Abstract
Anaplasma phagocytophilum, an unusual obligate intracellular pathogen that persists within neutrophils, causes human anaplasmosis (previously known as human granulocytic ehrlichiosis). To study the effects of this pathogen on the transcriptional profile of its host cell, we performed a comprehensive DNA microarray analysis of the early (4-h) transcriptional response of human neutrophils to A. phagocytophilum infection. A. phagocytophilum infection resulted in the up- and down-regulation of 177 and 67 neutrophil genes, respectively. These data were verified by quantitative reverse transcription-PCR of selected genes. Notably, the up-regulation of many antiapoptotic genes, including the BCL2A1, BIRC3, and CFLAR genes, and the down-regulation of the proapoptotic TNFSF10 gene were observed. Genes involved in inflammation, innate immunity, cytoskeletal remodeling, and vesicular transport also exhibited differential expression. Vascular endothelial growth factor was also induced. These data suggest that A. phagocytophilum may alter selected host pathways in order to facilitate its survival within human neutrophils. To gain further insight into the bacterium's influence on host cell gene expression, this report presents a detailed comparative analysis of our data and other gene expression profiling studies of A. phagocytophilum-infected neutrophils and promyelocytic cell lines.
Anaplasma phagocytophilum is the causative agent of human anaplasmosis (formerly human granulocytic ehrlichiosis), an emerging infectious disease in the United States, Europe, and Asia (10, 16, 22, 59). In nature, A. phagocytophilum cycles between its arthropod vector, Ixodes scapularis, and its primary mammalian reservoir, Peromyscus leucopus, the white-footed mouse (22, 58). Humans are accidental hosts.
The survival strategies employed by this gram-negative obligate intracellular bacterium are poorly understood (13). After entering its host neutrophil, A. phagocytophilum resides in a membrane-bound vacuole that does not fuse with lysosomes, thus avoiding phagocytic killing (25, 54). Other mechanisms exploited by this bacterium to survive within the hostile intraneutrophil environment include circumventing and directly inactivating neutrophil-killing mechanisms, delaying neutrophil apoptosis, and manipulating neutrophil chemokine expression (2, 4, 11, 12, 17, 55, 67, 77).
Currently, there is no reliable system for genetically manipulating A. phagocytophilum. However, as intracellular pathogens have been shown to subvert many host cell functions (64), an alternative approach for understanding the pathogenic strategies utilized by these organisms would be to look at changes in the host cell gene expression profile following infection. In the case of A. phagocytophilum, we have previously shown by use of macroarray analysis that the bacterium causes repression of rac2 mRNA expression in human promyelocytic HL-60 cells, with a resulting influence on the host cell capacity to generate a respiratory burst (12). Recent microarray studies by our group and others have analyzed the global gene expression responses of HL-60 cells (21), NB4 cells (60), and human neutrophils (8) to A. phagocytophilum infection.
In this report, we present an analysis of the early transcriptional response of human neutrophils to A. phagocytophilum (strain HZ) infection. While Borjesson et al. (8) used a gene array comprising 14,500 genes to study the neutrophil response to A. phagocytophilum strain NCH-1, our study relied upon a more comprehensive array consisting of 38,500 genes. We have chosen to examine the host cell transcriptional response at 4 h postinfection because we have previously shown that under the in vitro conditions employed, it takes approximately 4 h for ≥90% of the cell population to become infected (11). In addition to stimulating expression pathways typical of an antimicrobial response, our findings indicate that A. phagocytophilum promotes an antiapoptotic transcriptional profile and induces several additional pathways to collectively ensure a favorable environment for its survival and dissemination. Furthermore, this study also hints at the possible interactions of infected neutrophils with nonhematopoietic cells of the vascular system. The results of this study, in addition to confirming the results of Borjesson et al., provide notable and promising new information.
The availability of data from four previous studies that examined the transcriptional response of human neutrophils (8), HL-60 cells (12, 21), and NB4 cells (60) to A. phagocytophilum infection at various time points offers an excellent opportunity for comparison with the current study. Such a comparative analysis will contribute to the understanding of principles commonly involved in A. phagocytophilum survival within host cells.
MATERIALS AND METHODS
Cultivation of A. phagocytophilum.
A. phagocytophilum strain HZ was kindly provided by Ralph Horowitz of New York Medical College (Valhalla) and Yasuko Rikihisa of Ohio State University (Columbus). A. phagocytophilum was cultivated in HL-60 cells (240 CCL; American Type Culture Collection, Manassas, VA) as previously described (4, 26).
Isolation of neutrophils.
Human neutrophils were isolated as previously described (11). The neutrophil viability was determined to be ≥98% by trypan blue dye exclusion. The preparations contained >99% granulocytes, of which >95% were neutrophils, 1 to 3% were eosinophils, and <0.8% were monocytes as determined by Giemsa staining of cytocentrifuged (Thermo Electron, Pittsburgh, PA) samples. All studies with human blood were performed in accordance with protocols approved by the Human Investigation Committee at Yale University.
Isolation of A. phagocytophilum.
Host-cell-free A. phagocytophilum isolates were prepared as previously described (11). The number of host-cell-free A phagocytophilum isolates obtained was estimated according to the method of Kim and Rikihisa (38, 39).
Infection of neutrophils with A. phagocytophilum in vitro.
One ml of fresh neutrophils (106 neutrophils/ml) was added to individual wells of a six-well Ultra Low Attachment plate (Corning Inc., Corning, N.Y.). To these were added suspensions of freshly prepared A. phagocytophilum isolates that had been liberated from 5 × 106 (≥90%) infected HL-60 cells. The plates were incubated at 37°C in 5% CO2 for 4 h. The percentage of infected neutrophils was confirmed by immunofluorescence microscopy, and the number of A. phagocytophilum isolates per neutrophil was calculated as previously described (11). The ratio of bacteria to neutrophils was ∼5:1. At 4 h postinfection, the cells were recovered by centrifugation at 210 × g, and RNA was isolated as described below. For enzyme-linked immunosorbent assays (ELISA), the culture supernatants were collected by centrifugation and stored at −20°C until use.
RNA isolation, microarray hybridization, and statistical analysis.
Total RNA was isolated using TRIZOL (Invitrogen) according to the manufacturer's instructions. RNA samples were purified and DNase treated using the RNeasy mini kit (QIAGEN, Valencia, CA). Subsequent labeling reactions, hybridization, staining, and washing were performed according to the gene chip expression analysis technical manual (Affymetrix, Santa Clara) at the W. M. Keck Facility (Yale University). The Human Genome U133 Plus 2.0 array (Affymetrix) containing 54,675 probe sets was used. Image data were quantified using the Microarray Suite version 5.0 software package. Three independent experiments utilizing human neutrophils obtained from three independent donors were performed using separate gene chips per donor. There were two comparison groups (A. phagocytophilum-infected and noninfected neutrophils) with three replicates per group. A total of six CEL files were obtained by Microarray Suite version 5.0 analysis. The DNA chip analyzer (dChip, version 1.3) was applied to normalize the CEL files and to compute the model-based gene expression (Perfect Match-only model) (46). Genes were filtered out if the lower 90% confidence bound fold change was ≤1.5-fold (45). We used the Student's t test (P < 0.05) to identify differentially expressed genes between the groups. The false discovery rate of <5% was estimated by permutation. The final lists of genes were further classified based on function using the gene ontology (GO) classification (see Table S1 in the supplementary material). The GO classification was based on the GO slim format (http://www.geneontology.org/GO.slims.shtml) (see Table 2).
TABLE 2.
Genes differentially expressed in human neutrophils following 4 h of A. phagocytophilum infectiona
| Gene function and symbolb | GenBank IDc | Fold change | ||||
|---|---|---|---|---|---|---|
| Host defense, signal, transduction/reception, cell recognition | ||||||
| ANGPT4 | NM_015985 | 1.99 | ||||
| ANKRD15 | D79994 | 9.32 | ||||
| ARL8 | AW270158 | 5.33 | ||||
| ARL10C | NM_018184 | 2.02 | ||||
| ATP6V1C1 | AW024925 | 3.31 | ||||
| CCL3 | NM_002983 | 4.60 | ||||
| CCL4 | NM_002984 | 7.40 | ||||
| CCL20 | NM_004591 | 35.41 | ||||
| CCRL2 | AF015524 | 3.63 | ||||
| CD44 | AV700298 | 4.32 | ||||
| CD48 | NM_001778 | 2.35 | ||||
| CD83 | NM_004233 | 4.93 | ||||
| CDC42EP3 | AF104857 | 2.04 | ||||
| CEACAM1 | NM_001712 | 2.27 | ||||
| CLECSF9 | NM_014358 | 3.16 | ||||
| CPD | D85390 | 1.99 | ||||
| CXCL1 | NM_001511 | 3.02 | ||||
| CXCL2 | M57731 | 7.26 | ||||
| CXCL3 | NM_002090 | 22.14 | ||||
| CYBB | AI308863 | 3.34 | ||||
| DDIT4 | NM_019058 | 2.48 | ||||
| EHD1 | AF001434 | 3.25 | ||||
| GCH1 | NM_000161 | 8.80 | ||||
| GPR84 | AF237762 | 11.48 | ||||
| GPR108 | AL365404 | 2.20 | ||||
| HLA-DRA | M60334 | 2.12 | ||||
| ICAM1 | A1608725 | 5.87 | ||||
| IL1B | NM_000576 | 10.54 | ||||
| IL1RN | BE563442 | 10.42 | ||||
| IL8 | NM_000584 | 1.79 | ||||
| IL23A | NM_016584 | 2.25 | ||||
| IL12B | NM_002187 | 16.01 | ||||
| IRAK2 | NM_001570 | 5.25 | ||||
| LIMS1 | AL110164 | 3.22 | ||||
| MAP3K4 | AL109942 | 2.43 | ||||
| NBS1 | AK001017 | 4.08 | ||||
| NFKB1 | M55643 | 4.27 | ||||
| NFKB2 | NM_002502 | 4.10 | ||||
| NFKBIA | AI078167 | 2.13 | ||||
| NFKBIE | NM_004556 | 4.27 | ||||
| NFKBIZ | BE646573 | 5.03 | ||||
| ORM1 | NM_000607 | 3.33 | ||||
| PIK3AP1 | BC029917 | 4.82 | ||||
| PLAU | K03226 | 6.29 | ||||
| PLA2G7 | NM_005084 | 2.42 | ||||
| PLD1 | NM_002662 | 2.42 | ||||
| PPP1R15A | NM_014330 | 3.62 | ||||
| PPP1R15B | BF796046 | 1.88 | ||||
| P2RX4 | NM_002560 | 2.64 | ||||
| PTAFR | M80436 | 2.28 | ||||
| PTGER2 | NM_000956 | 4.05 | ||||
| PTX3 | NM_002852 | 12.96 | ||||
| RAB8B | NM_016530 | 2.81 | ||||
| RAB21 | NM_014999 | 3.34 | ||||
| REL | NM_002908 | 2.85 | ||||
| RELB | NM_006509 | 2.43 | ||||
| RIN2 | AL136924 | 4.49 | ||||
| RIPK2 | AF064824 | 3.12 | ||||
| SAMSN1 | AF519621 | 4.08 | ||||
| SCARF1 | NM_003693 | 2.04 | ||||
| SLAMF7 | AJ271869 | 4.66 | ||||
| SNX9 | BC005022 | 3.22 | ||||
| SPPL2A | AI674647 | 1.92 | ||||
| TAGAP | BF591040 | 2.05 | ||||
| TA-NFKBH | NM_032721 | 5.07 | ||||
| TANK | NM_004180 | 2.51 | ||||
| TBK1 | NM_013254 | 2.22 | ||||
| T2BP | AA195074 | 10.84 | ||||
| TLR2 | NM_003264 | 3.02 | ||||
| TLR4 | AF177765 | 1.95 | ||||
| TNFAIP6 | AW188198 | 1.99 | ||||
| TPRA40 | NM_016372 | 2.62 | ||||
| TRIF | NM_182919 | 4.99 | ||||
| CAMK2G | BF111268 | −2.91 | ||||
| DSIPI | AL110191 | −1.88 | ||||
| FLJ21047 | NM_024569 | −2.83 | ||||
| FRAT1 | NM_005479 | −3.28 | ||||
| FRAT2 | AB045118 | −2.56 | ||||
| HOM-TES-103 | AL080214 | −2.28 | ||||
| IL8RA | NM_000634 | −3.55 | ||||
| IL8RB | NM_001557 | −2.33 | ||||
| ITPK1 | AF279372 | −2.50 | ||||
| MMP25 | NM_022718 | −2.10 | ||||
| PECAM1 | AW574504 | −3.05 | ||||
| PRKCL2 | A1633689 | −2.99 | ||||
| PXN | NM_002859 | −2.04 | ||||
| RAB3D | NM_004283 | −2.87 | ||||
| RAB11-FIP4 | NM_032932 | −2.20 | ||||
| RAB37 | NM_001006637 | −3.21 | ||||
| SDC2 | AL577322 | −2.77 | ||||
| SELPLG | NM_003006 | −2.21 | ||||
| TGFBI | NM_000358 | −3.03 | ||||
| XPC | D21089 | −2.14 | ||||
| Cell cycle, growth, apoptosis | ||||||
| ADA | X02189 | 5.15 | ||||
| ADORA2A | NM_000675 | 6.09 | ||||
| BCL2A1 | NM_004049 | 2.26 | ||||
| BCL3 | NM_005178 | 2.41 | ||||
| BIRC3 | U37546 | 3.92 | ||||
| BTG2 | BG339064 | 3.88 | ||||
| BTG3 | NM_006806 | 4.12 | ||||
| CASP1 | U13699 | 2.54 | ||||
| CFLAR | U97075 | 3.45 | ||||
| EGLN2 | AW057545 | 2.07 | ||||
| EML4 | NM_019063 | 2.05 | ||||
| GADD45B | AF078077 | 8.02 | ||||
| G0S2 | NM_015714 | 3.32 | ||||
| INSIG1 | NM_005542 | 2.71 | ||||
| LIMK2 | AL117466 | 2.10 | ||||
| MAP3K8 | NM_005204 | 4.61 | ||||
| MARCKS | AW163148 | 3.68 | ||||
| MRPS24 | BF970023 | 2.59 | ||||
| PDCD1LG1 | AF233516 | 6.06 | ||||
| SERPINB9 | BC002538 | 3.07 | ||||
| SOD2 | AL050388 | 5.64 | ||||
| TFRC | NM_003234 | 3.54 | ||||
| TIEG | NM_005655 | 3.62 | ||||
| TNFAIP3 | AI738896 | 3.50 | ||||
| TRAF1 | NM_005658 | 12.09 | ||||
| VEGF | AF022375 | 3.15 | ||||
| APOL2 | BC004395 | −1.75 | ||||
| CBL | AV710415 | −2.17 | ||||
| CDK11 | AI738802 | −2.73 | ||||
| FKSG44 | BF435621 | −1.84 | ||||
| MYST3 | NM_006766 | −2.01 | ||||
| TNFSF10 | NM_003810 | −4.36 | ||||
| TNFSF13B | AF134715 | −2.96 | ||||
| Metabolism | ||||||
| ACSL5 | AW173691 | 3.87 | ||||
| APG7L | NM_006395 | 2.53 | ||||
| B4GALT5 | AL035683 | 4.96 | ||||
| DECR1 | AF049895 | 4.22 | ||||
| GK | X68285 | 2.51 | ||||
| LSS | AW084510 | 2.21 | ||||
| NDUFV2 | NM_021074 | 2.63 | ||||
| OAZIN | NM_015878 | 3.03 | ||||
| PSMA6 | BC002979 | 3.09 | ||||
| SLC7A11 | NM_014331 | 2.49 | ||||
| SLC11A2 | NM_000617 | 6.53 | ||||
| SLC17A7 | NM_020309 | 6.12 | ||||
| SLC39A8 | AB040120 | 9.59 | ||||
| SLC43A3 | BC003163 | 5.34 | ||||
| UBE2E1 | AL518159 | 2.74 | ||||
| BLVRA | NM_000712 | −2.28 | ||||
| CHST13 | AA677272 | −3.05 | ||||
| DGKZ | NM_003646 | −2.38 | ||||
| IDS | AF050145 | −1.97 | ||||
| ME2 | BC000147 | −2.44 | ||||
| MPPE1 | NM_023075 | −1.98 | ||||
| NT5C3 | AF312735 | −2.51 | ||||
| PFKFB4 | AL038787 | −2.43 | ||||
| PRKAG2 | AF087875 | −2.08 | ||||
| Transcription and translation | ||||||
| BAZIA | NM_013448 | 2.24 | ||||
| DNAJB9 | AL080081 | 2.01 | ||||
| DUSP2 | NM_004418 | 2.90 | ||||
| EGR1 | NM_001964 | 2.11 | ||||
| EYA3 | NM_172098 | 2.21 | ||||
| IBRDC2 | R83905 | 3.18 | ||||
| IER5 | NM_016545 | 2.46 | ||||
| IF1H1 | BC046208 | 2.71 | ||||
| IRLB | BC041706 | 5.43 | ||||
| MAFF | NM_012323 | 6.77 | ||||
| MTF1 | AW182367 | 3.07 | ||||
| PLAGL2 | NM_002657 | 2.71 | ||||
| RANBP2 | D42063 | 2.45 | ||||
| SLC7A5 | AB018009 | 4.80 | ||||
| TFEC | AI830469 | 9.05 | ||||
| TJP2 | NM_004817 | 2.72 | ||||
| XBP1 | NM_005080 | 5.76 | ||||
| ZNF272 | X78931 | 2.51 | ||||
| ANKFY1 | AK025960 | −2.28 | ||||
| BRD3 | NM_007371 | −3.21 | ||||
| CNOT6L | BF103856 | −1.87 | ||||
| DPEP2 | NM_022355 | −2.52 | ||||
| HSPC063 | AU144305 | −2.35 | ||||
| KIAA1718 | BE217882 | −2.64 | ||||
| PDLIM2 | NM_021630 | −2.07 | ||||
| PRKAG1 | AI394529 | −1.92 | ||||
| PTK9L | NM_007284 | −2.15 | ||||
| RERE | AB036737 | −4.54 | ||||
| SCAP | D83782 | −2.20 | ||||
| SLBP | NM_006527 | −4.09 | ||||
| TCBAP0758 | BC024312 | −2.46 | ||||
| TOR1B | AF317129 | −1.86 | ||||
| ZBP1 | NM_030776 | −2.10 | ||||
| ZCCHC2 | NM_017742 | −1.87 | ||||
| ZC3HAV1 | BG533558 | −2.29. |
Results from three separate experiments were compared (fold change, >1.5; P < 0.05) as described in Materials and Methods. The fold change data represent the mean fold up- or down-regulation of genes from three individuals.
Genes are classified by function (GO slim format). Gene symbol represents the symbol of the gene represented by the oligonucleotide in the spot of the array.
GenBank ID is the National Center for Biotechnology Information accession number of the gene represented by the oligonucleotide in the spot of the array.
QRT-PCR confirmation.
The RNA used as template for quantitative real-time (QRT)-PCR analysis was the same as used for the microarray analysis. Two micrograms were used to synthesize first-strand cDNA using the iScript cDNA synthesis kit (Bio-Rad, Richmond, CA). Quantitative PCR analysis of triplicate samples of cDNA from a single blood donor (experiment 3 of the three microarray experiments) was performed with the iCycler real-time detection system (Bio-Rad). Real-time PCR was performed according to the iQ SYBR Green Supermix (Bio-Rad) protocol. The selected host genes and their primer sequences are given in Table 1. Melting curve analysis and gel electrophoresis were used to check product specificity. The relative gene expression levels of each transcript were determined by comparison with a standard curve and normalized by dividing the relative gene expression by the expression value of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal standard (iCycler IQ software, version 3.1; Bio-Rad).
TABLE 1.
Gene-specific primers used for quantitative PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| CFLAR | GAGATTCATGCCCTTATCTAGC | TGAATTCCACATTCTTCATCGCT |
| BIRC3 | GGACAGGAGTTCATCCGTCAA | GGGCTGTCTGATGTGGATAGC |
| IL-8RA | CATGTCAAATATTACAGATCC | TACTTGTTGAGTGTCTCAGTTT |
| TNFSF10 | GTAGCAGCTCACATAACTG | AGTTCACCATTCCTCAAG |
| TRAF1 | GTGCCAGTTTGAATGCAGTC | TGGATGCCTCAGTTAATCAG |
| T2BP | TTAACTGCTCTTATTAATCTGTG | GAGAACTGTCATCTTAGGAGT |
| CXCL3 | AGAACAGCAGCTTTCTAGGGA | ACCCTGTCATTTATCAAGGTG |
| IL12B | TTCAGCTTTAGCTTCCATGGCA | TGTTAAGAAGCCACCTGCCATT |
| CCL20 | TCCTGGCTGCTTTGATGTCA | AGAATACGGTCTGTGTATCC |
| IL-8 | AACTAACAATCCTAGTTTGA | TTACTAATATTGACTGTGGA |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC |
ELISA.
Culture supernatants from one of three experiments used for microarray analysis was used for the assay. Aliquots (50 μl) of supernatant from uninfected or A. phagocytophilum-infected neutrophils were examined for levels of interleukin 8 (IL-8) and chemokine (C-C) ligand 20 (CCL20) using the Human Quantikine ELISA kit (R&D Systems), according to the manufacturer's instructions. Captured IL-8 and CCL20 were quantified at 450 nm. Results are presented as mean ± standard error of the mean. Statistical significance was evaluated by the Student's t test (P < 0.05).
RESULTS
Anaplasma phagocytophilum infection alters neutrophil gene expression.
To gain insight into the molecular processes that are modulated at the level of transcription by A. phagocytophilum in human neutrophils, we used the Human Genome U133 Plus 2.0 Array (Affymetrix), which is comprised of 54,675 probe sets encompassing 38,500 genes. We compared the gene expression patterns of uninfected and A. phagocytophilum-infected human neutrophils at 4 h postinfection. The percentage of infected neutrophils was confirmed to be between 85% and 90% with ∼5 organisms per cell (over three experiments) by immunofluorescent microscopy using polyclonal antiserum raised against A. phagocytophilum (data not shown). Experiments were performed on three sets of gene chips, each using RNA from a unique donor as a target. Transcripts that were found to be significantly up- or down-regulated (fold change, >1.5; P < 0.05; false discovery rate, <5%) in all three sets when comparing infected cells with uninfected cells comprised the final list of genes. We observed an A. phagocytophilum infection-dependent transcription profile that encompassed 244 differentially expressed genes. To facilitate subsequent analysis, the differentially expressed genes were divided into several categories based on function (GO slim format) (see Table S1 in the supplementary material). The summarized results are given in Table 2. The relative patterns of up- and down-regulated genes associated with various biological functions are also shown in Fig. 1.
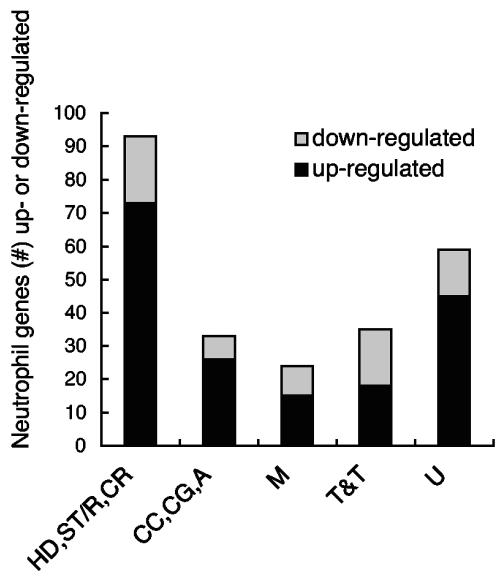
FIG. 1.
Differentially regulated genes in neutrophils following 4 h of A. phagocytophilum infection. The genes were assigned to the following categories according to their biological functions by using the Gene Ontology database: HD, host defense; ST/R, signal transduction/reception; CR, cell recognition; CC, cell cycle; CG, cell growth; A, apoptosis; M, metabolism; T&T, transcription and translation; U, unknown. The relative numbers of up- and down-regulated genes in neutrophils following A. phagocytophilum infection compared with uninfected neutrophils are shown for each gene category.
In general, microarray analysis of A. phagocytophilum-infected neutrophils revealed changes in the expression of genes that are integral for cell survival, cellular adhesion, host defense, inflammation, vesicular transport, and metabolism. Categories of particular interest are addressed in Discussion.
Comparison with other global gene expression studies of A. phagocytophilum-infected host cells.
Borjesson and colleagues have also studied the global changes in neutrophil transcription following A. phagocytophilum infection (8). We and de la Fuente et al. have also performed similar studies using the HL-60 and NB4 promyelocytic cell lines (12, 21, 60). The availability of data from these four global expression analyses presents an excellent opportunity for delineating common transcriptional profiles, if any, induced by A. phagocytophilum among different host cell types. However, it should be kept in mind that there are differences in the postinfection time points at which each of these experiments was performed as well as in the array probe sets and statistical methodologies used. Because the current study centered on the early transcriptional response of neutrophils to A. phagocytophilum infection, we have accordingly placed most of our focus for the comparative analyses on the results obtained by Borjesson et al. at 3 and 6 h postinfection.
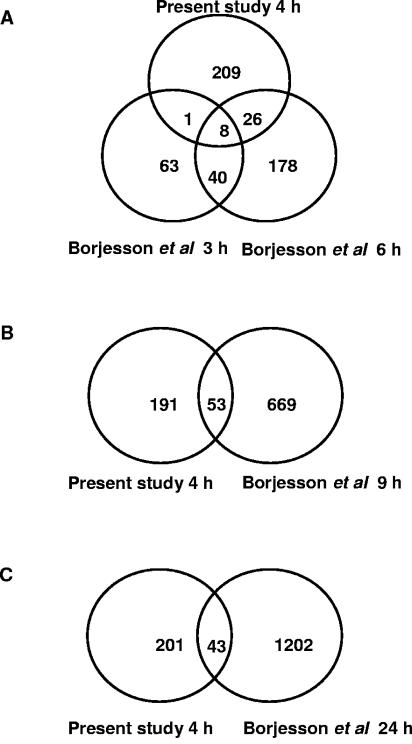
Supplementary Table S2 provides a comprehensive comparison of the differentially expressed genes studied by our group and Borjesson and colleagues. The results are summarized in Fig. 2. In our study, of the 244 genes altered in expression, 177 were up-regulated and 67 were down-regulated at 4 h postinfection. Borjesson et al. observed that out of a total of 112 differentially expressed genes at 3 h, 51 were up-regulated and 61 were down-regulated. At 6 h, 133 and 119 genes were up- and down-regulated, respectively. In our study, the up-regulated genes were far greater in number (73%) than the down-regulated genes at 4 h postinfection. This is in contrast to the results of Borjesson et al., who observed that at 3 h, up-regulated genes were only slightly more numerous than the down-regulated ones. Though the number of down-regulated genes had increased by 6 h, the number was relatively equal to and had not surpassed that of the up-regulated genes. A notable observation is that our data and those of Borjesson et al. had only a limited number of the altered genes in common. At 3 h, our results and those obtained by Borjesson et al. had nine differentially expressed genes in common, while there were 34 in common at 6 h (Fig. 2A). At 9 h and 24 h, our study and Borjesson's had 53 (Fig. 2B) and 43 (Fig. 2C) genes in common, respectively. We have also analyzed the relative number of up- and down-regulated genes in a biological function-specific manner (Fig. 1). There are fewer down-regulated genes than up-regulated ones in the categories of host defense/signal transduction/receptors/recognition, cell cycle/growth/apoptosis, metabolism, and unknown function. On the other hand, genes associated with transcription/translation functions have relatively comparable numbers of up- and down-regulated genes. Similarly, at 3 h Borjesson et al. observed more up-regulated genes than down-regulated genes in the categories of apoptosis and cell fate, cell adhesion and host defense, and metabolism and vesicular trafficking. On the other hand, down-regulated genes dominated in the classes of transcription and DNA binding and signal transduction. At 6 h, genes involved in apoptosis and cell fate and transcription and DNA binding were predominantly up-regulated, while those associated with signal transduction, metabolism, and vesicular trafficking were similar in numbers. The unknown function category contained more down-regulated genes at 3 h, 6 h, and 24 h. But at 24 h, notably, up-regulated genes dominated all categories except that of unknown function.
FIG. 2.
Comparison of gene expression profiles of A. phagocytophilum-infected neutrophils. The observed changes in neutrophil gene expression following 4 h of A. phagocytophilum infection as observed in this study were compared with those observed at (A) 3 h and 6 h; (B) 9 h; and (C) 24 h as observed by Borjesson and colleagues (8).
Another noteworthy study on the host response to A. phagocytophilum infection was by de la Fuente et al., who analyzed the expression profile of HL-60 cells at 3 days postinfection by using a synthetic polynucleotide microarray comprising 21,329 human genes (21). Out of approximately 200 genes whose expression was altered at least twofold during A. phagocytophilum infection of undifferentiated HL-60 cells, none were in common with our results. A recent report from our group on the transcriptional response of NB4 cells at 4 h post-A. phagocytophilum infection, which like the present study relied on the Human Genome U133 Plus 2.0 array (Affymetrix), identified differential transcription of 894 genes (60). However, only one of those genes, PLA2G7, was in common with differentially expressed genes identified in the current study. Consistent with the results in the present study, both de la Fuente et al. and Pedra et al. observed a greater proportion of up- versus down-regulated host cell genes upon A. phagocytophilum infection. While the results of Borjesson et al. at 6 h had three genes (MAF, ITGB1, and MCL-1) in common with data from a previous study using NB-4 cells (60), they had only one gene (UGCG) in common with de la Fuente and colleagues' study of HL-60 cells (21). Our initial hematologic and immunologic macroarray study (12) on the response of HL-60 cells to A. phagocytophilum infection identified eight differentially expressed genes. Two of these genes, IL-8 and IL-1β, were found to be up-regulated in the present study. Moreover, Borjesson et al. detected increased expression of both genes at late time points (8). However, neither the study by de la Fuente et al. (21) nor that by Pedra et al. (60) detected altered expression of IL-8 and IL-1β. Strikingly, among those genes altered in expression according to the host cell responses of two different promyelocytic cell lines, HL-60 (21) and NB-4 (60), only three were shared (GRM3, GNAS, and CTNND1) among them.
Validation of microarray results by QRT-PCR analysis.
In order to validate the gene chip data, QRT-PCR was performed using total RNA from donor 3. We chose eight genes (corresponding to the CASP8- and FADD-like apoptosis regulator [CFLAR], baculoviral IAP repeat-containing gene 3 [BIRC3], TRAF2 binding protein [T2BP], tumor necrosis factor receptor-associated factor 1 [TRAF1], natural killer cell stimulatory factor 2 [IL-12B], CCL20, chemokine [C-X-C motif] ligand 3 [CXCL3], and IL-8) that were up-regulated and two genes (corresponding to tumor necrosis factor [ligand]) superfamily member 10 [TNFSF10] and IL-8 receptor alpha [IL-8RA]) that were down-regulated following A. phagocytophilum infection. As presented in Table 3, the results obtained via QRT-PCR are consistent with the gene chip findings.
TABLE 3.
Validation of differential gene expression by use of real-time PCRa
| Gene | Fold induction of expression levels as determined by:
|
|
|---|---|---|
| Real-time PCR | Microarray analysis | |
| CFLAR | 5.5 | 3.45 |
| BIRC3 | 2.54 | 3.92 |
| IL-8RA | −1.8 | −3.55 |
| TNFSF10 | −5.31 | −4.36 |
| TRAF1 | 15.79 | 12.09 |
| T2BP | 4.39 | 10.84 |
| CXCL3 | 30.44 | 22.14 |
| IL12B | 9.56 | 16.01 |
| CCL20 | 24.12 | 35.41 |
| IL-8 | 4.08 | 1.79 |
Data are given as fold induction of expression levels in A. phagocytophilum-infected versus uninfected neutrophils.
Confirmation of microarray results by ELISA.
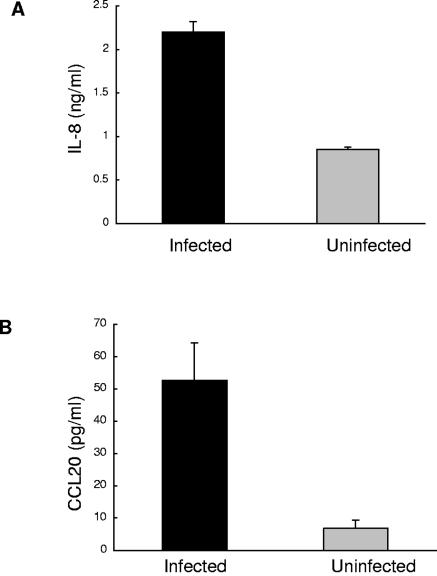
To determine whether the transcriptional induction detected by microarray analysis correlated with the expression of the corresponding protein, we assayed for differential expression of two proteins, IL-8 and CCL20, in infected culture supernatants. As shown in Fig. 3, the levels of both proteins support their corresponding mRNA levels.
FIG. 3.
Confirmation by ELISA of changes in observed differential transcription. The culture supernatants from one of the three independent experiments used for microarray analysis were used for the determination of IL-8 (A) and CCL20 (B) levels by ELISA. Results are expressed as means ± standard errors of triplicate samples.
DISCUSSION
In this study, we examined the early (4-h) transcriptional response by human neutrophils to infection with A. phagocytophilum (strain HZ). Three of the previous host response studies to A. phagocytophilum infection were incomplete due to their use of very small gene arrays (12) and/or nonneutrophil precursor cells (21, 60). A recent analysis by Borjesson et al. of the gene expression profiles of human neutrophils upon A. phagocytophilum infection, though very significant and informative, relied on a moderately sized array comprised of 14,500 genes (8). The current study constitutes a more representative analysis of the early transcriptional response of human neutrophils to A. phagocytophilum infection, using a comprehensive human DNA microarray having ∼2.5-fold more genes than the array used by Borjesson and colleagues. An additional feature of the present study is the comparative analysis of the similarities and differences among the responses by human neutrophils and promyelocytic cell lines to A. phagocytophilum infection encompassed by all gene expression profiling studies to date.
Anaplasma phagocytophilum infection alters neutrophil gene expression.
The results show that A. phagocytophilum infection alters the transcription of a large number of host genes within 4 h of infection. The host transcriptional response at this time point likely represents an early response to intracellular infection. On the other hand, it is also possible that the bacterium directly influences host gene transcription to facilitate its long-term survival. Previous studies on global changes in neutrophil transcriptional profiles following phagocytosis of a diverse group of bacterial pathogens identified the differential expression of 256 genes at 3 h postingestion (41). Also, the study by Borjesson and colleagues demonstrated the differential expression of 112 neutrophil genes at 3 h post-A. phagocytophilum infection (8). Our data, which identified 244 differentially expressed genes at 4 h postinfection, is consistent with these reports. The genes of particular interest are analyzed below according to functional classification.
Host proinflammatory cytokines and chemokines are considered to be responsible for many of the pathological and clinical manifestations associated with A. phagocytophilum infection (38-40). Previous reports have shown that A. phagocytophilum infection induces the secretion of several chemokines, including IL-8, MCP-1, CCL3 (MIP-1α), and CCL4 (MIP-1β) from neutrophils, bone marrow progenitors, and promyelocytic HL-60 cells (2, 40). Consistent with these data, our study also demonstrated highly up-regulated expression of genes encoding CXCL2 (MIP-2α), CXCL3 (MIP-2β), CCL20 (MIP-3α), and CCL4 with induction levels ranging from 7.2- to 35.4-fold. IL-8, CCL3, and CXCL-1 (GRO-α) demonstrated induction levels of 1.8- to 4.6-fold. We also found a 10.5-fold up-regulation of IL-1β. Previous reports have shown that A. phagocytophilum infection results in elevated expression of IL-1β by host cells (12, 39). Thus, A. phagocytophilum-infected neutrophils appear to mount a powerful inflammatory response to this pathogen. Alternatively, it can be hypothesized that A. phagocytophilum selectively up-regulates the expression of some of these genes for its own survival advantage. For example, we have previously shown that IL-8 secreted by infected neutrophils attracts naïve neutrophils, which consequently aids in dissemination of A. phagocytophilum infection (2). A. phagocytophilum also induced the expression of genes encoding cell surface molecules like CD44 and TNFAIP6, which may also contribute to the inflammatory process. The expression of CD44 has been shown to contribute to several types of inflammatory diseases, such as arthritis and colitis (63). TNFAIP6 is a member of a family of hyaluronate binding proteins closely related to adhesion receptor CD44 and is often associated with extracellular matrix remodeling (43). In order to prevent the tissue damage caused by the production of excessive inflammatory cytokines, a balance has to be maintained by anti-inflammatory molecules and/or inhibitors of inflammatory signal transduction pathways. Indeed, anti-inflammatory cytokines, such as IL-1RA, and the G-protein-coupled receptor ADORA2A showed increased expression upon A. phagocytophilum infection.
Contrary to the up-regulation of chemokines, we found that genes encoding key surface chemokine receptor molecules including IL-8RA (CXCR1), IL-8RB (CXCR2), and PECAM1 (CD31) were down-regulated in A. phagocytophilum-infected neutrophils. A recent report has shown that Helicobacter pylori regulates neutrophil activation and migration in gastric mucosa by simultaneously up-regulating IL-8 expression and down-regulating the expression of IL-8RA (CXCR1) and IL-8RB (CXCR2) on neutrophils (68). Thus, the down-regulation of chemokine receptors IL-8RA and IL-8RB may be a pathogenic mechanism of A. phagocytophilum.
Innate immunity has evolved a number of arms to recognize extracellular and intracellular pathogens. Toll-like receptors (TLRs) constitute a major family of extracellular pathogen detection proteins, while the cytosolic NOD (nucleotide binding oligomerization domain) proteins are involved in the detection of intracellular pathogens (62). TLR2 and TLR4, both of which are known to interact with various bacterial products (71), were found to be up-regulated upon A. phagocytophilum infection. It is important to note, however, that a previous study suggested that TLRs do not play a crucial role in subduing A. phagocytophilum infection (75). RIPK2, an integral component of the NOD pathway (32), was up-regulated in A. phagocytophilum-infected neutrophils, thereby hinting at the possible involvement of this pathway in the immune system's clearance of A. phagocytophilum infection. PTX3, which is proposed to act as a soluble pattern recognition receptor (23), was also found to be up-regulated.
Gamma interferon was reported to have a role in the clearance of A. phagocytophilum infection (1). The cytokine IL-12 is shown to induce secretion of gamma interferon (14). IL-12B, a component of IL-12, was up-regulated in this study.
A poorly investigated area in A. phagocytophilum pathogenesis is the role of the bacterium in eliciting tissue damage. It has been suggested previously that A. phagocytophilum-induced neutrophil degranulation may have a role in the proinflammatory tissue injury associated with human anaplasmosis (19). Our microarray analysis showed the up-regulation of the expression of plasminogen activator urokinase, which supports tissue remodeling by breakdown of fibrin clots and degradation of extracellular matrix (7, 52). Thus, the increased levels of plasminogen activator urokinase may also contribute to the tissue damage associated with A. phagocytophilum infection (44).
Though many intracellular bacterial pathogens have been shown to modify the actin cytoskeleton of their host cells (65), there has been no definitive demonstration of this phenomenon in the invasion and intraneutrophil survival of A. phagocytophilum. A recent study implicated the involvement of caveola-dependent mechanisms in the entry of A. phagocytophilum (47). Involvement of actin polymerization in caveola-mediated entry of simian virus 40 is well documented (61). Notably, the neutrophil response to A. phagocytophilum at 4 h postinfection showed the up-regulation of many genes that promote actin polymerization. These include: Lin11, Isl-1, and Mec-3 (LIM) kinase 2 (LIMK2), which phosphorylates and inactivates actin depolymerizing factor/cofilin to promote actin filament formation (3); phospholipase D1 (PLD1), which is involved in actin fiber formation (50) as well as phagocytosis (34); myristoylated alanine-rich C kinase substrate, which is seen on nascent and older phagosomes alike and is an inducer of actin polymerization (79); and Rab8b, which is known to colocalize with actin (42). Contrary to the induction of many actin polymerization-promoting genes, we also observed down-regulation of Rab3D, an exocytosis regulator previously shown to be involved in the polymerization of actin around zymogen granules (73).
While it has been shown that A. phagocytophilum-containing vacuoles do not fuse with lysosomes (54, 77), little is known about the molecular mechanisms associated with this phenomenon. In the present study, two small GTPases involved in vesicular transport were found to be up-regulated concomitant with A. phagocytophilum infection. These are Rab21, which plays a role in early endosomal dynamics (69), and Rab8b, another secretory pathway associated GTPase (15). On the other hand, the expression of some of the other small GTPases was found to be down-regulated following A. phagocytophilum infection. These are Rab3D and Rab37, which are associated with exocytosis of secretory vesicles (51, 73) and secretory granules (49), respectively, and Rab11-FIP4, which has been shown to mediate endosomal recycling (48). Transferrin receptor, a marker of early endocytic compartments, was found to be elevated in the present study. However, a previous report showed that A. phagocytophilum-containing vacuoles do not colocalize with transferrin receptor (5). While all these genes are reported to be involved in regulation of vesicular transport, further experiments are required to elucidate their involvement in the intracellular survival of A. phagocytophilum.
Pathogen-mediated enhancement of host cell survival has been widely documented (29, 35, 74, 78, 80). Overall, a prosurvival response encompasses up-regulation of antiapoptotic pathways, down-regulation of proapoptotic components, and clearance of toxic metabolites. The gene expression pattern of A. phagocytophilum-infected neutrophils indicates the activation of prosurvival signaling. Induction of the transcription factor NF-κB is generally associated with a prosurvival response (70). The neutrophil response to A. phagocytophilum infection showed a remarkable elevation in the expression of NF-κB signaling with several family members exhibiting up-regulation, including NF-κB1, NF-κB2, RELB, REL, BCL3, and NF-κB1A. Another survival-promoting factor exhibiting increased expression is IL-1β, which has been linked to protection against cell death (9). Removal of free radicals is extremely important for minimizing cytotoxicity. Consistent with this notion, we have observed an increase in the transcription of the free radical scavenger manganese superoxide dismutase (SOD2).
A. phagocytophilum delays apoptosis of its host (67, 81), which likely serves as a mechanism for providing the bacterium with sufficient time for multiplication. A recent study has indicated that the organism inhibits neutrophil apoptosis via up-regulation of the antiapoptotic factor bfl-1 (24). The recent microarray analysis by Borjesson et al. (8) reported the differential expression of many apoptotic pathway genes indicative of a delay of cell death after A. phagocytophilum infection. The present investigation also presents a gene expression profile consistent with an antiapoptotic effect. BCL2A1 (BFL1), as well as many other antiapoptotic genes, is up-regulated with concomitant inhibition of proapoptotic genes. This is in contrast to the neutrophil expression profile following infection with Streptococcus pyogenes, a pathogen that represses many survival pathways (41). Two additional classes of antiapoptotic molecules that are up-regulated in A. phagocytophilum-infected neutrophils are the inhibitor-of-apoptosis (IAP) family of proteins and CFLAR. CFLAR is reported to promote cell survival by both inhibiting initiator caspases and activating survival signals (37). BIRC3 (cIAP2) can block apoptosis at multiple levels, including inhibition of caspases (66). Up-regulation of IAPs, which have been implicated in Toxoplasma gondii-mediated apoptosis inhibition (53), further supports the notion that A. phagocytophilum actively delays neutrophil apoptosis. TRAF1, another antiapoptotic gene (33), was also found to be up-regulated. Also interesting is the down-regulation of the proapoptotic gene TNFSF10 (TRAIL [tumor necrosis factor-related apoptosis-inducing ligand]), which has been shown to be a very potent proapoptotic gene in some tumor cells (27) and has recently been implicated in the regulation of neutrophil apoptosis (36). Also, it has been demonstrated that the antiapoptotic activity of IL-1β can prolong the otherwise short life of peripheral neutrophils (76). In accordance with the elevated expression of IL-1β, the expression of caspase 1, which converts pro-IL-1β to active IL-1β (6), was also up-regulated in our current microarray analysis. Collectively, these data convincingly demonstrate the selective modulation of neutrophil apoptosis by A. phagocytophilum. In contrast to that observed for neutrophils, it has been shown that A. phagocytophilum-infected HL-60 cells are considerably more apoptotic than uninfected cells (30). Thus, A. phagocytophilum-induced apoptosis delay is a neutrophil-specific process and not a global consequence of infection (13).
Infection with A. phagocytophilum modifies the binding of neutrophils to endothelial cells (18). A. phagocytophilum-infected neutrophils are reported to lose surface expression of P-selectin glycoprotein ligand-1 (PSGL-1/SELPLG) and L-selectin. It is hypothesized that reduced PSGL-1 expression may contribute to the reduced interaction of neutrophils with vascular endothelial cells, which may thereby increase the availability of infected neutrophils in peripheral blood for acquisition by feeding ticks (18). Consistent with this report, our study also detected a decrease in PSGL-1 expression upon A. phagocytophilum infection.
A. phagocytophilum has been shown to infect human (HMEC-1, MVEC) as well as bovine (BCE C/D-1b) endothelial cells in vitro, and it is proposed that vascular endothelium may mediate A. phagocytophilum infection of blood cells (57). Previous reports have shown that some pathogens known to interact with vascular tissue also modulate their growth properties. For example, Bartonella henselae, an intraerythrocytic pathogen, often causes vascular proliferation by inducing the secretion of vascular endothelial growth factor (VEGF) by infected macrophages (20). An interesting similarity has been observed with A. phagocytophilum-infected neutrophils in this study, which demonstrates elevated levels of VEGF transcript.
The effects of A. phagocytophilum on NADPH oxidase activation (11, 31, 55, 56) as well as the expression of its individual components have been studied by several laboratories. We (11) and others (8, 31) have shown that postbinding invasion of neutrophils is a lengthy process that lasts for up to 4 h. This prolonged association with the outer membrane of the neutrophil stimulates at least some degree of NADPH oxidase assembly, as indicated by the colocalization of Rac2 and p22phox and the mobilization of gp91phox and p22phox to the cell surface (11). Consistent with this activation profile, our array results indicate a 3.44-fold increase in expression of the CYYB gene, which encodes gp91phox, at 4 h postinfection. No change in rac2 transcription was detected. Similarly, Borjesson and colleagues reported a 1.6-fold rise in CYYB levels, while rac2 levels were unchanged at 6 h of infection. The authors extended their analyses to 24 h of infection and observed that expression of all oxidase components either remained unchanged or was elevated. Based on these data, they concluded that the loss of superoxide production by long-term infected neutrophils is not due to repression of genes encoding NADPH oxidase components (8). On the contrary, we have shown that such repression in HL-60 cells requires at least 48 h of infection (4, 12). Moreover, we have demonstrated that CYYB gene repression in A. phagocytophilum infected cells is linked to altered expression of the genes encoding interferon regulatory factor 1 and PU.1 with a concomitant increase in binding of the transcriptional repressor CCAAT displacement protein to the CYYB gene promoter (72). As these studies were performed using HL-60 cells, they should be repeated using long-term (≥48 h) infected neutrophils to resolve these discrepancies.
An obvious remaining question is whether the neutrophil transcriptional profile described above is specific to A. phagocytophilum infection or is a generalized response to infection. It has been reported by Borjesson et al. that heat-killed A. phagocytophilum also elicited a human neutrophil response somewhat similar to that of live bacteria. Both responses, however, were different from that elicited by Staphylococcus aureus (8). Thus, the transcriptional profile of A. phagocytophilum-infected neutrophils is specific for the bacterium and is partially elicited in response to interactions with Anaplasma surface structures. These reports validate the reliability of the current report as a study of the specific response of human neutrophils to A. phagocytophilum infection.
In summary, many of the differentially expressed genes identified in the current microarray analysis can be attributed to having potentially specific functional advantages for the intraneutrophil survival of A. phagocytophilum. While some of these differentially expressed genes may provide new clues to the possible mechanisms A. phagocytophilum employs to subvert neutrophil defenses, another subset of genes represents common host cell responses to bacterial infection. Our data also suggest that A. phagocytophilum may alter the functional aspects of nonneutrophil host cells like endothelial cells. This study provides the impetus for further experimentation to understand the complex interaction of this unique bacterium with its primary host cell, the neutrophil.
Comparative analysis of investigations on host cell responses to A. phagocytophilum infection.
Comparative analysis of our data with the early (3-h and 6-h) and late (9-h and 24-h) time point results of Borjesson et al. is shown in Table S2 in the supplemental material. Our discussion primarily focuses on comparing early response results. Out of the 112 (at 3 h) and 252 (at 6 h) differentially expressed genes in the study by Borjesson et al., only 9 (from 3 h) and 34 (from 6 h) genes were common with the 244 genes identified in our investigation (Fig. 2A). Approximately 78% of the differentially expressed genes identified in the present study were represented in the array probe set used Borjesson et al. The discrepancies between the two studies could have arisen from a variety of factors, including differences in bacterial strain and/or load, time course of the response, criteria adopted to define the results, and experimental handling.
Most of the genes for which similar results were found in both studies are involved in cell survival/apoptosis and inflammatory response, though notable differences exist in these categories as well. For example, in our study, many of the genes involved in the regulation of apoptosis are found to have altered expression favoring an antiapoptotic direction as early as 4 h. In contrast, Borjesson et al. did not detect altered expression of the apoptosis-regulating genes GADD45B, BIRC3, and CFLAR until 6 to 24 h. Moreover, the potent proapoptotic gene TNFSF10, which was found to be down-regulated in our study, showed no alteration in expression in the study by Borjesson and colleagues. However, many members of the NF-κB family were found to be up-regulated in both studies, though the observed time of onset of differential expression varied.
Expression of the genes of inflammatory response pathway also showed a common trend between our and Borjesson and colleagues' experiments, albeit with some important differences. The major differences include the time required to manifest a change, or the magnitude of the change (Table S2). For instance, we observed an early induction of many of the genes encoding proinflammatory molecules such as CCL3, CCL4, IL-1β, and CXCL-1 by 4 h. Our results, however, do not fully comply with Borjesson et al., who did not detect significant increases in proinflammatory gene transcription until 9-24 h. As such, they concluded that there is a delay of the neutrophil proinflammatory response upon A. phagocytophilum infection. An additional difference between our studies is that we observed decreased expression of the gene encoding transforming growth factor β1, while their data showed no differential expression of this gene. On the other hand, expression of the gene encoding the anti-inflammatory molecule IL-1RN was up-regulated in both studies at all time points. Notably, while ADORA2A was up-regulated only at 24 h in Borjesson and colleagues' study, we observed its induction at 4 h. Another marked difference between the results of the present study and theirs concerns IL-8 expression. While we observed its up-regulation (at both the mRNA and protein levels) at 4 h, Borjesson et al. observed a down-regulation at 6 h followed by up-regulation at 9 h, after which there was no detectable change in its expression. IL-8 has been reported to be up-regulated after A. phagocytophilum invasion of neutrophils (2).
The SELPLG (PSGL-1) gene, which encodes the dominant ligand for binding and entry of A. phagocytophilum into neutrophils (28), was found to be down-regulated in our array study, while Borjesson et al. detected no change in its expression at any time point. Our results are consistent with a previous report that PSGL-1 expression is down-regulated in neutrophils following A. phagocytophilum infection (18).
Comparisons of transcriptional profiles of A. phagocytophilum-infected neutrophils with those of infected promyelocytic cell lines present very limited similarities. For example, IL-8 and IL-1β were the only two differentially expressed genes common to the present study and our earlier macroarray analysis of infected HL-60 cells (12). Moreover, among the differentially expressed genes identified in our study, none were common with those observed in infected HL-60 cells by de la Fuente and colleagues (21). Only one differentially expressed gene was common to the study of NB4 cells (60) and our study. Similarly, at 6 h Borjesson et al. found one and three differentially expressed genes common to the data of de la Fuente et al. (21) and Pedra et al. (60), respectively. These drastic differences likely stem from differences between the transcriptional responses of undifferentiated promyelocytic cell lines and mature neutrophils. Yet, the similarity in expression profiles among different precursor cell lines are different, as the results of de la Fuente et al. (21) and Pedra et al. (60) identified only three common differentially expressed genes upon A. phagocytophilum infection. While the array probe sets used by Pedra et al. were the same as those used in our study, de la Fuente et al. used a synthetic polynucleotide microarray representing 21,329 human genes.
In conclusion, while comparison of our results with those of Borjesson et al. denote common trends in differential expression by A. phagocytophilum-infected neutrophils, the majority of identified altered genes were unique to each study. Moreover, many of the common genes varied in the onset of differential expression. Notably, observations of the transcriptional profiles of A. phagocytophilum-infected neutrophils drastically differed from those of infected HL-60 and NB4 cells. Thus, this study provides further insight into the effects of A. phagocytophilum on host cell expression profiles and highlights the limitations of comparing results from independent microarray studies.
Supplementary Material
Acknowledgments
We thank Ralph Horwitz of New York Medical College and Yasuko Rikihisa of Ohio State University for providing A. phagocytophilum strain HZ; Venetta Thomas and Joao H. F. Pedra for helpful discussions; Manoj Krishnan for critical reading of the manuscript; and the W. M. Keck Foundation Biotechnology Resource laboratory at Yale University for performing the microarray analysis.
This work was supported by grants from the NIH. E.F. is the recipient of a Burroughs Wellcome Clinical Scientist Award in Translational Research.
Editor: D. L. Burns
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akkoyunlu, M., and E. Fikrig. 2000. Gamma interferon dominates the murine cytokine response to the agent of human granulocytic ehrlichiosis and helps to control the degree of early rickettsemia. Infect. Immun. 68:1827-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkoyunlu, M., S. E. Malawista, J. Anguita, and E. Fikrig. 2001. Exploitation of interleukin-8-induced neutrophil chemotaxis by the agent of human granulocytic ehrlichiosis. Infect. Immun. 69:5577-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano, T., K. Tanabe, T. Eto, S. Narumiya, and K. Mizuno. 2001. LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem. J. 354:149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, R., J. Anguita, D. Roos, and E. Fikrig. 2000. Cutting edge: infection by the agent of human granulocytic ehrlichiosis prevents the respiratory burst by down-regulating gp91phox. J. Immunol. 164:3946-3949. [DOI] [PubMed] [Google Scholar]
- 5.Barnewall, R. E., N. Ohashi, and Y. Rikihisa. 1999. Ehrlichia chaffeensis and E. sennetsu, but not the human granulocytic ehrlichiosis agent, colocalize with transferrin receptor and up-regulate transferrin receptor mRNA by activating iron-responsive protein 1. Infect. Immun. 67:2258-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, R. A., S. R. Kronheim, J. E. Merriam, C. J. March, and T. P. Hopp. 1989. A pre-aspartate-specific protease from human leukocytes that cleaves pro-interleukin-1 beta. J. Biol. Chem. 264:5323-5326. [PubMed] [Google Scholar]
- 7.Blasi, F., and P. Carmeliet. 2002. uPAR: a versatile signalling orchestrator. Nat. Rev. Mol. Cell Biol. 3:932-943. [DOI] [PubMed] [Google Scholar]
- 8.Borjesson, D. L., S. D. Kobayashi, A. R. Whitney, J. M. Voyich, C. M. Argue, and F. R. Deleo. 2005. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J. Immunol. 174:6364-6372. [DOI] [PubMed] [Google Scholar]
- 9.Bossu, P., E. del Grosso, M. P. Cesaroni, G. Maurizi, N. Balestro, A. Stoppacciaro, E. del Giudice, P. Ruggiero, and D. Boraschi. 2001. Balance between autocrine interleukin-1beta and caspases defines life versus death of polymorphonuclear cells. Eur. Cytokine Netw. 12:177-186. [PubMed] [Google Scholar]
- 10.Cao, W. C., Q. M. Zhao, P. H. Zhang, J. S. Dumler, X. T. Zhang, L. Q. Fang, and H. Yang. 2000. Granulocytic ehrlichiae in Ixodes persulcatus ticks from an area in China where Lyme disease is endemic. J. Clin. Microbiol. 38:4208-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlyon, J. A., D. Abdel-Latif, M. Pypaert, P. Lacy, and E. Fikrig. 2004. Anaplasma phagocytophilum utilizes multiple host evasion mechanisms to thwart NADPH oxidase-mediated killing during neutrophil infection. Infect. Immun. 72:4772-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlyon, J. A., W. T. Chan, J. Galan, D. Roos, and E. Fikrig. 2002. Repression of rac2 mRNA expression by Anaplasma phagocytophila is essential to the inhibition of superoxide production and bacterial proliferation. J. Immunol. 169:7009-7018. [DOI] [PubMed] [Google Scholar]
- 13.Carlyon, J. A., and E. Fikrig. 2003. Invasion and survival strategies of Anaplasma phagocytophilum. Cell. Microbiol. 5:743-754. [DOI] [PubMed] [Google Scholar]
- 14.Chan, S. H., B. Perussia, J. W. Gupta, M. Kobayashi, M. Pospisil, H. A. Young, S. F. Wolf, D. Young, S. C. Clark, and G. Trinchieri. 1991. Induction of interferon gamma production by natural killer cell stimulatory factor: characterization of the responder cells and synergy with other inducers. J. Exp. Med. 173:869-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, S., M. C. Liang, J. N. Chia, J. K. Ngsee, and A. E. Ting. 2001. Rab8b and its interacting partner TRIP8b are involved in regulated secretion in AtT20 cells. J. Biol. Chem. 276:13209-13216. [DOI] [PubMed] [Google Scholar]
- 16.Chen, S.-M., J. S. Dumler, J. S. Bakken, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi, K. S., and J. S. Dumler. 2003. Early induction and late abrogation of respiratory burst in A. phagocytophilum-infected neutrophils. Ann. N. Y. Acad. Sci. 990:488-493. [DOI] [PubMed] [Google Scholar]
- 18.Choi, K. S., J. Garyu, J. Park, and J. S. Dumler. 2003. Diminished adhesion of Anaplasma phagocytophilum-infected neutrophils to endothelial cells is associated with reduced expression of leukocyte surface selectin. Infect. Immun. 71:4586-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi, K. S., D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum infection induces protracted neutrophil degranulation. Infect. Immun. 72:3680-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehio, C. 2003. Recent progress in understanding Bartonella-induced vascular proliferation. Curr. Opin. Microbiol. 6:61-65. [DOI] [PubMed] [Google Scholar]
- 21.de la Fuente, J., P. Ayoubi, E. F. Blouin, C. Almazan, V. Naranjo, and K. M. Kocan. 2005. Gene expression profiling of human promyelocytic cells in response to infection with Anaplasma phagocytophilum. Cell. Microbiol. 7:549-559. [DOI] [PubMed] [Google Scholar]
- 22.Dumler, J. S., and J. S. Bakken. 1998. Human ehrlichioses: newly recognized infections transmitted by ticks. Annu. Rev. Med. 49:201-213. [DOI] [PubMed] [Google Scholar]
- 23.Garlanda, C., E. Hirsch, S. Bozza, A. Salustri, M. De Acetis, R. Nota, A. Maccagno, F. Riva, B. Bottazzi, G. Peri, A. Doni, L. Vago, M. Botto, R. De Santis, P. Carminati, G. Siracusa, F. Altruda, A. Vecchi, L. Romani, and A. Mantovani. 2002. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature 420:182-186. [DOI] [PubMed] [Google Scholar]
- 24.Ge, Y., K. Yoshiie, F. Kuribayashi, M. Lin, and Y. Rikihisa. 2005. Anaplasma phagocytophilum inhibits human neutrophil apoptosis via upregulation of bfl-1, maintenance of mitochondrial membrane potential and prevention of caspase 3 activation. Cell. Microbiol. 7:29-38. [DOI] [PubMed] [Google Scholar]
- 25.Gokce, H. I., G. Ross, and Z. Woldehiwet. 1999. Inhibition of phagosome-lysosome fusion in ovine polymorphonuclear leucocytes by Ehrlichia (Cytoecetes) phagocytophila. J. Comp. Pathol. 120:369-381. [DOI] [PubMed] [Google Scholar]
- 26.Goodman, J. L., C. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 27.Griffith, T. S., W. A. Chin, G. C. Jackson, D. H. Lynch, and M. Z. Kubin. 1998. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J. Immunol. 161:2833-2840. [PubMed] [Google Scholar]
- 28.Herron, M. J., C. M. Nelson, J. Larson, K. R. Snapp, G. S. Kansas, and J. L. Goodman. 2000. Intracellular parasitism by the human granulocytic ehrlichiosis bacterium through the P-selectin ligand, PSGL-1. Science 288:1653-1656. [DOI] [PubMed] [Google Scholar]
- 29.Heussler, V. T., J. Machado, Jr., P. C. Fernandez, C. Botteron, C. G. Chen, M. J. Pearse, and D. A. Dobbelaere. 1999. The intracellular parasite Theileria parva protects infected T cells from apoptosis. Proc. Natl. Acad. Sci. USA 96:7312-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh, T. C., M. E. Aguero-Rosenfeld, J. M. Wu, C. Ng, N. A. Papanikolaou, S. A. Varde, I. Schwartz, J. G. Pizzolo, M. Melamed, H. W. Horowitz, R. B. Nadelman, and G. P. Wormser. 1997. Cellular changes and induction of apoptosis in human promyelocytic HL-60 cells infected with the agent of human granulocytic ehrlichiosis (HGE). Biochem. Biophys. Res. Commun. 232:298-303. [DOI] [PubMed] [Google Scholar]
- 31.IJdo, J. W., and A. C. Mueller. 2004. Neutrophil NADPH oxidase is reduced at the Anaplasma phagocytophilum phagosome. Infect. Immun. 72:5392-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inohara, N., and G. Nunez. 2003. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 33.Irmler, M., V. Steiner, C. Ruegg, H. Wajant, and J. Tschopp. 2000. Caspase-induced inactivation of the anti-apoptotic TRAF1 during Fas ligand-mediated apoptosis. FEBS Lett. 468:129-133. [DOI] [PubMed] [Google Scholar]
- 34.Iyer, S. S., J. A. Barton, S. Bourgoin, and D. J. Kusner. 2004. Phospholipases D1 and D2 coordinately regulate macrophage phagocytosis. J. Immunol. 173:2615-2623. [DOI] [PubMed] [Google Scholar]
- 35.Joshi, S. G., C. W. Francis, D. J. Silverman, and S. K. Sahni. 2003. Nuclear factor κB protects against host cell apoptosis during Rickettsia rickettsii infection by inhibiting activation of apical and effector caspases and maintaining mitochondrial integrity. Infect. Immun. 71:4127-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamohara, H., W. Matsuyama, O. Shimozato, K. Abe, C. Galligan, S. Hashimoto, K. Matsushima, and T. Yoshimura. 2004. Regulation of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) and TRAIL receptor expression in human neutrophils. Immunology 111:186-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kataoka, T., R. C. Budd, N. Holler, M. Thome, F. Martinon, M. Irmler, K. Burns, M. Hahne, N. Kennedy, M. Kovacsovics, and J. Tschopp. 2000. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr. Biol. 10:640-648. [DOI] [PubMed] [Google Scholar]
- 38.Kim, H.-Y., and Y. Rikihisa. 2000. Expression of interleukin-1β, tumor necrosis factor alpha, and interleukin-6 in human peripheral blood leukocytes exposed to human granulocytic ehrlichiosis agent or recombinant major surface protein P44. Infect. Immun. 68:3394-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, H.-Y., and Y. Rikihisa. 2002. Roles of p38 mitogen-activated protein kinase, NF-κB, and protein kinase C in proinflammatory cytokine mRNA expression by human peripheral blood leukocytes, monocytes, and neutrophils in response to Anaplasma phagocytophila. Infect. Immun. 70:4132-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein, M. B., S. Hu, C. C. Chao, and J. L. Goodman. 2000. The agent of human granulocytic ehrlichiosis induces the production of myelosuppressing chemokines without induction of proinflammatory cytokines. J. Infect. Dis. 182:200-205. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi, S. D., K. R. Braughton, A. R. Whitney, J. M. Voyich, T. G. Schwan, J. M. Musser, and F. R. DeLeo. 2003. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc. Natl. Acad. Sci. USA 100:10948-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau, A. S., and D. D. Mruk. 2003. Rab8B GTPase and junction dynamics in the testis. Endocrinology 144:1549-1563. [DOI] [PubMed] [Google Scholar]
- 43.Lee, T. H., H. G. Wisniewski, and J. Vilcek. 1992. A novel secretory tumor necrosis factor-inducible protein (TSG-6) is a member of the family of hyaluronate binding proteins, closely related to the adhesion receptor CD44. J. Cell Biol. 116:545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lepidi, H., J. E. Bunnell, M. E. Martin, J. E. Madigan, S. Stuen, and J. S. Dumler. 2000. Comparative pathology, and immunohistology associated with clinical illness after Ehrlichia phagocytophila-group infections. Am. J. Trop. Med. Hyg. 62:29-37. [DOI] [PubMed] [Google Scholar]
- 45.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:research0032.1-research0032.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, C., and W. Wong. 2003. DNA-chip analyzer (dChip). Springer, New York, N.Y.
- 47.Lin, M., and Y. Rikihisa. 2003. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cell. Microbiol. 5:809-820. [DOI] [PubMed] [Google Scholar]
- 48.Lindsay, A. J., and M. W. McCaffrey. 2004. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J. Cell Sci. 117:4365-4375. [DOI] [PubMed] [Google Scholar]
- 49.Masuda, E. S., Y. Luo, C. Young, M. Shen, A. B. Rossi, B. C. Huang, S. Yu, M. K. Bennett, D. G. Payan, and R. H. Scheller. 2000. Rab37 is a novel mast cell specific GTPase localized to secretory granules. FEBS Lett. 470:61-64. [DOI] [PubMed] [Google Scholar]
- 50.McDermott, M., M. J. Wakelam, and A. J. Morris. 2004. Phospholipase D. Biochem. Cell Biol. 82:225-253. [DOI] [PubMed] [Google Scholar]
- 51.Millar, A. L., N. J. Pavios, J. Xu, and M. H. Zheng. 2002. Rab3D: a regulator of exocytosis in non-neuronal cells. Histol. Histopathol. 17:929-936. [DOI] [PubMed] [Google Scholar]
- 52.Moir, E., N. A. Booth, B. Bennett, and L. A. Robbie. 2001. Polymorphonuclear leucocytes mediate endogenous thrombus lysis via a u-PA-dependent mechanism. Br. J. Haematol. 113:72-80. [DOI] [PubMed] [Google Scholar]
- 53.Molestina, R. E., T. M. Payne, I. Coppens, and A. P. Sinai. 2003. Activation of NF-kappaB by Toxoplasma gondii correlates with increased expression of antiapoptotic genes and localization of phosphorylated IkappaB to the parasitophorous vacuole membrane. J. Cell Sci. 116:4359-4371. [DOI] [PubMed] [Google Scholar]
- 54.Mott, J., R. E. Barnewall, and Y. Rikihisa. 1999. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect. Immun. 67:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mott, J., and Y. Rikihisa. 2000. Human granulocytic ehrlichiosis agent inhibits superoxide anion generation by human neutrophils. Infect. Immun. 68:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mott, J., Y. Rikihisa, and S. Tsunawaki. 2002. Effects of Anaplasma phagocytophila on NADPH oxidase components in human neutrophils and HL-60 cells. Infect. Immun. 70:1359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Munderloh, U. G., M. J. Lynch, M. J. Herron, A. T. Palmer, T. J. Kurtti, R. D. Nelson, and J. L. Goodman. 2004. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet. Microbiol. 101:53-64. [DOI] [PubMed] [Google Scholar]
- 58.Ogden, N. H., Z. Woldehiwet, and C. A. Hart. 1998. Granulocytic ehrlichiosis: an emerging or rediscovered tick-borne disease? J. Med. Microbiol. 47:475-482. [DOI] [PubMed] [Google Scholar]
- 59.Park, J. H., E. J. Heo, K. S. Choi, J. S. Dumler, and J. S. Chae. 2003. Detection of antibodies to Anaplasma phagocytophilum and Ehrlichia chaffeensis antigens in sera of Korean patients by Western immunoblotting and indirect immunofluorescence assays. Clin. Diagn. Lab. Immunol. 10:1059-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedra, J. H., B. Sukumaran, J. A. Carlyon, N. Berliner, and E. Fikrig. 2005. Modulation of NB4 promyelocytic leukemic cell machinery by Anaplasma phagocytophilum. Genomics 86:365-377. [DOI] [PubMed] [Google Scholar]
- 61.Pelkmans, L., D. Puntener, and A. Helenius. 2002. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science 296:535-539. [DOI] [PubMed] [Google Scholar]
- 62.Philpott, D. J., and S. E. Girardin. 2004. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol. Immunol. 41:1099-1108. [DOI] [PubMed] [Google Scholar]
- 63.Pure, E., and C. A. Cuff. 2001. A crucial role for CD44 in inflammation. Trends Mol. Med. 7:213-221. [DOI] [PubMed] [Google Scholar]
- 64.Ren, Q., S. J. Robertson, D. Howe, L. F. Barrows, and R. A. Heinzen. 2003. Comparative DNA microarray analysis of host cell transcriptional responses to infection by Coxiella burnetii or Chlamydia trachomatis. Ann. N. Y. Acad. Sci. 990:701-713. [DOI] [PubMed] [Google Scholar]
- 65.Rottner, K., S. Lommel, J. Wehland, and T. E. Stradal. 2004. Pathogen-induced actin filament rearrangement in infectious diseases. J. Pathol. 204:396-406. [DOI] [PubMed] [Google Scholar]
- 66.Roy, N., Q. L. Deveraux, R. Takahashi, G. S. Salvesen, and J. C. Reed. 1997. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 16:6914-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scaife, H., Z. Woldehiwet, C. A. Hart, and S. W. Edwards. 2003. Anaplasma phagocytophilum reduces neutrophil apoptosis in vivo. Infect. Immun. 71:1995-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmausser, B., C. Josenhans, S. Endrich, S. Suerbaum, C. Sitaru, M. Andrulis, S. Brandlein, P. Rieckmann, H. K. Muller-Hermelink, and M. Eck. 2004. Downregulation of CXCR1 and CXCR2 expression on human neutrophils by Helicobacter pylori: a new pathomechanism in H. pylori infection? Infect. Immun. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simpson, J. C., G. Griffiths, M. Wessling-Resnick, J. A. Fransen, H. Bennett, and A. T. Jones. 2004. A role for the small GTPase Rab21 in the early endocytic pathway. J. Cell Sci. 117:6297-6311. [DOI] [PubMed] [Google Scholar]
- 70.Sinai, A. P., T. M. Payne, J. C. Carmen, L. Hardi, S. J. Watson, and R. E. Molestina. 2004. Mechanisms underlying the manipulation of host apoptotic pathways by Toxoplasma gondii. Int. J. Parasitol. 34:381-391. [DOI] [PubMed] [Google Scholar]
- 71.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 72.Thomas, V., S. Samanta, C. Wu, N. Berliner, and E. Fikrig. 2005. Anaplasma phagocytophilum modulates gp91phox gene expression through altered interferon regulatory factor 1 and PU.1 levels and binding of CCAAT displacement protein. Infect. Immun. 73:208-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Valentijn, J. A., K. Valentijn, L. M. Pastore, and J. D. Jamieson. 2000. Actin coating of secretory granules during regulated exocytosis correlates with the release of rab3D. Proc. Natl. Acad. Sci. USA 97:1091-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Zandbergen, G., J. Gieffers, H. Kothe, J. Rupp, A. Bollinger, E. Aga, M. Klinger, H. Brade, K. Dalhoff, M. Maass, W. Solbach, and T. Laskay. 2004. Chlamydia pneumoniae multiply in neutrophil granulocytes and delay their spontaneous apoptosis. J. Immunol. 172:1768-1776. [DOI] [PubMed] [Google Scholar]
- 75.von Loewenich, F. D., D. G. Scorpio, U. Reischl, J. S. Dumler, and C. Bogdan. 2004. Frontline: control of Anaplasma phagocytophilum, an obligate intracellular pathogen, in the absence of inducible nitric oxide synthase, phagocyte NADPH oxidase, tumor necrosis factor, Toll-like receptor (TLR)2 and TLR4, or the TLR adaptor molecule MyD88. Eur. J. Immunol. 34:1789-1797. [DOI] [PubMed] [Google Scholar]
- 76.Watson, R. W., O. D. Rotstein, J. Parodo, R. Bitar, and J. C. Marshall. 1998. The IL-1 beta-converting enzyme (caspase-1) inhibits apoptosis of inflammatory neutrophils through activation of IL-1 beta. J. Immunol. 161:957-962. [PubMed] [Google Scholar]
- 77.Webster, P., J. W. IJdo, L. M. Chicoine, and E. Fikrig. 1998. The agent of human granulocytic ehrlichiosis resides in an endosomal compartment. J. Clin. Investig. 101:1932-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yanai, A., Y. Hirata, Y. Mitsuno, S. Maeda, W. Shibata, M. Akanuma, H. Yoshida, T. Kawabe, and M. Omata. 2003. Helicobacter pylori induces antiapoptosis through nuclear factor-kappaB activation. J. Infect. Dis. 188:1741-1751. [DOI] [PubMed] [Google Scholar]
- 79.Yarmola, E. G., A. S. Edison, R. H. Lenox, and M. R. Bubb. 2001. Actin filament cross-linking by MARCKS: characterization of two actin-binding sites within the phosphorylation site domain. J. Biol. Chem. 276:22351-22358. [DOI] [PubMed] [Google Scholar]
- 80.Yilmaz, O., T. Jungas, P. Verbeke, and D. M. Ojcius. 2004. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect. Immun. 72:3743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshiie, K., H. Y. Kim, J. Mott, and Y. Rikihisa. 2000. Intracellular infection by the human granulocytic ehrlichiosis agent inhibits human neutrophil apoptosis. Infect. Immun. 68:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.