Abstract
Telomeres are protein–DNA complexes that cap chromosome ends and protect them from being recognized and processed as DNA breaks. Loss of capping function results in genetic instability and loss of cellular viability. The emerging view is that maintenance of an appropriate telomere structure is essential for function. Structural information on telomeric proteins that bind to double and single-stranded telomeric DNA shows that, despite a lack of extensive amino-acid sequence conservation, telomeric DNA recognition occurs via conserved DNA-binding domains. Furthermore, telomeric proteins have multidomain structures and hence are conformationally flexible. A possibility is that telomeric proteins take up different conformations when bound to different partners, providing a simple mechanism for modulating telomere architecture.
Introduction
Telomeres are specific protein–DNA complexes that provide a protective cap at the ends of linear eukaryotic chromosomes. Their main function is to prevent chromosome ends from being recognized and processed as DNA breaks (reviewed in Lundblad, 2000). Loss of capping function results in genetic instability and loss of cellular viability.
The discovery, over 20 years ago, that telomeric DNA commonly consists of conserved sequence repeats opened the way for identifying specific protein components of the telomeric cap and the replication machinery unique to telomeres (reviewed in Blackburn, 2001). Telomeric DNA consists of tandemly repeated, often short, sequence motifs that typically contain clusters of three or four guanines (e.g. TTGGGG in Tetrahymena and TTAGGG in humans). These telomeric repeats are added by the enzyme telomerase, a ribonucleoprotein complex consisting of a reverse transcriptase-like subunit (TERT) and a large RNA subunit (TER) that contains the template for the telomeric DNA repeat. The G-rich strand extends in the 3′ direction, forming a singlestranded overhang that acts as the substrate for telomerase-mediated elongation. In vitro, these G-rich telomeric single strands can fold into DNA-quadruplex structures (Parkinson et al., 2002), which may inhibit telomerase activity (Zahler et al., 1991). Recent evidence suggests that DNA quadruplexes are present in vivo (Schaffitzel et al., 2001), but their functional significance has yet to be established.
The terminal regions of the telomeric double-stranded DNA repeats in humans (and other species) are not thought to be packaged into nucleosomes (Tommerup et al., 1994), but together with the singlestranded overhang serve the crucial function of providing binding sites for sequence-specific DNA-binding proteins. These DNA-binding proteins in turn recruit other proteins to the telomere, forming a multiprotein telomeric cap (reviewed in Blackburn, 2001; Kim et al., 2002). Figure 1 shows a partial summary of the known components of human telomeres. Results from a large number of in vivo experiments in which either the protein components were overexpressed, or telomeric proteins or DNA-binding sites were mutated, show that changes in the composition of the telomeric complex lead to telomere dysfunction (reviewed in Shore, 1997; De Lange, 2002). Maintenance of the correct telomere structure is important for appropriate function.
Figure 1.
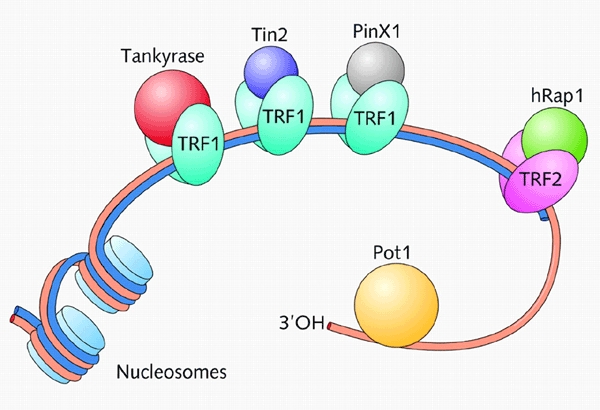
The human telomeric complex consists of DNA and a number of associated proteins (reviewed in Blackburn, 2001; Kim et al., 2002). Human telomeres consist of 7000 to 10 000 bp of doublestranded DNA. Most of this DNA is packaged into nucleosomes, but at the extreme ends of each chromosome a few 100 bp of telomeric DNA are thought to be bound specifically by two related DNA-binding proteins, TRF1 and TRF2. TRF1 forms complexes with two other telomeric proteins, Tin2 and the PARP-containing tankyrase. TRF2 recruits Rap1, which is distantly related to the budding yeast Rap1. However, unlike the human orthologue, the yeast protein binds directly to telomeric double-stranded DNA. In addition to the components shown here, other proteins such as Ku and the Mre11–Rad50–NSB1 complex that are involved in double-stranded DNA repair are also localized at telomeres. The single-stranded G-rich overhang (several 100 nucleotides in length) binds Pot1, which is distantly related to the α-subunit of the end-binding complex of O. nova and also to the budding yeast Cdc13.
Recent discoveries suggest that telomeres exist in at least two different states or architectures: an 'open' and a 'closed' complex. The 'closed' complex is thought to represent the state that caps and protects chromosome ends. In mammals and some other species a lasso-like structure, called the t-loop, has been observed (Griffith et al., 1999; Munoz-Jordan et al., 2001). In the t-loop model for this higher order 'closed' complex structure, the 3′ singlestranded overhang is tucked into the double-stranded telomeric tracts, providing one solution to the end-protection problem (reviewed in De Lange, 2002). In budding yeast, telomeres are also thought to fold into a hairpin-like higher order structure involving the interaction of the telomere and adjacent nucleosomes (Zaman et al., 2002). The 'open' complex is probably required for the telomerase and other enzymes that function at telomeres to access the end of the telomeric DNA, and hence it is likely that different telomere states are cell cycle-dependent. One very interesting question is how telomeres switch from one structure to another. This could happen at the DNA replication stage, by modification of telomeric proteins, or by different complexes being formed at telomeres at different stages of the cell cycle. In order to gain insight into telomere architecture, three-dimensional structural information on telomeric complexes is required. This review focuses on recent structural information that provides important insights into the way in which telomeric proteins recognize telomeric DNA repeats.
Double-stranded telomeric DNA is recognized by Myb/homeodomains
The double-stranded TTAGGG repeats of mammals are bound by two related proteins, TRF1 (Chong et al., 1995) and TRF2 (Bilaud et al., 1997; Broccoli et al., 1997). TRF1 is a negative regulator of telomere maintenance. Overexpression of TRF1 results in telomere shortening, whereas mutants that are defective in DNA binding give rise to telomere lengthening (van Steensel and de Lange, 1997; Smogorzewska et al., 2000). In budding yeast, the telomeric protein ScRap1 binds specifically to the doublestranded telomeric DNA and is also involved in controlling telomere length (Marcand et al., 1997). In vitro, the formation of the t-loop is crucially dependent on TRF2 (reviewed in De Lange, 2002). If TRF2 is removed from telomeres in human cell lines the chromosome ends are immediately recognized as sites of DNA breaks, leading to chromosome end-to-end fusions, the activation of the p53 and p16/RB pathways, and the induction of senescence or apoptosis (van Steensel et al., 1998; Karlseder et al., 1999; Smorgorzewska and de Lange, 2002). Similarly, fission yeast telomeres are protected from end-to-end fusions by the TRF-related protein Taz1 (Cooper et al., 1997).
Both the TRFs and ScRap1 have multidomain structures in which different domains have distinct functions. The most striking feature of these telomeric proteins is that they bind to double-stranded DNA via a structurally conserved DNA-binding domain of the Myb/homeodomain type (Bilaud et al., 1996; König et al., 1996, 1998; Nishikawa et al., 1998). Myb/homeodomains belong to the common helix–turn–helix (HTH) family of DNA-recognition motifs found in many transcription regulators, including bacterial DNA-binding proteins. The conservation was not apparent from amino-acid sequence comparisons between members of the TRF family and the yeast ScRap1 as the overall homology is low or non-existent, but instead emerged when structural information became available. ScRap1 binds to DNA as a monomer, via a DNA-binding domain consisting of two homeodomains repeated in tandem at the centre of the protein (König et al., 1996). By contrast, both TRF1 and TRF2 contain a single motif at their C-termini that shares significant sequence homology with the Myb DNA-binding motif (Chong et al., 1995; Bilaud et al., 1996). Both TRF1 (Bianchi et al., 1997) and TRF2 (Broccoli et al., 1997) function as preformed homodimers, and hence sequencespecific recognition of double-stranded telomeric DNA involves the juxtaposition of two Myb/homeodomains. Dimerization of the TRFs involves the TRF-homology domain (Bianchi et al., 1997). The crystal structure (Fairall et al., 2001) of this domain from both hTRF1 and hTRF2 shows that, despite limited sequence similarity, they have the same horseshoeshaped α-helical architecture (Figure 2A). The two dimer interfaces, consisting of three α-helices from each monomer, feature unique interactions that prevent heterodimerization between TRF1 and TRF2 (Fairall et al., 2001). Furthermore, despite the conservation in architecture, the surfaces of the two dimerization domains are different, consistent with the separate roles of TRF1 and TRF2 in recruiting different partners to the telomere. For TRF1 it has been shown that the DNA-binding domain is linked to the dimerization domain via a long, flexible linker (Figure 2A), allowing it to bind not only to adjacent telomeric repeats but also to binding sites spaced far apart (Bianchi, 1999), and even to two different DNA molecules (Griffith et al., 1998). The multidomain architecture of these telomeric proteins, as well as their DNA-binding flexibility, is likely to be relevant for the dynamic structure of telomeres.
Figure 2.
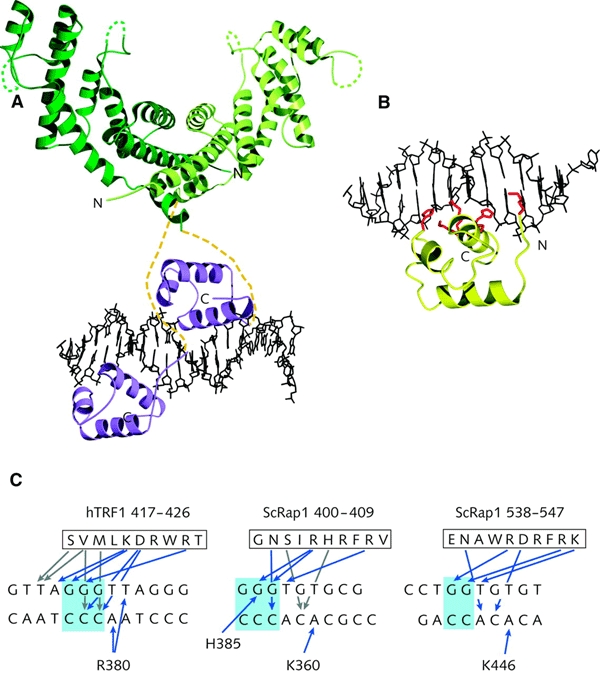
Recognition of double-stranded telomeric DNA. (A) A model of hTRF1 bound to telomeric DNA. The model is a composite of the crystal structure of the dimerization domain (green) and the NMR structure of the DNA-binding domain (purple) of TRF1 in complex with telomeric DNA. Yellow dashed lines represent the flexible linkers. (B) The ScRap1 domain 1 in complex with telomeric DNA. Side chains involved in DNA recognition are shown in red. (C) Comparison of the contacts made by ScRap1 and hTRF1 with telomeric DNA repeats. Only the contacts made by the DNA-recognition helix are shown. Blue arrows depict hydrogen bonds and grey arrows hydrophobic interactions.
The X-ray crystal structure of the DNA-binding domain of the yeast ScRap1 bound to a telomeric DNA-binding site (König et al., 1996) and the NMR structure of the Myb motif from hTRF1 in complex with a human telomeric DNA fragment (Nishikawa et al., 2001) permit a direct comparison of how telomeric proteins recognize doublestranded telomeric DNA repeats (Figure 2). The core of the two ScRap1 sub-domains and the TRF1 Myb motif have the same overall architecture, consisting of a bundle of three α-helices in which the second and third helices form the 'helix–turn–helix' motif. The third α-helix is the DNA-recognition helix, which makes basespecific contacts in the major groove of DNA (Figure 2B and C). In addition, all three motifs have an N-terminal arm that makes basespecific contacts in the minor groove of DNA (Figure 2A and B). The N-terminal arm extends the length of the DNA-binding sites from ∼3 bp as seen for Myb motifs to 5 or 6 bp, and hence increases stability and specificity. This mode of DNA-binding classifies both the Rap1 and TRF DNA-binding domains as homeodomains (König and Rhodes, 1997; König et al., 1998).
In ScRap1, the two subdomains in the DNA-binding domain are tandemly arranged on the DNA so that each is aligned and makes very similar contacts with a GGTGT sequence present in the binding site for ScRap1 d(TGGTGTGTGGGTGT) (König et al., 1996). The spacing between the two binding sites within the DNA appears to be determined by the linker between the two ScRap1 subdomains (Taylor et al., 2000). Similarly, hTRF1 interacts with the sequence AGGGTT (König et al., 1998; Bianchi, 1999; Nishikawa et al., 2001). Figure 2C summarizes the protein–DNA interactions observed in the recognition of doublestranded telomeric DNA. Significantly, the G-clusters that characterize telomeric DNA repeats play a central role in recognition. Despite some limitations arising from comparing the crystal and NMR structures, the patterns of sequence-specific contacts made by hTRF1 and ScRap1 are very similar. In both complexes, arginine and lysine side chains from similar positions in the DNA-recognition helix make hydrogen bond contacts to guanines in the G-rich strand. In addition, a number of hydrophobic interactions are seen, particularly by residues in the N-terminus of the hTRF1 DNA-recognition helix. Residues from the DNA-recognition helix also contact the C-rich strand (Figure 2C). Whereas in the crystal structure of the ScRap1–DNA complex the protein–DNA interface is well ordered and includes a number of water molecules involved in bridging interactions (König et al., 1996), the solution structure of the TRF1 homeodomain–DNA complex shows that long side chains can take up alternative conformations, switching interactions between an adjoining A and G in the DNA-binding site (Figure 2C; Nishikawa et al., 2001). The N-terminal arm of all three domains crosses the ribose-phosphate backbone, making contacts in the minor groove: in ScRap1, both subdomains use a lysine to contact an A in the C-rich strand; in hTRF1, an arginine contacts an A on the C-rich strand and a T on the G-rich strand (Figure 2C). In each of these domains, the binding is further stabilized by multiple interactions with the ribose-phosphate backbone.
The single-stranded telomeric overhang is recognized by OB-folds
The ciliate Oxytricha nova telomere end-binding protein (OnTEBP) was the first protein to be identified that specifically recognizes the single-stranded G-rich overhang (Gottschling and Zakian, 1986; Price and Cech, 1987). It is composed of two subunits, OnTEBPα and OnTEBPβ. These two proteins can form two alternative complexes that bind specifically but differently to the telomeric overhang of the macronuclear chromosomes. A single copy of the α–β heterodimeric complex is sufficient for binding the 16 nucleotide O. nova singlestranded overhang in vitro (Fang and Cech, 1993), whereas two αsubunit homodimers can bind to the same overhang (Peersen et al., 2002). Due to the different modes of DNA binding by the two complexes, it has been proposed that they might be involved in the assembly and disassembly of higher order telomeric structures (Peersen et al., 2002). Furthermore, the α–β heterodimer inhibits the action of telomerase whereas the α-homodimer does not, and hence the two different complexes must have separate functions at telomeres (Froelich-Ammon et al., 1998). More recently, Pot1 from both fission yeast and humans was identified through a weak sequence similarity to the N-terminal region of OnTEBPα (Baumann and Cech, 2001). Deletion of the Pot1 gene in fission yeast leads to rapid loss of telomeric DNA and to chromosome circularization, providing evidence that Pot1 has a crucial role in telomere capping (Baumann and Cech, 2001). Whether higher eukaryotes have a βsubunit homologue is not known. In budding yeast, the single-stranded telomeric DNA is bound specifically by Cdc13 (Lin and Zakian, 1996; Nugent et al., 1996). Cdc13 has two separate functions: it is involved both in chromosome end protection and the recruitment of the telomerase, and hence in telomere replication (Nugent et al., 1996).
As is the case for telomeric proteins that bind to double-stranded telomeric repeats, recent information suggests that proteins that bind to the single-stranded G-rich overhang do so via a conserved DNA-binding domain, the OB-fold. The OB-fold is a small structural domain, binding substrates as diverse as oligonucleotides, oligosaccharides and peptides (Murzin, 1993). Again, three-dimensional structural information was crucial for discovering the structural and functional conservation, as OB-folds are difficult to identify from amino-acid sequence analyses alone. The role of the OB-fold in the recognition of singlestranded telomeric DNA was observed in the crystal structure of the OnTEBPα–OnTEBPβ–ssDNA complex (Horvath et al., 1998), and has more recently been found in the structure of the DNA-binding domain of the budding yeast Cdc13 (Mitton-Fry et al., 2002). Amino-acid sequence alignments also suggest that Pot1 proteins from both fission yeast and humans contain OB-folds (Baumann and Cech, 2001), which consist of a fivestranded β-sheet coiled to form a closed β-barrel and bind ligands primarily via the loops connecting the β-strands (Figure 3C and D). Recognition via flexible loops gives rise to extended and adaptable surfaces for singlestranded DNA recognition.
Figure 3.
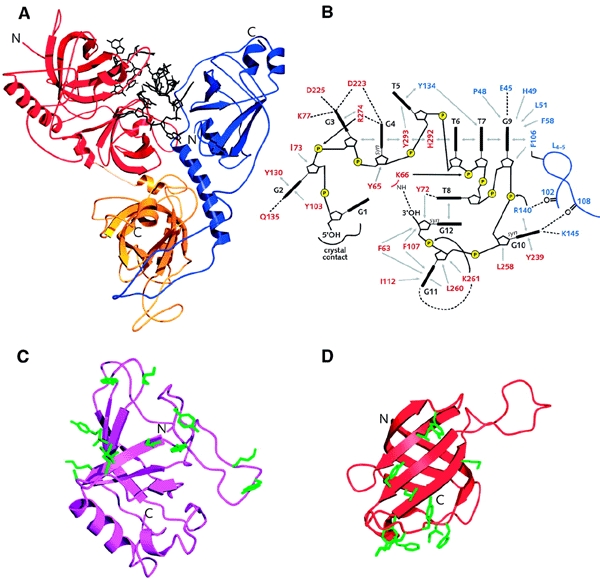
Recognition of single-stranded telomeric DNA. (A) Structure of the OnTEBPα–OnTEBPβ–d(GGGGTTTTGGGG) complex. The α-subunit is shown in red and yellow and the β-subunit is shown in blue. (B) Schematic representation of the protein–ssDNA interaction in the OnTEBPα–OnTEBPβ–d(GGGGTTTTGGGG) complex. Bases are shown in black, amino acid side chains from the α-subunit are indicated in red, and those from the β-subunit in blue. Hydrogen bonds are depicted as dotted lines and hydrophobic interactions by grey arrows. (C) Structure of the Cdc13 OB-fold. Amino acid side chains involved in ssDNA recognition are shown in green. (D) Structure of the first OnTEBPα OB-fold (residues 37–150). Side chains involved in ssDNA recognition are shown in green.
Two crystal structures, of the OnTEBPα–OnTEBPβ heterodimer and the OnTEBPα homodimer, and the NMR structure of the DNA-binding domain of ScCdc13 reveal the details of telomeric single-stranded DNA recognition by OB-folds, and show that telomeric proteins can take up different conformations when bound to different partners. Figure 3 summarizes the versatility of OB-folds in singlestranded DNA recognition. The crystal structure of the OnTEBPα–OnTEBPβ–d(GGGGTTTTGGGG) 1:1:1 complex (Horvath et al., 1998) contains four OB-folds: three are used in singlestranded DNA binding and the fourth for protein–protein interactions between the α- and β-subunits (Figure 3A). The αsubunit consists of two structural domains: an N-terminal domain that consists of two OB-folds that bind DNA, and a C-terminal domain containing a third OB-fold that interacts with the β-subunit. The OB-fold present in the β-subunit also interacts with DNA. Together, the three OB-folds of the α–β heterodimer form a deep DNA-binding cleft, which is likely to form by co-folding of protein and DNA (Figure 3A; Horvath et al., 1998). The two OB-folds in the N-terminal domain of the αsubunit pack tightly together and the loops of the two domains co-operate to interact with G1 to T5, T8 and G10 to G12, whereas the OB-fold of the β-subunit interacts with T5 to G10 (Figure 3B). The singlestranded DNA adopts an irregular non-helical folded-back path, in which the ribose-phosphate backbone is solvent-exposed and the bases are completely buried in the protein cleft. A wealth of different interactions is made, including the stacking of most of the bases either with aromatic amino acid side chains or with another base of the single-stranded DNA (Figure 3B). All of the G nucleotides in the 12-mer sequence are involved in hydrogen bonding interactions. Significantly, G12, the 3′-terminal guanine, is buried deep within the complex, making it inaccessible to the telomerase.
The crystal structure of the OnTEBPα–d(TTTTGGGG) 2:2 complex revealed a homodimeric head-to-tail arrangement, in which each α-subunit binds one telomeric single-stranded DNA overhang on opposite sides of the dimer, suggesting a mechanism for protein-mediated telomere–telomere association (Peersen et al., 2002). Comparison of the structures of the homodimeric and heterodimeric complexes shows a reorientation of domains. The OnTEBPα C-terminal domain in the heterodimeric complex is tilted by ∼45° and rotated clockwise by ∼90° relative to its position in the homodimer. This is made possible by distortion of the flexible linker that connects the two domains in the αsubunit. This has consequences for nucleic acid recognition, since part of the nucleic acid-binding surface is involved in protein–protein interactions in the α-homodimer. Consequently, dimerization of the α-subunit and single-stranded DNA binding in the α–β complex are mutually exclusive. The α-subunit also interacts differently with the 8-mer d(TTTTGGGG) in the homodimer than it does with the 12-mer d(GGGGTTTTGGGG) in the heterodimer. The telomeric single-stranded DNA is shifted in register by one repeat, so that the 3′-terminal G-cluster in the homodimeric complex occupies the same binding site as the 5′-terminal G-cluster in the heterodimeric complex. In the homodimer, nucleotides T1 to G6 are recognized by the first OB-fold, and G7 and G8 by the second OB-fold in the N-terminus of the α-subunit. The recognition of guanines is essentially conserved between the homodimeric and heterodimeric complexes. Significantly, and contrary to the situation in the heterodimeric complex, in the α-subunit homodimer the 3′ hydroxyl group of the terminal guanine is solvent-exposed and accessible to the telomerase, consistent with functional differences between the TEBP homodimeric and heterodimeric complexes.
The solution structure of the DNA-binding domain of Cdc13 in complex with its telomeric DNA recognition sequence, dGTGTGGGTGTG, revealed that despite a lack of any significant sequence similarity to other known telomeric proteins, it also contains an OB-fold (Mitton-Fry et al., 2002). Although in this NMR structure the path of the singlestranded DNA was not determined, it was possible to identify the amino acid side chains involved in DNA binding (Figure 3C), and these show a broadly similar pattern of contacts to those seen for the OnTEBP (Figure 3D). The Cdc13 OB-fold contains an unusually long loop between βstrands 2 and 3, which packs tightly over these two strands (Figure 3C) and extends the DNA interaction surface of the OB-fold.
Conclusions and prospectives
The information on the three-dimensional structures of telomeric proteins has started to give insights into the precise architecture of these proteins and their function in telomeric DNA recognition. Despite the surprising overall divergence in the sequence of telomeric proteins that have a similar function in different organisms, structural similarities in their DNA-binding domains have emerged. This stresses the power of structure-based sequence alignments to reveal homologies and functional insights (König and Rhodes, 1997; Mitton-Fry et al., 2002). The information available so far indicates that the recognition of telomeric DNA, both singlestranded and double-stranded, is conserved. Given that a large number of other DNA-binding motifs are found in nature, the selection of the Myb/homeodomain for double-stranded DNA recognition (König, 1996, 1998; Nishikawa et al., 2001) and the OB-fold for singlestranded DNA recognition (Horvath et al., 1998; Baumann and Cech, 2001; Mitton-Fry, 2002; Peersen et al., 2002) suggests that each of the two classes of telomeric DNA-binding proteins has a common ancestor. This could be the consequence of selective pressure imposed by the target of recognition, the telomeric DNA, which contains highly conserved sequence features such as repeated clusters of three or four guanines. As the information on the amino-acid sequence and structure of more telomeric proteins becomes available, it will be interesting to see if this apparent conservation is widespread.
The mechanistically attractive 'open' and 'closed' telomeric complexes—linear and t-loop—need to be elucidated at the structural level. The structural transitions that have to take place when switching from one structure to the other probably involve the formation of different complexes, for instance through DNA-binding proteins binding to different partners at different stages of the cell cycle. The participation of different known telomere-associated proteins throughout the cell cycle has yet to be established. Furthermore, since most human telomeric DNA is organized into nucleosomes (Tommerup et al., 1994), the involvement of higher order telomere structures such as t-loops needs to be investigated. With regard to a dynamic telomere structure, it is significant that many of the known telomeric proteins such as ScRAP1 (König, 1996), SpTaz1, hTRF1, hTRF2 (Li et al., 2000; Fairall et al., 2001) and OnTEBPs (Horvath et al., 1998) are all multidomain proteins. In these proteins, functional domains are joined by flexible linkers, and hence have the potential to adopt different conformations when bound to different partners. This kind of protein design provides an elegant mechanism for modulating the overall architecture of telomeres or, as illustrated by the OnTEBP complexes, a mechanism for telomere assembly and disassembly (Peersen et al., 2002). Structural information on various telomeric multiprotein complexes will be required to understand fully how the various telomeric proteins function in assembling and regulating telomere structure.

References
- Baumann P. and Cech T.R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science, 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Bianchi A., Smith S., Chong L., Elias P. and de Lange T. (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J., 16, 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Stansel R.M., Fairall L., Griffith J.D., Rhodes D. and de Lange T. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J., 18, 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T., Koering C.E., Binet-Brasselet E., Ancelin K., Pollice A., Gasser S.M. and Gilson E. (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res., 24, 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T. and Gilson E. (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L. and de Lange T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P. and de Lange T. (1995) A human telomeric protein. Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Nimmo E.R., Allshire R.C. and Cech T.R. (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature, 385, 744–747. [DOI] [PubMed] [Google Scholar]
- De Lange T. (2002) Protection of mammalian telomeres. Oncogene, 21, 532–540. [DOI] [PubMed] [Google Scholar]
- Fairall L., Chapman L., Moss H., de Lange T. and Rhodes D. (2001) Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell, 8, 351–361. [DOI] [PubMed] [Google Scholar]
- Fang G. and Cech T.R. (1993) Oxytricha telomere-binding protein: DNA-dependent dimerization of the α and β subunits. Proc. Natl Acad. Sci. USA, 90, 6056–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich-Ammon S.J., Dickinson B.A., Bevilacqua J.M., Schultz S.C. and Cech T.R. (1998) Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev., 12, 1504–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D.E. and Zakian V.A. (1986) Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell, 47, 195–205. [DOI] [PubMed] [Google Scholar]
- Griffith J., Bianchi A. and de Lange T. (1998) TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol., 278, 79–88. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H. and de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Horvath M.P., Schweiker V.L., Bevilacqua J.M., Ruggles J.A. and Schultz S.C. (1998) Crystal structure of the Oxytricha nova telomere end-binding protein complexed with single strand DNA. Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Broccoli D., Dai Y., Hardy S. and de Lange T. (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science, 283, 1321–1325. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Kaminker P. and Campisi J. (2002) Telomeres, aging and cancer: in search of a happy ending. Oncogene, 21, 503–511. [DOI] [PubMed] [Google Scholar]
- König P. and Rhodes D. (1997) Recognition of telomeric DNA. Trends Biochem. Sci., 22, 43–47. [DOI] [PubMed] [Google Scholar]
- König P., Giraldo R., Chapman L. and Rhodes D. (1996) The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell, 85, 125–136. [DOI] [PubMed] [Google Scholar]
- König P., Fairall L. and Rhodes D. (1998) Sequence specific DNA recognition by the Myb-like domain of the human telomere binding protein TRF1: a model for the protein–DNA complex. Nucleic Acids Res., 26, 1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Oestreich S. and de Lange T. (2000) Identification of human Rap1: implications for telomere evolution. Cell, 101, 471–483. [DOI] [PubMed] [Google Scholar]
- Lin J.J. and Zakian V.A. (1996) The Saccharomyces CDC13 protein is a singlestrand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. (2000) DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res., 451, 227–240. [DOI] [PubMed] [Google Scholar]
- Marcand S., Wotton D., Gilson E. and Shore D. (1997) Rap1p and telomere length regulation in yeast. Ciba Found. Symp., 211, 76–93. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry R.M., Anderson E.M., Hughes T.R., Lundblad V. and Wuttke D.S. (2002) Conserved structure for singlestranded telomeric DNA recognition. Science, 296, 145–147. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan J.L., Cross G.A.M., de Lange T. and Griffith J.D. (2001) T-loops at trypanosome telomeres. EMBO J., 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G. (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T., Nagadoi A., Yoshimura S. and Nishimura Y. (1998) Solution structure of the DNA-binding domain of human telomeric protein, hTRF1. Structure, 6, 1057–1065. [DOI] [PubMed] [Google Scholar]
- Nishikawa T., Okamura H., Nagadoi A., König P., Rhodes D. and Nishimura Y. (2001) Solution structure of a telomeric DNA complex of human TRF1. Structure, 9, 1237–1251. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes T.R., Lue N.F. and Lundblad V. (1996) Cdc13p: a singlestrand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- Parkinson G.N., Lee M.P. and Neidle S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- Peersen O.B., Ruggles J.A. and Schultz S.C. (2002) Dimeric structure of the Oxytricha nova telomere end-binding protein αsubunit bound to ssDNA. Nat. Struct. Biol., 9, 182–187. [DOI] [PubMed] [Google Scholar]
- Price C.M. and Cech T.R. (1987) Telomeric DNA–protein interactions of Oxytricha macronuclear DNA. Genes Dev., 1, 783–793. [DOI] [PubMed] [Google Scholar]
- Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J. and Pluckthun A. (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA, 98, 8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. (1997) Telomere length regulation: getting the measure of chromosome ends. Biol. Chem., 378, 591–597. [PubMed] [Google Scholar]
- Smogorzewska A. and de Lange T. (2002) Different telomere damage signaling pathways in human and mouse cells. EMBO J., 21, 4338–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogozewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G. and de Lange T. (2000) Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol., 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor H.O., O'Reilly M., Leslie A.G. and Rhodes D. (2000) How the multifunctional yeast Rap1p discriminates between DNA target sites: a crystallographic analysis. J. Mol. Biol., 303, 693–707. [DOI] [PubMed] [Google Scholar]
- Tommerup H., Dousmanis A. and de Lange T. (1994) Unusual chromatin in human telomeres. Mol. Cell. Biol., 14, 5777–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B. and de Lange T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature, 385, 740–743. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A. and de Lange T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Zahler A.M., Williamson J.R., Cech T.R. and Prescott D.M. (1991) Inhibition of telomerase by G-quartet DNA structures. Nature, 350, 718–720. [DOI] [PubMed] [Google Scholar]
- Zaman Z., Heid C. and Ptashne M. (2002) Telomere looping permits repression 'at a distance' in yeast. Curr. Biol., 12, 930–933. [DOI] [PubMed] [Google Scholar]


