.
GUS staining flowers of background plant with AP3∷GUS only and double transgenic plant with both AP3∷GUS and activation construct AP1∷ZFPAP3-VP64//nos. (A). Predicted GUS staining patterns of AP3∷GUS and AP1∷GUS (based on personal communication with Martin Yanofsky, University of California, San Diego). (B) Flowers from background plant with AP3∷GUS only stained for GUS activity. GUS signal is detected only in the petal (p) and stamen (not visible here, see Fig. 6A), but not in the carpal (not visible) and sepal (se). Picture taken directly after staining procedure. (C) Flowers from double transgenic plant expressing AP3∷GUS and AP1∷ZFPAP3-VP64//nos simultaneously. GUS signal is increased in petal and is detected throughout the young flower primordia. (D) Mounted flowers from background plant with AP3∷GUS only stained for GUS activity. GUS signal is detected only in the petal (p) and stamen (not visible here, see Fig. 7A) and not in the carpal and sepal (se). (E) Mounted flowers from double transgenic plant expressing AP3∷GUS and AP1∷ZFPAP3-VP64//nos simultaneously. GUS signal is increased in petal (p), extended to sepal (se), and detected throughout the young flower primordia (arrow).
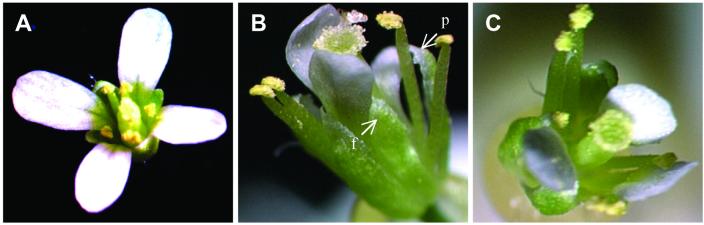
Figure 4.
Floral phenotypic changes in double transgenic plant expressing AP3∷GUS and AP1∷ZFPAP3-VP64//nos simultaneously. A seven-petal flower is shown here. Two extra petals are fully converted (f), and the third one is partially converted (p).
In Planta Repression of Endogenous AP3.
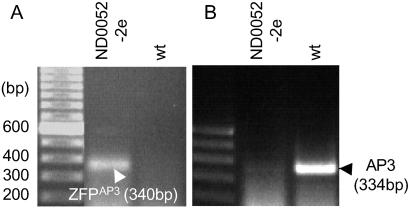
For in planta repression, a constitutive UBQ3 promoter was used to drive the expression of an AP3-specific ZFP fused to a human transcriptional repression domain (19). This construct (UBQ3∷Sid-ZFPAP3//nos) was transformed into wild-type Arabidopsis. We found that both TFsZF and endogenous AP3 expression levels varied over a wide range among independent transgenic events. However, endogenous AP3 gene expression levels were repressed in most transgenic lines. Higher TFsZF expression in line ND0052–2e (Fig. 5A) produced a marked down regulation of endogenous AP3 expression (Fig. 5B). Quantitative RT-PCR analysis revealed a nearly 50-fold AP3 repression in this line (data not shown). We also observed that most of the transgenic plants having reduced AP3 expression were sterile and had unopened flowers at maturity. Dissection of flowers from ND0052–2e plants revealed petals that were shorter and narrower than wild-type flowers of the same maturity. In addition, the lower portions of the petals were converted partially to sepal-like structures, and the stamens were greatly reduced in size compared with wild type. The floral phenotypes of the strongly AP3-repressed plants are similar to the flowers of the characterized ap3 and sap mutants (44, 45).
Figure 5.
Repression of endogenous AP3 expression by the constitutive repression construct UBQ3∷Sid-ZFPAP3//nos in transgenic plant ND0052–2e. (A) RT-PCR identification of transgene ZFPAP3 in transgenic event ND0052–2e and wild-type control plant. (B) RT-PCR evaluation of endogenous gene AP3 expression level in transgenic event ND0052–2e and wild-type plant. In plant ND0052–2e, the expression of AP3 is significantly repressed by the expression of repressor Sid-ZFPAP3 fusion protein. Quantitative PCR indicated 46-fold repression.
Floral tissue-specific repression, the construct AP1∷ Sid-ZFPAP3//nos and control construct AP1∷ZFPAP3//nos, lacking a repression domain, were transformed into Arabidopsis plants already carrying an AP3∷GUS (25) transgene stably integrated into the genome. Because AP1 is expressed in whorls 1 and 2 (sepals and petals), whereas AP3 is expressed in whorls 2 and 3 (petals and stamens), we predicted that the GUS signal should be eliminated from whorl 2 but not whorl 3 in the double transgenic plant-expressing construct AP1∷Sid-ZFPAP3//nos in the presence of AP3∷GUS. Fig. 6 shows this expected pattern of stamen-only GUS expression. Petals from some of these flowers are absent, as expected if the endogenous AP3 gene is repressed (Fig. 6C). Two types of petal morphology were observed in these double transformants: missing petals (Figs. 6C and 7C) and partial conversion of petals to sepals (sepaloid petals) (Fig. 7B). These plants were fully fertile despite the floral alterations. Only the constitutive expressed ZFPAP3 plants were sterile.
Figure 6.
GUS staining flowers of background plant with AP3∷GUS only and double transgenic plant with both AP3∷GUS and repression construct AP1∷Sid-ZFPAP3//nos. (A) Flowers from background plant with AP3∷GUS only stained for GUS activity. GUS signal is detected only in the petal (p) and stamen (st) and not in the carpal and sepal (se). Picture taken directly after staining procedure. (B) Flowers from double transgenic plant expressing AP3∷GUS and AP1∷Sid-ZFPAP3//nos simultaneously. GUS signal disappeared from petals but is detectable in stamens. (C) Flowers from a different double transgenic plant expressing AP3∷GUS and AP1∷Sid-ZFPAP3//nos simultaneously. Low level GUS activity is detectable in stamens. In addition, the petals were absent in this flower.
Figure 7.
Floral phenotypic changes in double transgenic plant expressing AP3∷GUS and AP1∷Sid-ZFPAP3//nos simultaneously. (A) Flowers from background plant with AP3∷GUS only. (B) Flowers from double transgenic plant expressing AP3∷GUS and AP1∷Sid-ZFPAP3//nos simultaneously. A three-petal flower is shown here. In this flower, an extra sepal is fully replaced by one petal (f), and another petal is partially replaced (p). (C) Flowers from a different double transgenic plant expressing AP3∷GUS and AP1∷Sid-ZFPAP3//nos simultaneously with one missing petal.
Discussion
We have shown that ZFP-based artificial transcription factors can be designed and synthesized to manipulate transgene and endogenous gene expression levels in transgenic plants. Although regulation of only one transgene (GUS) and one endogenous gene (AP3) are presented here, we believe this approach will be generally applicable to all genes. This study supports and extends previous studies of TFsZF that have been designed to activate and repress endogenous genes in mammalian cells (8, 21–24). Therefore, we believe that this approach should be viable in other transgenic organisms as well.
Our studies indicate that transcriptional activation and repression with our TFsZF are specific for the targeted AP3 gene. Preliminary GeneChip (47) analysis of transgenic plants with or without ZFPAP3-effector fusion revealed no significant changes in gene expression in the 8,000 nontargeted floral or nonfloral genes examined (data not shown). Despite this apparent specificity, recovery of plant lines that constitutively expressed TFsZF activators (UBQ3∷ZFPAP3-VP64//nos) was inefficient. In addition, we observed that the VP64 tetrameric repeat was subject to somatic rearrangement in plant cells. This result parallels difficulties encountered in the generation of transgenic animals and cell lines expressing the VP16 activation domain or designed versions of this domain like VP64, suggesting that this activation domain itself carries with it an intrinsic toxicity (48).
We believe that our TFsZF approach to gene regulation can be further enhanced by combination with other gene regulation technologies, such as inducible gene expression, which has been recently adapted to create chemically regulated TFsZF (49). Tissue-specific expression of the TFsZF provides for another level of control as demonstrated here. Jack and colleagues have shown that overexpression of AP3 causes conversion of sepals to petals, carpels to stamens, as well as loss of fertility (32, 44). Presumably, tissue-specific activation of AP3 in our AP1-driven activation plants allows them to maintain their fertility. Likewise, transcriptional repression with the human Sid domain in plants was potent and yielded phenotypes analogous to those observed in plants harboring a temperature-sensitive allele of AP3 (46), suggesting that both gain of function and loss-of-function phenotypes are accessible by using zinc finger technology in whole organisms.
Abbreviations
- ZFP
zinc finger protein
- TFsZF
zinc finger transcription factors
- VP64
tetrameric repeat of herpes simplex VP16's minimal activation domain
- Sid
mammalian mSin3 interaction domain
- GUS
β-glucuronidase
- ZFPAP3
AP3-specific zinc finger protein
Note Added in Proof.
In a companion article, we have demonstrated the efficacy of our TFZF approach in transgenic tobacco plants maintained over several generations (50).
References
- 1.The Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.Goff S, Ricke D, Lan T, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 3.Riechmann J, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe O J, Samaha R R, et al. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 4.Berg J M. Annu Rev Biophys Biophys Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- 5.Ashworth A, Denny P. Mamm Genome. 1991;1:196–200. doi: 10.1007/BF00351068. [DOI] [PubMed] [Google Scholar]
- 6.Pieler T, Bellefroid E. Mol Biol Rep. 1994;20:1–8. doi: 10.1007/BF00999848. [DOI] [PubMed] [Google Scholar]
- 7.Takatsuji H. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beerli R, Barbas C F., III Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 9.Elrod-Erickson M, Rould M A, Nekludova L, Pabo C O. Structure (London) 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 10.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Yang W, Barbas C F., III Proc Natl Acad Sci USA. 1995;92:344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rebar E J, Pabo C O. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 13.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe S A, Grant R A, Elrod-Erickson M, Pabo C O. Structure (London) 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 15.Greisman H A, Pabo C O. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 16.Segal D J, Dreier B, Beerli R R, Barbas C F., III Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreier B, Segal D J, Barbas C F., III J Mol Biol. 2000;303:489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Segal D J, Ghiara J B, Barbas C F., III Proc Natl Acad Sci USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beerli R R, Segal D J, Dreier B, Barbas C F., III Proc Natl Acad Sci USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang J S, Kim J. J Biol Chem. 2000;275:8742–8748. doi: 10.1074/jbc.275.12.8742. [DOI] [PubMed] [Google Scholar]
- 21.Beerli R R, Dreier B, Barbas C F., III Proc Natl Acad Sci USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Spratt S K, Liu Q, Johnstone B, Qi H, Raschke E E, Jamieson A C, Rebar E J, Wolffe A P, Case C C. J Biol Chem. 2000;275:33850–33860. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]
- 23.Dreier B, Beerli R, Segal D, Flippin J, Barbas C F., III J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Rebar E J, Zhang L, Liu Q, Jamieson A C, Liang Y, Qi H, Li P X, Chen B, Mendel M C, et al. J Biol Chem. 2001;276:11323–11334. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- 25.Weigel D. Annu Rev Genet. 1995;29:19–39. doi: 10.1146/annurev.ge.29.120195.000315. [DOI] [PubMed] [Google Scholar]
- 26.Theissen G, Saedler H. Curr Opin Genet Dev. 1995;5:628–639. doi: 10.1016/0959-437x(95)80032-8. [DOI] [PubMed] [Google Scholar]
- 27.Bowman J L, Baum S F, Eshed Y, Putterill J, Alvarez J. Curr Top Dev Biol. 1999;45:155–205. doi: 10.1016/s0070-2153(08)60316-6. [DOI] [PubMed] [Google Scholar]
- 28.Yanofsky M. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:167–188. [Google Scholar]
- 29.Irish V F. Dev Biol. 1999;209:211–220. doi: 10.1006/dbio.1999.9226. [DOI] [PubMed] [Google Scholar]
- 30.Jenik P D, Irish V F. Development (Cambridge, UK) 2001;128:13–23. doi: 10.1242/dev.128.1.13. [DOI] [PubMed] [Google Scholar]
- 31.Jack T, Brockman L L, Meyerowitz E M. Cell. 1992;68:683–687. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 32.Jack T, Fox G L, Meyerowitz E M. Cell. 1994;76:703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- 33.Irish V F, Yamamoto Y T. Plant Cell. 1995;7:1635–1644. doi: 10.1105/tpc.7.10.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barbas, C. F., III, Stege, J., Guan, X. & Dalmia, B. (2001) PCT WO 01/52620.
- 35.Norris S R, Meyer S E, Callis J. Plant Mol Biol. 1993;21:895–906. doi: 10.1007/BF00027120. [DOI] [PubMed] [Google Scholar]
- 36.Callis J, Raasch J A, Vierstra R D. J Biol Chem. 1990;265:12486–12493. [PubMed] [Google Scholar]
- 37.Bechtold N, Pelletier G. In: Arabidopsis Protocols. Martinez-Zapater J, Salinas J, editors. Clifton, NJ: Humana; 1998. pp. 259–266. [Google Scholar]
- 38.Bowman J, editor. Arabidopsis, An Atlas of Morphology and Development. New York: Springer; 1993. pp. 135–145. [Google Scholar]
- 39.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sessions A, Weigel D, Yanofsky M. Plant J. 1999;20:259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- 41.Ruzin S E, editor. Plant Microtechnique and Microscopy. New York: Oxford Univ. Press; 1999. pp. 57–86. [Google Scholar]
- 42.Mandel A M, Gustafson-Brown C, Savidge B, Yanofsky M F. Nature (London) 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 43.Gustafson-Brown C, Savidge B, Yanofsky M F. Cell. 1994;76:131–143. doi: 10.1016/0092-8674(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 44.Byzova M V, Franken J, Aarts M G, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne M M, Angenent G C. Genes Dev. 1999;13:1002–1014. doi: 10.1101/gad.13.8.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi Y, Jack T. Plant Cell. 1998;10:1465–1477. doi: 10.1105/tpc.10.9.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren D, Collingwood T N, Rebar E J, Wolffe A P, Camp H S. Genes Dev. 2002;16:27–32. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu T, Wang X. Plant Physiol. 2000;124:1472–1476. doi: 10.1104/pp.124.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akagi K, Kanai M, Saya H, Kozu T, Berns A. Nucleic Acids Res. 2001;29:1–5. doi: 10.1093/nar/29.4.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beerli R R, Schopfer U, Dreier B, Barbas C F., III J Biol Chem. 2000;275:32617–32627. doi: 10.1074/jbc.M005108200. [DOI] [PubMed] [Google Scholar]
- 50.Ordiz M I, Barbas C F, III, Beachy R N. Proc Natl Acad Sci USA. 2002;99:13290–13295. doi: 10.1073/pnas.202471899. [DOI] [PMC free article] [PubMed] [Google Scholar]