Abstract
The adenovirus early gene product Gam1 is crucial for virus replication and induces certain cellular genes by inactivating histone deacetylase 1 (HDAC1). We demonstrate that Gam1 (i) destroys promyelocitic leukemia nuclear bodies, (ii) delocalizes SUMO-1 into the cytoplasm and (iii) influences the SUMO-1 pathway. In addition, we show that Gam1 counteracts HDAC1 sumoylation both in vivo and in vitro. Sumoylation of HDAC1 does not seem to be absolutely required for HDAC1 biological activity but is part of a complex regulatory circuit that also includes phosphorylation of the deacetylase. Our data demonstrate that Gam1 is a viral protein that can affect simultaneously two signaling pathways: sumoylation and acetylation.
Introduction
Gam1 is an adenovirus early gene product essential for viral replication (Glotzer et al., 2000) with no homology to known proteins. Although originally identified in a screen for viral genes that could promote cell survival (Chiocca et al., 1997), it is now clear that its major role is to influence gene expression. Gam1 expression increases transcription from a variety of eukaryotic promoters most likely by inactivating histone deacetylase 1 (HDAC1) (Chiocca et al., 2002). Our evidence that replication of a Gam1-deleted virus is restored by adding the HDAC inhibitor trichostatin A (TSA) (Chiocca et al., 2002) is in agreement with recent findings showing that deacetylases may serve as a cellular defense against viruses (Meier, 2001; Shestakova et al., 2001; Zhang and Jones, 2001; Merezak et al., 2002; Murphy et al., 2002).
Acetylation of histones has been known to closely correlate with transcriptional activation (Grunstein, 1990; Struhl, 1998). Two types of enzymes, the histone acetyltransferases (HATs) and the histone deacetylases (HDACs) control the acetylation of histones and other substrates. Several cellular proteins have been shown to have intrinsic HAT activity (Sterner and Berger, 2000), whereas HDACs include a family of enzymes that have been grouped into three classes based upon their homology to yeast proteins (Khochbin et al., 2001).
A number of viruses, including herpes simplex virus type 1 (HSV-1), Epstein–Barr virus (EBV) and cytomegalovirus (CMV), disperse nuclear structures known as promyelocitic leukemia (PML) nuclear bodies (Everett and Maul, 1994; Korioth et al., 1996; Ahn and Hayward, 1997; Adamson and Kenney, 2001). The viral proteins responsible for PML body dispersion are immediate-early proteins and, in the case of HSV-1 immediate-early protein ICP0, the viral ICP0 mutants that cannot disperse PML bodies cannot replicate efficiently, suggesting a link between viral replication and PML bodies (Everett et al., 1995). In the present study, we show that Gam1 alters these nuclear domains and causes a cytoplasmic diffusion of SUMO-1, a small protein with significant homology to ubiquitin, recently found conjugated with a growing list of proteins with distinct functions (Hochstrasser, 2000; Hay, 2001). In agreement with this, we noticed a reduction of global sumoylation in the presence of Gam1. A Gam1 mutant (Gam1mt) that is unable to activate transcription (Chiocca et al., 2002) has no effect on sumoylation. Very recently, HDACs such as HDAC1 and HDAC4 have been shown to be modified by sumoylation (David et al., 2002; Kirsh et al., 2002). We also present evidence that HDAC1 is modified by SUMO-1, identify Lys444 and Lys476 as HDAC1 sumoylation sites and demonstrate that Gam1 interferes with sumoylation of HDAC1.
Results and Discussion
Gam1 destroys PML-containing nuclear bodies
ND10 (or PML-containing nuclear bodies) has gained increasing interest as the nuclear site where DNA viruses start transcription and replication (Ishov and Maul, 1996). Several DNA viruses such as HSV-1 or HCMV dismantle or modify PML-containing nuclear bodies through the action of their immediate-early proteins (Everett and Maul, 1994; Korioth et al., 1996; Ahn and Hayward, 1997; Adamson and Kenney, 2001). Furthermore, it has been demonstrated that PML represses transcription by interacting with HDACs (Wu et al., 2001) and that Daxx, another protein found in the nuclear bodies (Yang et al., 1997), binds to HDAC1 (Li et al., 2000). We therefore decided to test whether Gam1, being a DNA virus immediate-early protein, could alter these nuclear domains. HeLa cells were transfected with myc-tagged Gam1 and analyzed for localization of PML and Daxx, (Figure 1A and B). As shown in Figure 1A (b and j), we observed a dispersal of PML bodies and Daxx in Gam1-expressing HeLa cells. In addition, we infected WI38 fibroblasts with a recombinant adenoviral construct directing Gam1 expression (AdGam1) and obtained the same result (see supplementary figure 1 available at EMBO reports Online). We also tested the ability of Gam1mt to affect these nuclear structures. Gam1mt, a mutant with no cell survival function (Chiocca et al., 1997), is unable to activate transcription and to inactivate HDAC1 (Chiocca et al., 2002). As shown in Figure 1B (n and v), Gam1mt does not affect PML and Daxx.
Figure 1.
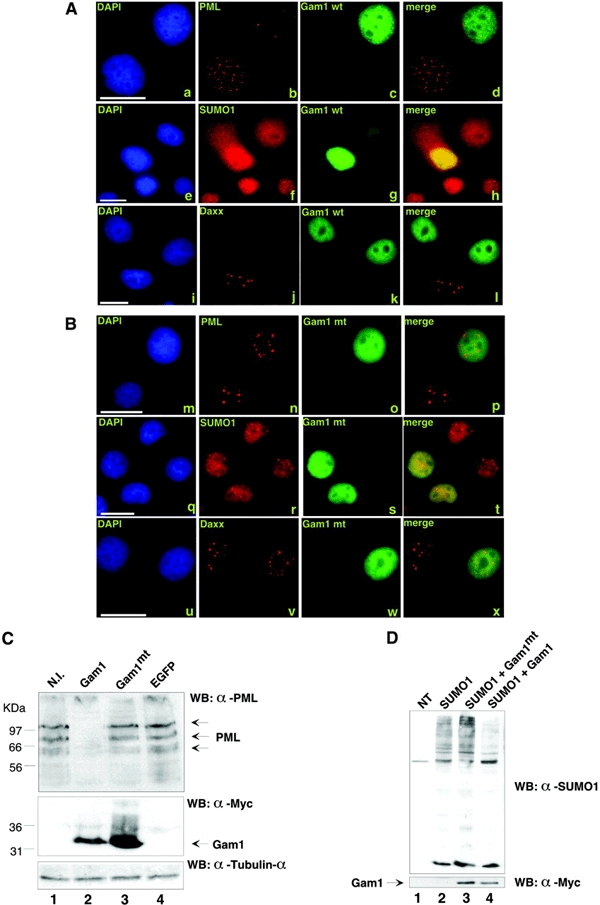
(A and B) Gam1 wt destroys PML-containing nuclear bodies. HeLa cells were treated with 1000 U/ml of human γ-interferon to upregulate the number of PML nuclear bodies and transfected with plasmids directing expression of (A) myc-Gam1 wt or (B) myc-Gam1mt (Chiocca et al., 1997). Cells were stained as described in Methods with antibodies against the myc epitope (c, g, k, o, s and w), against PML (b and n), against Daxx (j and v) or against SUMO-1 (f and r). Scale bars, 15 μm. (C) Gam1 destroys endogenous PML isoforms in WI38 cells. WI38 cells were non-infected (lane 1) or infected with adenoviral recombinant constructs expressing myc-Gam1 wt (lane 2), myc-Gam1mt (lane 3) and EGFP (lane 4) (Glotzer et al., 2000; Chiocca et al., 2002). Lysates (100 μg) were loaded on a 10% SDS–polyacrylammide gel and, after western blotting, Gam1 was visualized with an anti-myc antibody, endogenous PML with a mouse polyclonal and lysates were normalized with α-tubulin. (D) Gam1 wt reduces global sumoylation. HeLa cells were transfected with the indicated plasmids (5 μg of myc-Gam1 wt or myc-Gam1mt and 3 μg of SUMO-1; NT, non-transfected cells).
Gam1 induces loss of PML and delocalizes SUMO-1 into the cytoplasm
The PML IV protein has a molecular mass of about 70 KDa; however, there are several higher-molecular-mass isoforms of PML, which are modified by SUMO-1 (Ishov et al., 1999; Zhong et al., 2000). Interestingly, expression of Gam1 led to diffusion of SUMO-1 into the cytoplasm (Figure 1A, f). In contrast, Gam1mt had no effect on SUMO-1 localization (Figure 1B, r; and see supplementary figure 2). To determine whether Gam1 disrupts SUMO-1-modified PML isoforms, we expressed Gam1 in WI38 human fibroblasts using the AdGam1 construct and immunoblotted the cellular lysates with an anti-PML antibody (Figure 1C). Gam1-expressing cells lost both unmodified and the higher-molecular-weight PML species. In contrast, there was no change in PML expression between non-infected cells and cells infected with AdGam1mt or AdGFP. A similar delocalization of SUMO-1 by Gam1 was also observed in PML−/− cells, indicating that the effect of Gam1 on SUMO-1 was PML-independent (see supplementary figure 3).
Gam1 influences the SUMO-1 pathway
The observation that Gam1 delocalizes SUMO-1 both in the presence and in the absence of PML suggested that Gam1 could influence the sumoylation pathway and affect other sumoylated targets besides PML. To assess the effect of Gam1 expression on global sumoylation, HeLa cells were transfected with Gam1 and Gam1mt. As shown in Figure 1D, western blotting analysis with antisUMO-1 revealed a reduction in total sumoylation only in the presence of Gam1 wt (lane 4), consistent with the immunofluorescence data.
Since we have recently described the ability of Gam1 to inhibit histone deacetylation by HDAC1 (Chiocca et al., 2002), we hypothesized that HDAC1 might be a target for SUMO-1 conjugation and that Gam1 could affect its sumoylation. HDAC1 has been recently shown to be a substrate for SUMO-1 modification in vitro and in vivo (David et al., 2002). We also determined that HDAC1 was conjugated to SUMO-1 in vitro (see Figure 3C, lane 3) and in vivo, by expressing myc-HDAC1 together with HAsUMO-1 and UBC9 in HeLa cells (Figure 2A and B). Western blotting analysis revealed at least three bands in the presence of HAsUMO-1 (Figure 2A, lanes 5 and 6) that, upon UBC9 expression, increased in intensity together with other higher-molecular-weight species (Figure 2A, lane 6), as expected in all SUMO-1-modified substrates (Hay, 2001; Wilson and Rangasamy, 2001). Curiously, unmodified HDAC1 was immunoprecipitated, indicating its possible interaction with the SUMO-1-modified isoform (Figure 2A, lanes 5 and 6), in agreement with published data (Taplick et al., 2001). Similarly, we detected the same HDAC1 modified forms upon performing the converse immunoprecipitation, confirming that these were SUMO-1-modified forms of HDAC1 (Figure 2B). Transfection efficiency was evaluated by western immunoblotting of total cellular proteins (data not shown).
Figure 3.
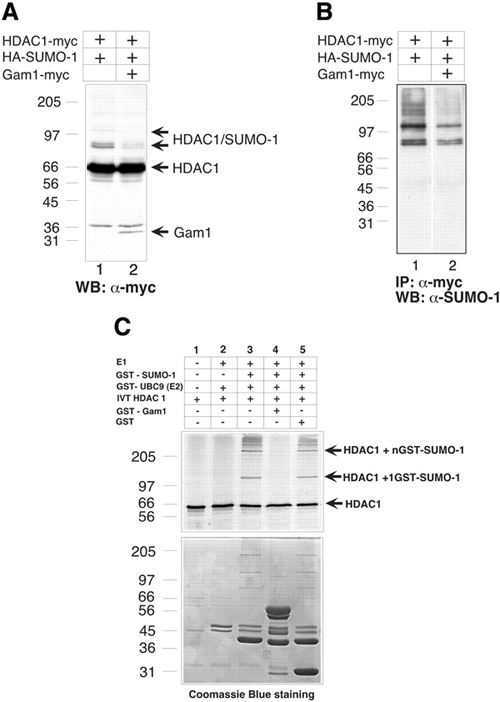
The SUMO-1-modified forms of HDAC1 are influenced by the presence of Gam1. (A) SDS lysed HeLa cells (30 μg) transfected with the indicated plasmids were immunoblotted with the indicated antibody (WB) in a 10% SDS–polyacrilamide gel. The arrows indicate HDAC1 and SUMO-1-modified HDAC1 (lane 1 and 2) and the presence of Gam1 (lane 2). (B) HeLa cells (500 μg) transfected with the indicated plasmids and lysed in SDS were immunoprecipitated (IP), electrophoresed on a 10% SDS–polyacrylamide gel and immunoblotted (WB) with the indicated antibodies. (C). In vitro reaction. HDAC1 was in vitro translated and incubated with the indicated GST proteins as described previously (Gostissa et al., 1999). Lower panel, Coomassie Blue staining.
Figure 2.
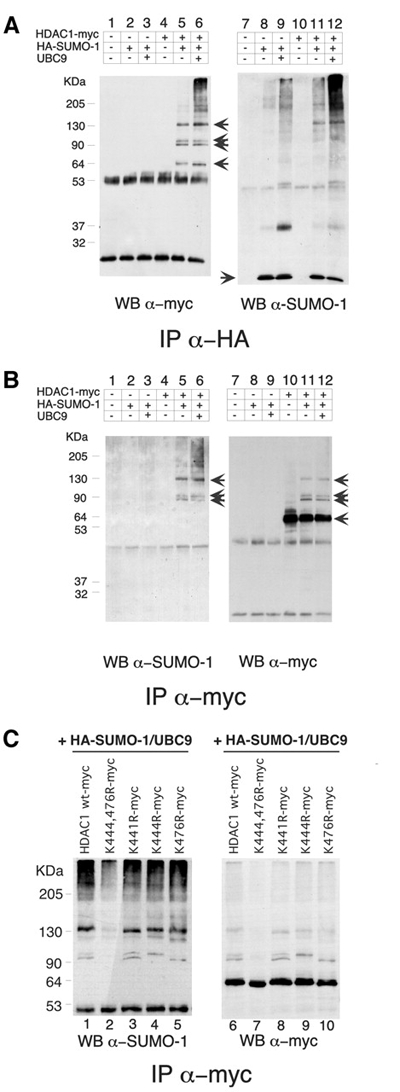
(A and B) HDAC1 is modified by SUMO-1 in vivo. HeLa cells (500 μg) transfected with 5 μg of the indicated plasmids and lysed in SDS were immunoprecipitated (IP), electrophoresed on a 10% SDS–polyacrylamide gel and immunoblotted (WB) with the indicated antibodies. The three upper arrows indicate the SUMO-1-modified forms. (C) Lys444 and Lys476 are the major SUMO-1 modification sites in HDAC1. HeLa cells (500 μg) transfected with 5 μg of the indicated plasmids were lysed in SDS, immunoprecipitated (IP) with the indicated antibody, electrophoresed on a 10% SDS–polyacrylamide gel and immunoblotted (WB) with the indicated antibodies.
HDAC1 is modified by SUMO-1 on Lys444 and Lys476
The consensus ΦKXE motif is a well-conserved pattern in almost all SUMO-1-modified substrates (Rodriguez et al., 2001). There are two ΦKXE motifs at positions 444 and 476 in HDAC1 that represent potential SUMO-1 attachment sites. To determine whether they could be modified by SUMO-1, these candidate lysine residues were mutated to arginine, and the resulting mutants were tested for SUMO-1 modification by expressing them together with HAsUMO-1 and UBC9 in HeLa cells. Immunoprecipitation of the cell lysates with the anti-myc antibody and western blot analysis with both anti-HA antibody and anti-myc showed that K444R and K476R single point mutants lost one species each (Figure 2C, lanes 4, 5, 9 and 10); in the double mutant, HDAC1 K 444 476 R, the SUMO-1-modified forms of HDAC1 were strongly reduced (Figure 2C, lane 2 and 7). This was also confirmed with the reverse analysis (data not shown). Since there was the possibility that mutations at these lysine residues disrupted the nuclear localization signal (NLS) sequence, we also mutated K441, which as K444 is found in the NLS but does not represent a potential SUMO-1 attachment site. K441R had the same sumoylation pattern of HDAC1 wt (Figure 2C, lane 3 and 8). We thus conclude that K444 and K476 are the predominant SUMO-1 attachment sites. These findings are in agreement with data published recently (David et al., 2002).
Gam1 diminishes the SUMO-1-modified forms of HDAC1
Since Gam1 was previously shown to inhibit transcriptional repression by HDAC1, we wanted to assess whether Gam1 could influence HDAC1 sumoylation. HeLa cells were transfected with constructs for myc-HDAC1, HA-SUMO1 and myc-Gam1, and cell lysates were analyzed for the presence of sumoylated HDAC1. As shown in Figure 3A, in the absence of Gam1, anti-myc antibody recognizes at least three bands corresponding to sumoylated HDAC1. However, in the presence of Gam1, these modified forms of HDAC1 were significantly diminished (Figure 3A, lane 2), in agreement with our data in Figure 1D showing that the global level of sumoylation seemed to be affected by Gam1. In addition, upon myc immunoprecipitation, we observed a specific reduction in HDAC1 sumoylation in the presence of Gam1 (Figure 3B, lane 2).
To further address the role of Gam1 in regulating HDAC1, we determined its ability to inhibit HDAC1 SUMO modification in vitro. Figure 3C shows that HDAC1 was efficiently conjugated to SUMO-1 when incubated with E1 enzyme activity, ATP, an ATP regenerating system and UBC9 (E2) (Figure 3C, lanes 3 and 5). These modified species disappeared in the presence of Gam1 (lane 4).
Contribution of sumoylation to HDAC1 functional properties
We then tested the effect of sumoylation on HDAC1 repressing activity and enzymatic function. As reported previously (Hassig et al., 1998), HDAC1 fused to GAL4 DNA binding domain acts as a transcriptional repressor of the luciferase gene under the control of Gal4 binding sites (Figure 4A, lane 3). The repressor activity of the mutant HDAC1 K 444 476 R was similar (Figure 4A, lane 5), indicating that SUMO-1 modification is not necessary for the full repressive activity of HDAC1. Also, TSA addition reduced both HDAC1 wt and HDAC1 K 444 476 R- mediated transcriptional repression to the same extent (Figure 4A, lanes 4 and 6), whereas addition of SUMO-1 had no effect (Figure 4A, lanes 7 and 8). Similarly, the enzymatic activity of HDAC1 wt and the sumoylation-defective mutant were comparable, whereas HDAC1 S 421 423 A, a phoshorylation-deficient mutant, showed strongly reduced activity as described previously (Pflum et al., 2001) (Figure 4B). Interestingly, this phoshorylation-deficient mutant with impaired enzymatic activity was found to be heavily sumoylated (Figure 4C, compare lanes 1 and 3; and Figure 4B, compare lanes 5 and 7). Our results are consistent with the view that HDAC activity is responsible for HDAC1-mediated repression (Hassig et al., 1998) and indicate that sumoylation is not required for transcriptional repression by HDAC1 or its enzymatic activity. While this manuscript was in preparation, Ronald De Pinho's group showed that SUMO-1 modification of HDAC1 modulates HDAC1-mediated transcriptional repression (David et al., 2002). Although we do not know the reasons for this discrepancy, our data point towards a more complicated scenario where different interdependent modifications might modulate the biological function of HDAC1.
Figure 4.
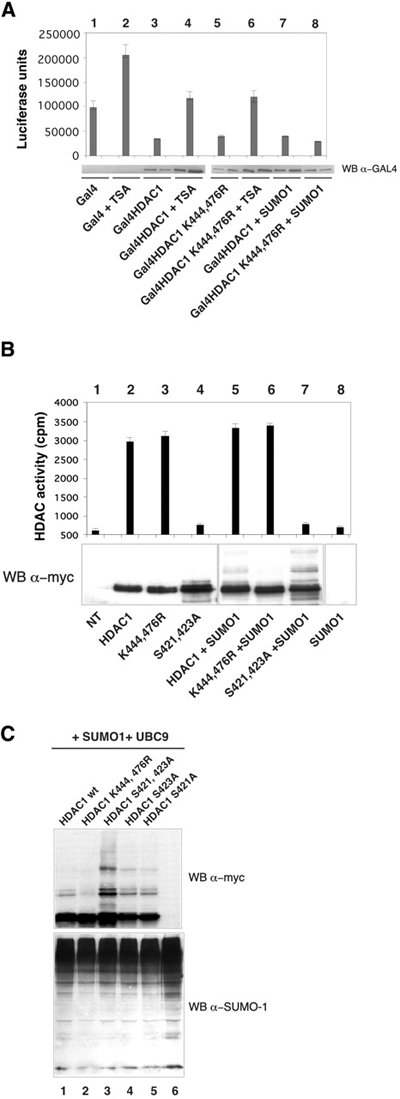
Lys444 and Lys476 on HDAC1 are not critical for HDAC1 repression and enzymatic function. (A) NIH 3T3 cells were transfected with a 5 GAL4 binding site-driven luciferase reporter plasmid plus the indicated plasmids with or without TSA (see Methods). Results are the means of two independent experiments, and the lower panel shows the expression levels of Gal4–HDAC1 fusion proteins detected by western blot analysis. This experiment was repeated several times with consistent results. (B) HDAC1 wt or mutant fused to a myc tag were expressed in HeLa cells plus or minus SUMO-1, immunoprecipitated using α-myc antibody and tested for deacetylase activity as described previously (Chiocca et al., 2002). The SD of three different experiments is shown. Protein quantities were assessed by western blot analysis (gel image). (C) SDS lysed HeLa cells (40 μg) transfected with the indicated plasmids were immunoblotted with the indicated antibody (WB) in a 10% SDS–polyacrylamide gel. HDAC1 S 421 423 A, HDAC1 S 423 A and HDAC1 S 421 A have been described previously (Pflum et al., 2001).
Conclusions
In summary, we found that Gam1 destroys PML-containing nuclear bodies and causes a PML-independent diffusion of SUMO-1 into the cytoplasm. Furthermore, Gam1 counteracts the sumoylation of HDAC1, PML and probably other substrates. HSV-1 ICP0 was the first viral protein shown to abrogate PML (Everett and Maul, 1994), and its bovine functional homolog has also been shown to interact directly with HDAC1 (Zhang and Jones, 2001). ICP0 alone is sufficient to abrogate the conjugation of SUMO-1 to PML and to induce its degradation (Muller and Dejean, 1999; Parkinson and Everett, 2000), leading then to the disruption of PML nuclear bodies. Similarly, Gam1 disrupts PML, but, unlike ICP0 or other viral proteins, it also influences global sumoylation. Viral interference with sumoylation of host proteins could confer a more favorable environment for viral propagation. A decrease in sumoylated host proteins could be accomplished either by preventing de novo sumoylation or enhancing desumoylation, and we are currently defining which of these two scenarios reflects the Gam1 mode of action. We have recently identified Gam1 as a negative regulator of HDAC1 enzymatic function. However, it is not clear how Gam1 inhibits HDAC1 enzymatic activity. Here, we show that Gam1 has the ability to negatively affect HDAC1 sumoylation. Since the role of SUMO-modified HDAC1 is not yet defined, we still do not understand whether this is linked to the ability of Gam1 to inactivate HDAC1. Undoubtedly, Gam1 inactivates HDAC1 regardless of its sumoylation state. We present here a viral protein with the potential to target simultaneously two signaling pathways: sumoylation and acetylation.
Methods
Immunoprecipitations and antibodies.
Whole-cell extracts were prepared and immunoprecipitated as described previously (Chiocca et al., 2002); to detect SUMO-1-modified forms of HDAC1, an SDS buffer was used to lyse the cells (Adamson and Kenney, 2001). Briefly, after sonication, incubation at 95°C for 10 min and centrifugation, the cellular lysate was diluted 1:10 in E1A buffer (50mM HEPES pH 7.0, 250mM NaCl, 0.1% NP-40, and protease inhibitors) and immunoprecipitated. The following antibodies were used in this study: anti-myc epitope (9E10; (Evan et al., 1985), anti-α-tubulin (sc-8035, Santa Cruz), anti-HA epitope (12CA5), anti-PML (polyclonal murine antibody and goat polyclonal A-20, Santa Cruz), antisUMO-1 (sc-9060, Santa Cruz), anti-Daxx (sc-7152, Santa Cruz), anti-Gal4 (sc-510, Santa Cruz).
In vitro sumoylation assay.
Full-length HDAC1 was in vitro translated in a rabbit reticulocyte lysate system (Promega), (35S)Met-labeled (Amersham) and then subjected to in vitro SUMO-1 modification as described previously (Gostissa et al., 1999).
Cell culture, transfection and infection.
HeLa, WI38 and NIH 3T3 cell lines were grown in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% fetal calf serum. Cells were transfected overnight with a calcium precipitation method or with lipofectamine for 3 h. WI38 were infected with 10 000 particles per cell of recombinant adenovirus, constructed as described previously (Glotzer et al., 2000). For reporter assays, 1.5 × 105 cells were plated in 6-well dishes and cells were transfected with 0.2 μg of tk-(GAL4)5-luc reporter plasmid, 0.2 μg of pGAL4 or the pGAL4 constructs. Cells were treated with 100 ng/ml TSA for 12–16 h. Luciferase activity was determined as described previously (Chiocca et al., 2002) and normalized for transfection efficiency.
Immunofluorescence.
Cells were plated on glass cover slips (12 mm × 12 mm) in 6-well dishes 24 h before transfection or infection. Cells were fixed 2 days after transfection or 1 day after infection and stained as described previously (Glotzer et al., 2000). Images were acquired with a cooled CCD camera (Hamamatsu C5985) mounted on an AX70 Provis microscope (Olympus).
Plasmids.
All HDAC1 mutants were generated using standard cloning procedures and verified by DNA sequencing. Mutations were introduced using PCR mutagenesis. Details are available upon request.
Supplementary Material
Supplementary data
Acknowledgments
We thank Dr Matt Cotten for recombinant Adenoviruses, G. Brosch for labeled chicken histones, and Dr P.G. Pelicci for PML−/− fibroblasts and anti-PML antibody. We thank Dr Francesca Fiore and Dr Mario Faretta for helpful discussions. This work was supported by grants from AIRC (Associazione Italiana per la Ricerca sul Cancro), CNR (Consiglio Nazionale delle Ricerche), FIRC (Fondazione Italiana per la Ricerca sul Cancro) and Telethon. C.S. was supported by the Austrian FWF (P13068-GEN and P14909-GEN).
References
- Adamson A.L. and Kenney S. (2001) Epstein–Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol., 75, 2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J.H. and Hayward G.S. (1997) The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol., 71, 4599–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocca S., Baker A. and Cotten M. (1997) Identification of a novel antiapoptotic protein, GAM-1, encoded by the CELO adenovirus. J. Virol., 71, 3168–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocca S., Kurtev V., Colombo R., Boggio R., Sciurpi M.T., Brosch G., Seiser C., Draetta G.F. and Cotten M. (2002) Histone deacetylase 1 inactivation by an adenovirus early gene product. Curr. Biol., 12, 594–598. [DOI] [PubMed] [Google Scholar]
- David G., Neptune M.A. and DePinho R.A. (2002) SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem., 17, 17. [DOI] [PubMed] [Google Scholar]
- Evan G.I., Lewis G.K., Ramsay G. and Bishop J.M. (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol., 5, 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R.D. and Maul G.G. (1994) HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J., 13, 5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R., O'Hare P., O'Rourke D., Barlow P. and Orr A. (1995) Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J. Virol., 69, 7339–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer J.B., Saltik M., Chiocca S., Michou A.I., Moseley P. and Cotten M. (2000) Activation of heatshock response by an adenovirus is essential for virus replication. Nature, 407, 207–211. [DOI] [PubMed] [Google Scholar]
- Gostissa M., Hengstermann A., Fogal V., Sandy P., Schwarz S.E., Scheffner M. and Del Sal G. (1999) Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J., 18, 6462–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1990) Nucleosomes: regulators of transcription. Trends Genet., 6, 395–400. [DOI] [PubMed] [Google Scholar]
- Hassig C.A., Tong J.K., Fleischer T.C., Owa T., Grable P.G., Ayer D.E. and Schreiber S.L. (1998) A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl Acad. Sci. USA, 95, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.T. (2001) Protein modification by SUMO. Trends. Biochem. Sci., 26, 332–333. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (2000) Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol., 2, E153–157, . [DOI] [PubMed] [Google Scholar]
- Ishov A.M. and Maul G.G. (1996) The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol., 134, 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A.M., Sotnikov A.G., Negorev D., Vladimirova O.V., Neff N., Kamitani T., Yeh E.T., Strauss J.F. III and Maul G.G. (1999) PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol., 147, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khochbin S., Verdel A., Lemercier C. and Seigneurin-Berny D. (2001) Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev., 11, 162–166. [DOI] [PubMed] [Google Scholar]
- Kirsh O. et al. (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J., 21, 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korioth F., Maul G.G., Plachter B., Stamminger T. and Frey J. (1996) The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res., 229, 155–158. [DOI] [PubMed] [Google Scholar]
- Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E.J. and Chen J.D. (2000) Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol., 20, 1784–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J.L. (2001) Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol., 75, 1581–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merezak C., Reichert M., Van Lint C., Kerkhofs P., Portetelle D., Willems L. and Kettmann R. (2002) Inhibition of histone deacetylases induces bovine leukemia virus expression in vitro and in vivo. J. Virol., 76, 5034–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S. and Dejean A. (1999) Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol., 73, 5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J.C., Fischle W., Verdin E. and Sinclair J.H. (2002) Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J., 21, 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. and Everett R.D. (2000) Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol., 74, 10006–10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum M.K., Tong J.K., Lane W.S. and Schreiber S.L. (2001) Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem., 276, 47733–47741. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C. and Hay R.T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem., 276, 12654–12659. [DOI] [PubMed] [Google Scholar]
- Shestakova E., Bandu M.T., Doly J. and Bonnefoy E. (2001) Inhibition of histone deacetylation induces constitutive derepression of the β interferon promoter and confers antiviral activity. J. Virol., 75, 3444–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E. and Berger S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Taplick J., Kurtev V., Kroboth K., Posch M., Lechner T. and Seiser C. (2001) Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J. Mol. Biol., 308, 27–38. [DOI] [PubMed] [Google Scholar]
- Wilson V.G. and Rangasamy D. (2001) Intracellular targeting of proteins by sumoylation. Exp. Cell Res., 271, 57–65. [DOI] [PubMed] [Google Scholar]
- Wu W.S., Vallian S., Seto E., Yang W.M., Edmondson D., Roth S. and Chang K.S. (2001) The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol., 21, 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Khosravi-Far R., Chang H.Y. and Baltimore D. (1997) Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell, 89, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Jones C. (2001) The bovine herpesvirus 1 immediate-early protein (bICP0) associates with histone deacetylase 1 to activate transcription. J. Virol., 75, 9571–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Muller S., Ronchetti S., Freemont P.S., Dejean A. and Pandolfi P.P. (2000) Role of SUMO-1-modified PML in nuclear body formation. Blood, 95, 2748–2752. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


