Abstract
We report that a lipoprotein-based nanoplatform generated by conjugating tumor-homing molecules to the protein components of naturally occurring lipoproteins reroutes them from their normal lipoprotein receptors to other selected cancer-associated receptors. Multiple copies of these targeting moieties may be attached to the same nanoparticle, or a variety of different targeting moieties can be attached. Such a diverse set of tumor-homing molecules could be used to create a variety of conjugated lipoproteins as multifunctional, biocompatible nanoplatforms with a broad application to both cancer imaging and treatment. The same principle can be applied to imaging and treatment of other diseases and for monitoring specific tissues. To validate this concept, we prepared a low-density lipoprotein (LDL)-based folate receptor (FR)-targeted agent by conjugating folic acid to the Lys residues of the apolipoprotein B (apoB)-100 protein. To demonstrate the ability of the lipoprotein-based nanoplatform to deliver surfaceloaded and core-loaded payloads, the particles were labeled either with the optical reporter 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine that was intercalated in the phospholipid monolayer or with the lipophilic photodynamic therapy agent, tetra-t-butyl-silicon phthalocyanine bisoleate, that was reconstituted into the lipid core. Cellular localization of the labeled LDL was monitored by confocal microscopy and flow cytometry in FR-overexpressing KB cells, in FR-nonexpressing CHO and HT-1080 cells, and in LDL receptor-overexpressing HepG2 cells. These studies demonstrate that the folic acid conjugation to the Lys side-chain amino groups blocks binding to the normal LDL receptor and reroutes the resulting conjugate to cancer cells through their FRs.
Keywords: folate receptor, low-density lipoprotein receptor, biocompatible, optical imaging, photodynamic therapy
Nanoplatforms are nanoscale structures designed as general platforms for multifunctional diagnostic and therapeutic devices. Most of the reported nanoplatforms are synthetic, consisting of structures such as dendrimers (1), silica-coated micelles (2), polymeric (3, 4) and ceramic (5, 6) nanoparticles, perfluorocarbon emulsions (7), cross-linked liposomes (8, 9), and magnetic surface-coated (10) and semiconductor (11) nanoparticles. Most of these nanostructures are easily manipulated, providing the opportunity to add a variety of molecules (payloads) to the surface and/or interior of the nanoparticle. These nanoparticles can be targeted to cancer cells or the tumor vasculature by attaching monoclonal antibodies or cell-surface receptor/transporter ligands that bind specifically to molecules found on the surfaces of the target cells. One common concern is the biocompatibility of these elegant nanoplatforms, which is closely related to their short- and long-term toxicity.
Lipoproteins are natural nanostructures that transport cholesterol and other lipids in the blood. Lipoproteins share a common structure consisting of an apolar core surrounded by a shell composed of a phospholipid monolayer containing unesterified cholesterol and one or more apolipoproteins. The five main lipoprotein classes with characteristic sizes, densities, lipids, and apolipoproteins are listed in descending order of size and ascending order of density (the diameter range is indicated in parentheses): chylomicrons (75-1,200 nm), very low-density lipoprotein (LDL) (30-80 nm), intermediate LDL (25-35 nm), LDL (18-25 nm), and high-density lipoprotein (HDL) (8-12 nm). Being endogenous carriers, lipoproteins escape recognition as foreign entities by the human immune system and escape absorption by the reticuloendothelial system (12). Thus, lipoprotein nanoplatforms may provide a solution to the biocompatibility problem associated with most synthetic nanodevices.
The overexpression of the LDL receptor (LDLR) relative to normal liver and adrenals by various tumor cell lines (13) has been attributed to the large quantities of cholesterol and fatty acids required for sustaining rapid proliferation. Consequently, LDL particles (≈22 nm in diameter) have long been used as vehicles for the selective delivery of diagnostic and therapeutic agents to tumor cells by LDLR-mediated endocytosis (12, 14-17). There are three ways to incorporate diagnostic and therapeutic agents into LDL particles: (i) by intercalation into the phospholipid monolayer (surface-loading) (18), (ii) by reconstitution into the apolar core (core-loading) by a procedure introduced by Krieger et al. (19, 20), and (iii) by covalent conjugation to the amino acid residues of apolipoprotein B (apoB)-100 (21).
Although LDL has proven to be a useful vehicle for delivery of lipophilic drugs and diagnostic agents to tumors (22), its application is largely limited to LDLR-related diseases. Specifically, it has a limited use for cancer therapy because many tumors do not overexpress the LDLR, whereas some normal tissues do. Moreover, a number of other receptors, called “cancer signatures,” have proven to be more tumor-specific than LDLR, including Her2/neu (23), ΔEGF (24), somatostatin (25), folate (FA) (26), and αvβ3 integrins (27). In this article, we report the development and validation of a general strategy for rerouting the LDL particle to desired receptors, which not only provides a method for targeting cancer cells with greater specificity but also facilitates selective delivery of selected agents to other diseased or normal cells.
The rerouting strategy takes advantage of a highly basic domain that contains Lys residues with an anomalously low pKa value in the receptor-targeting moiety of apoB-100. Of the 357 Lys residues in apoB-100, 225 are exposed on the protein surface, and the remaining 132 Lys side-chain nitrogens are shielded by either lipid-protein or protein-protein interactions (28). Of the 225 exposed Lys residues, 53 Lys ε-amino groups are “active,” with a low pKa value of 8.9, and 172 Lys are “normal,” with a pKa value of 10.5 (28). As noted, the Lys residues in the receptor-binding region have active ε-amino groups (pKa = 8.9). Thus, if one titrates apoB-100 with agents that alkylate the Lys ε-amino groups, the active Lys residues are alkylated first, and when ≈20% of the Lys residues are modified, the binding capability of this protein to the LDLR is essentially abolished (28). By using alkylating groups that target non-LDL receptors [e.g., FA (26), peptides or peptidomimetics that bind to somatostatin (25), Her2/neu (29), ΔEGF (24), and αvβ3 integrins (27), etc.], one could both abolish the LDLR-targeting capability of LDL and simultaneously reroute the particle to these alternate receptors.
In the present study, we validate this concept in the case of rerouting the lipoprotein-based nanoplatform (LBNP) (Fig. 1) to folate receptors (FRs) on KB cells (26) that naturally overexpress these receptors. As controls, we use CHO (30) and HT-1080 (31) cells that lack FR, and HepG2 cells (32) that overexpress LDLR. We have chosen FA as the tumor-homing molecule because it has a high affinity (Kd ≤ 10-9 M) for FRs, which are overexpressed in epithelial malignancies such as ovarian, breast, and colorectal cancers (26). Moreover, when FA is covalently linked to a macromolecule (≤60 nm) via its γ-carboxyl moiety, it retains its high affinity for FR (33).
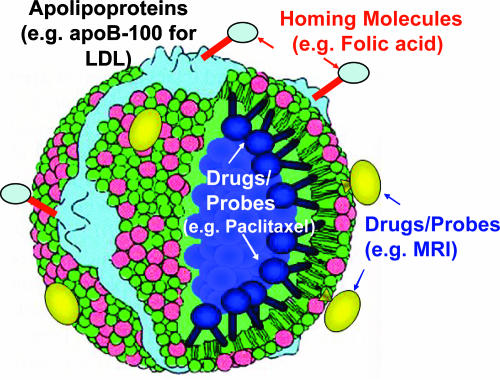
Fig. 1.
The LBNP concept.
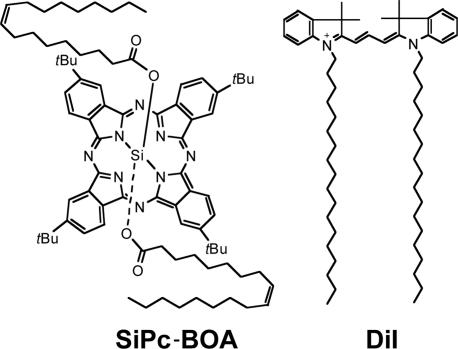
To demonstrate the ability of LBNP to deliver surface- and core-loaded payloads, we have incorporated tetra-t-butyl-silicon phthalocyanine bisoleate (SiPc-BOA) (34) and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine (DiI) (35), respectively, into the nanoparticles. The structures of SiPc-BOA and DiI are shown in Fig. 2. SiPc-BOA is a new photodynamic therapy agent for LDL core-loading and is a neutral, lipophilic analog of phthalocyanine 4, a well-known photodynamic therapy agent in clinical trials (36). Because its central silicon atom allows axial coordination of two oleate moieties, SiPc-BOA is nonaggregatable and highly lipophilic; both properties are essential for achieving high payloads by LDL reconstitution (core-loading) (34). DiI is a lipid-anchored, carbocyanine-based optical probe known to intercalate into the LDL phospholipid monolayer (surface-loading) (35). Thus, we were able to demonstrate that LDL nanoparticles carrying diagnostic/therapeutic agents can be delivered to a cancer-specific receptor that is distinct from its normal receptor, LDLR, and simultaneously abolish delivery to LDLR.
Fig. 2.
Structures of SiPc-BOA and DiI.
Materials and Methods
Synthesis of Folate-Conjugated LDL. Synthesis of folate-N-hydroxysuccinimide ester. FA dihydrate (54 mg, 113 μmol) was dissolved in 1 ml of anhydrous DMSO followed by the addition of 20 μl of triethylamine. Then, N-hydroxysuccinimide (NHS) (20 mg, 170 μmol) and dicyclohexylcarbodiimide (46 mg, 226 μmol) were added, and the reaction mixture was kept at room temperature under argon for 40 h. After removing by-product dicyclohexy-lurea by filtration, folate-N-hydroxysuccinimide ester (FA-NHS) was precipitated from the concentrated filtrate by using a 5-fold volume excess of anhydrous ether. The yellow FA-NHS precipitate was washed several times with anhydrous ether, dried under high-vacuum conditions, and stored as a powder.
Synthesis of r-Pc-LDL and DiI-LDL. LDL, isolated from fresh plasma of healthy donors by sequential ultracentrifugation, as described in ref. 37, was purchased from Sissel Lund-Katz's laboratory at the Children's Hospital of Philadelphia (Philadelphia). The SiPc-BOA synthesis, its reconstitution into the LDL core, and the characterization of the resulting SiPc-BOA reconstituted LDL (r-Pc-LDL), including reconstitution efficiency, payload, particle size measurement, and LDLR-mediated intracellular uptake, are described in ref. 34. For the preparation of the LDL surface-loaded with DiI (DiI-LDL), DiI (Molecular Probes) was surface-loaded into LDL by using the method described in refs. 35 and 38. The molar ratios of adduct to protein of Pc/LDL and DiI/LDL were 300:1 and 50:1, respectively.
Synthesis of the r-Pc-LDL-FA and DiI-LDL-FA conjugate. The pH of r-Pc-LDL or DiI-LDL solutions (5 ml) was increased to 9.4 by dialyzing against the 0.1 M NaH2PO4/0.1MH3BO3 buffer at 4°C overnight. FA-NHS in anhydrous DMSO was slowly added to this LDL solution (molar ratio of FA-NHS/LDL = 200:1). The reaction mixture was kept at 4°C for 40 h, centrifuged at 50 × g rpm at 4°C to remove any precipitate from degraded LDL, and subsequently dialyzed against EDTA buffer (0.3 mM EDTA/0.9% NaCl, pH 7.4) at 4°C overnight. Over the course of the dialysis, the pH of the r-Pc-folate-conjugated LDL (r-Pc-LDL-FA) solution or the folate-conjugated DiI-LDL (DiI-LDL-FA) solution returned to 7.4, and unreacted FA-NHS starting materials were eliminated. This dialysis procedure was repeated until no spectrophotometrically detectable free FA was present. The resulting two conjugates, DiI-LDL-FA and r-Pc-LDL-FA, were obtained in 75% and 90% yield, respectively.
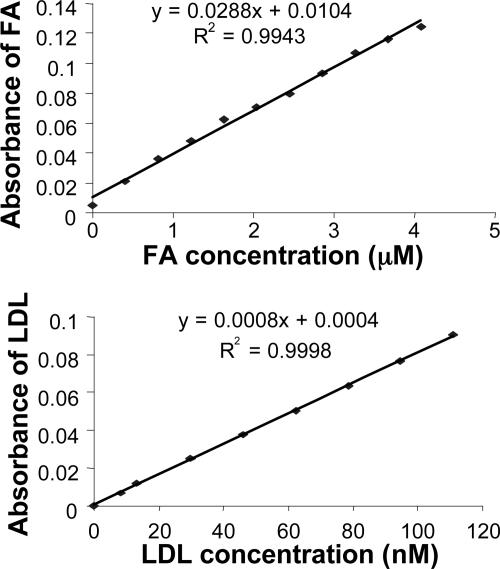
Determination of FA Payload of DiI-LDL-FA and r-Pc-LDL-FA. To determine the number of FA molecules attached to each LDL nanoparticle, two standard absorbance curves were generated for FA and LDL in saline at 280 nm. Absorbance measurements were performed using a PerkinElmer Lambda 2 spectrophotometer with standard solutions of free FA (0-4.2 μM) and LDL (0-110 nM), respectively. As shown in Fig. 3, both LDL and free FA exhibited linear correlations between their respective concentrations and absorbances. Because the protein content of LDL-FA particles was determined by Lowry's method (39), the LDL absorbance (ALDL) was derived on the basis of the LDL standard curve. With ALDL known, the FA absorbance (AFA) was calculated by using the formula AFA = ALDL-FA - ALDL, where ALDL-FA is the absorbance of LDL-FA. The FA concentration was derived from the standard curve, and the FA payload was calculated.
Fig. 3.
The standard curves of the absorbance of FA (Upper) and LDL (Lower) vs. concentration.
Cell Lines. KB cells (human epidermoid carcinoma cells; FR+) (26), HT-1080 cells (human fibrosarcoma cells; FR-) (31), and CHO cells (FR-) (30) were purchased from the American Type Culture Collection. Both KB and HT-1080 cells were cultured in Eagle's MEM supplemented with 2 mM l-glutamine, 17.9 mM sodium bicarbonate, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 10% FBS. CHO cells were cultured in Ham's F-12K medium with 2 mM l-glutamine, 17.9 mM sodium bicarbonate, and 10% FBS. HepG2 cells (human hepatoblastoma G2; LDLR+) (32), a generous gift from Theo van Berkel's laboratory at the University of Leiden (Leiden, The Netherlands), were cultured in DMEM supplemented with 10% FBS and 100 units/ml penicillin-streptomycin. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
Confocal Microscopic Studies. Cells were grown for 3 days in four-well Lab-Tek chamber slides. After three quick washes with ice-cold PBS, experiments were initiated by adding FA-deficient RPMI medium 1640 (Invitrogen) containing 0.38 μM FA-conjugated LDL. After a 4-h incubation at 37°C, cells were washed five times with ice-cold PBS containing 0.8% BSA and two times with PBS alone and then fixed for 20 min with 2% formaldehyde in PBS at room temperature. Chamber slides were then mounted and sealed for confocal microscopic analysis with a Leica TCS SPII laser scanning confocal microscope. Filter settings were 633 nm for r-Pc-LDL and 488 nm for DiI-LDL, respectively.
Flow Cytometry Studies. KB cells (≈5 × 105) were seeded in each T25 flask 2 days before the experiments were performed. On the day of the experiment, cells were washed twice with FA-deficient RPMI medium 1640. Then, 1.5 ml of FA-deficient medium was added to each flask. Cells were then incubated with LDL-FA under these conditions, except for the competitive inhibition assay, which required adding different concentrations of free FA or LDL to the medium as inhibitors. At the end of the incubation period for all experiments, the medium was aspirated, and the cells were washed three consecutive times with 2 ml of ice-cold PBS. Subsequently, cells were harvested by adding 0.25% trypsin-EDTA and resuspended in PBS. Suspensions were examined by flow cytometry with a FACSort flow cytometer (Becton Dickinson).
Electron Microscopy Studies. Electron microscopy was used to determine the morphology and the size of the aqueous dispersion of nanoparticles with a JEOL JEM 1010 electron microscope equipped with a charge-coupled device camera (Hamamatsu, Middlesex, NJ) at 80 kv by using amt 12-hr software (Advanced Microscopy Technique, Danvers, MA). Thus, 5 μl of lipoprotein nanoparticle suspension was placed on carbon-coated 200-mesh copper grids and allowed to stand for 5 min. The excess sample was removed with lens paper, and 2% saturated aqueous uranyl acetate was applied to the grid in five consecutive drops within 20 s. The stain was then drained off with filter paper, and the grid was air dried before taking the digital images. All electron microscopy supplies were purchased from Electron Microscopy Sciences (Fort Washington, PA).
Results
To synthesize LDL-FA conjugates, LDL was first labeled with DiI by surface-loading or with SiPc-BOA by core-loading. This labeling produced two functionalized LDL particles, DiI-LDL (molar ratio, 50:1) and r-Pc-LDL (molar ratio, 300:1). Conjugation of FA to the Lys residues of apoB-100 yielded two LBNP particles, DiI-LDL-FA and r-Pc-LDL-FA, with molar ratios of 50:1:170 and 300:1:170, respectively. Electron microscopy studies revealed a moderate increase in mean particle size (26.12 ± 3.00 nm for DiI-LDL-FA and 24.01 ± 4.30 nm for r-Pc-LDL-FA) compared with native LDL (20.00 ± 2.70 nm).
Attaching multiple copies of FA to the Lys residues was critical because it is known that 20% side-chain alkylation (by reductive methylation) of the Lys residues of apoB-100 is required to abolish the binding of these modified LDL particles to the LDLR. Thus, attachment of 170 FA molecules per LDL particle to both DiI-LDL-FA and r-Pc-LDL-FA, which is equivalent to ≈50% modification of total Lys residues, should effectively block LDLR binding. Moreover, because FA is known for its ability to direct macromolecules (≤60 nm) to cancer cells via FR (33), it is anticipated that FA can also direct both LBNP nanoparticles to FR-expressing cancer cells.
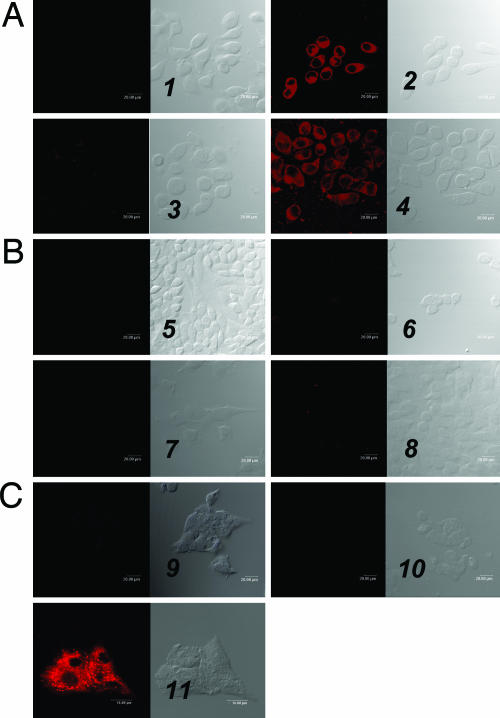
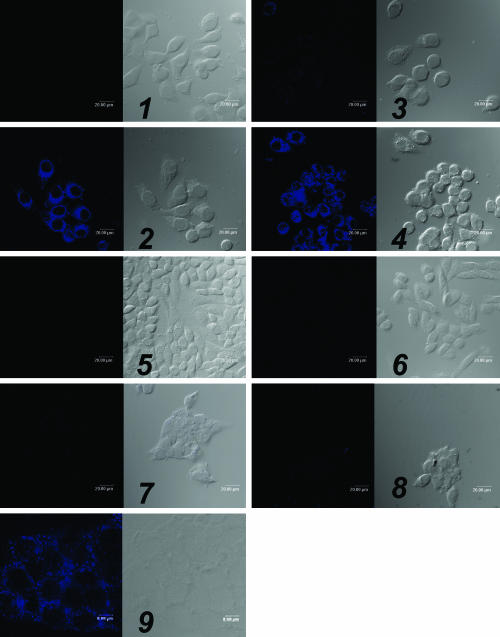
To validate the LDL rerouting strategy, confocal microscopy and flow cytometry studies were performed. First, FR-mediated uptake of DiI-LDL-FA was tested in KB cells (FR+). As expected, confocal images showed strong accumulation of DiI-LDL-FA throughout the whole cell except for the nucleus (Fig. 4A2). To determine the FR-specificity of DiI-LDL-FA, three sets of control experiments were performed yielding the following data: (i) An excess of free FA, the native ligand of FR, completely blocked the uptake of DiI-LDL-FA by FR+ KB cells (Fig. 4A3), whereas an excess of native LDL had no effect on DiI-LDL-FA uptake (Fig. 4A4); (ii) no DiI-LDL-FA uptake was observed in both FR- CHO and HT-1080 cells (Fig. 4 B6 and B8); and (iii) in sharp contrast to the strong accumulation of DiI-LDL in HepG2 cells (LDLR+) (Fig. 4C11), no accumulation of DiI-LDL-FA was observed in HepG2 cells (Fig. 4C10), indicating blockage of DiI-LDL-FA binding to LDLR.
Fig. 4.
Confocal microscopy studies. The numbers refer to each pair of images, with the fluorescence image (Left) and the bright field image (Right) shown. (A) DiI-LDL-FA uptake by the FR+ cell line, which shows KB cells alone (1), KB cells + 0.38 μM DiI-LDL-FA (2), KB cells + 0.38 μM DiI-LDL-FA + the 250-fold excess of free FA (3), and cells + 0.38 μM DiI-LDL-FA + the 50-fold excess of native LDL (4). (B) DiI-LDL-FA uptake by the FR- cell line, which shows CHO cells alone (5), CHO cells + 0.38 μM DiI-LDL-FA (6), HT-1080 cells alone (7), and HT-1080 cells + 0.38 μM DiI-LDL-FA (8). (C) DiI-LDL-FA uptake by HepG2 cells (LDLR+), which shows HepG2 cells alone (9), HepG2 cells + 0.38 μM DiI-LDL-FA (10), and HepG2 cells + 0.04 μM DiI-LDL (11).
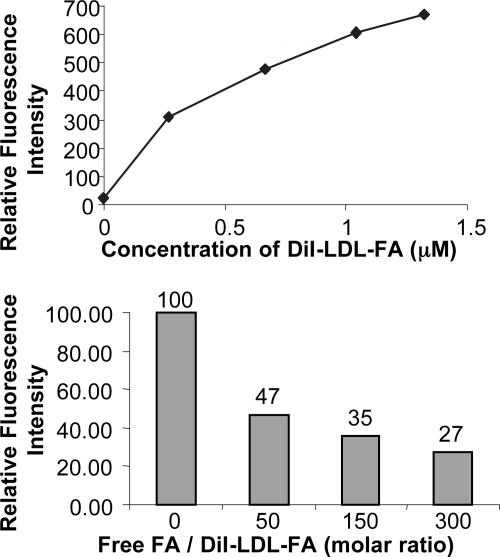
To evaluate the binding specificity of DiI-LDL-FA for FR, flow cytometric assays were performed on the KB cells (FR+). The results demonstrated both a concentration-dependent uptake of DiI-LDL-FA (Fig. 5 Upper) and a concentration-dependent inhibition of DiI-LDL-FA uptake by free FA (Fig. 5 Lower). Taken together, these data indicate that DiI-LDL-FA uptake in KB cells (FR+) is FR-specific, and FA conjugation reroutes LDL from LDLR to FR.
Fig. 5.
Flow cytometric study of DiI-LDL-FA in KB cells provides two lines of quantitative evidence of the FR-mediated uptake mechanism: the concentration dependence of DiI-LDL-FA uptake in KB cells (Upper) and the concentration-dependent inhibition of DiI-LDL-FA uptake in KB cells by free FA (Lower).
Similarly, FR-specific uptake of r-Pc-LDL-FA by KB cells was also confirmed. Briefly, r-Pc-LDL-FA emitted strong fluorescence in FR+ cells (KB) (Fig. 6, 2) but not in FR- cells (CHO) (Fig. 6, 6). This fluorescent signal was inhibited by free FA (Fig. 6, 3) but not by native LDL (Fig. 6, 4). Although r-Pc-LDL produced strong fluorescence in LDLR+ cells (HepG2) (Fig. 6, 9), incorporating multiple FA moieties in the LDL led to a diminished fluorescence signal in these cells (Fig. 6, 8). These data demonstrate that FA conjugation reroutes r-Pc-LDL-FA from LDLR to FR.
Fig. 6.
Confocal microscopy study of r-Pc-LDL-FA Uptake in KB (FR+), CHO (FR-), and HepG2 cells (LDLR+). The numbers refer to each pair of images, with the fluorescence image (Left) and the bright field image (Right) shown. (1) KB cells alone. (2) KB cells + 0.58 μM r-Pc-LDL-FA. (3) KB cells + 0.58 μM r-Pc-LDL-FA + the 250-fold excess of free FA. (4) KB cells + 0.58 μM r-Pc-LDL-FA + the 25-fold excess of native LDL. (5) CHO cells alone. (6) CHO cells + 0.58 μM r-Pc-LDL-FA. (7) HepG2 cells alone. (8) HepG2 cells + 0.58 μM r-Pc-LDL-FA. (9) HepG2 cells + 0.29 μM r-Pc-LDL. Similar to what was observed for DiI-LDL-FA, these images confirm the rerouting of r-Pc-LDL-FA from LDLR to FR.
To validate the LBNP concept in vivo, preliminary imaging studies of the near-infrared, dye-labeled LDL-FA were performed on a nude mouse bearing KB and HT-1080 tumors on its opposite flanks. Selective accumulation of LBNP in the KB tumor compared with the HT-1080 tumor was observed (data not shown). Further detailed in vivo studies are needed.
Discussion
This study demonstrates that alkylation of the Lys side-chain amino groups of apoB-100 with FA groups can be used to block delivery of LDL to its natural receptor and reroute this protein to the FR. Presumably, a similar strategy could be used to reroute other lipoprotein conjugates to tumor-specific or other disease-specific receptors.
Mechanism Underlying Selective Alkylation of Lys ε-Amino Groups. ApoB-100 of LDL is among the largest proteins known, with 4,536 amino acids and a molecular mass of 550 kDa. Two clusters of basic amino acids (site A, residues 3147-3157; site B, 3359-3367) appear to be involved in binding to anionic sites on the receptors (40). There is a general consensus that site B is involved in binding, but some controversy exists over the role of site A (41). The highly conserved B site has a striking homology to the receptor-binding site of apoE, a much smaller apolipoprotein (299 residues) in which the structure and interactions with the LDLR are currently being defined by physicochemical methods (42-46).
The sequences of these putative receptor-binding sites suggest that the anomalously low pKa value of the active Lys residues at the receptor-binding site can be attributed to their proximity to nearby cationic Lys and Arg side chains in which positive charges facilitate proton release from the Lys ε-amino groups. A decrease in the local dielectric constant could also lower the pKa value of the Lys side chains, but this mechanism seems unlikely in view of the surface exposure of these residues. Similar electrostatic effects are believed to be responsible for the low pKa value of the other active Lys residues of apoB-100. Both apoB-100 and apoE bind to the same receptor site on LDLR. Recombinant LDL retained normal receptor binding when site B of apoB-100 was replaced by the receptor-binding sequence of apoE (47). Electrostatic interactions with anionic domains on the receptor appear to be essential for binding to the LDLR (48, 49). These clusters of cationic side chains on both apoB-100 and apoE can be blocked by binding heparin (28, 47), which could provide another pathway for blocking LDLR binding. It is also noteworthy that familial defective apoB-100, one of the most common genetic abnormalities known, is caused by a mutation of apoB-100 (R3500Q) that does not involve a cationic cluster but is in proximity to site B (50). This site appears to form a salt-bridge to the C-terminal group that blocks access of the receptor to site B when Arg 3500 is replaced by glutamine (Q) or tryptophan (W) (47, 51). The interaction with the C-terminal group seems stereospecific because the replacement of Arg by Lys did not restore receptor binding and Arg is highly conserved at this sequence position over many species (47). In contrast, familial hypercholesteremia is caused by mutations in LDLR.
Potential Applications of Directed LBNPs. The lipoproteins offer a number of key advantages as natural nanoparticle platforms: they are completely biodegradable and nonimmunogenic; they come in various sizes and densities; and they contain components that can be chemically modified or to which drugs or diagnostic agents can readily be attached by physical or chemical manipulation. The key limitation of lipoproteins is that they are targeted to lipoprotein receptors that are systemically distributed among various organs and tissues. Consequently, specificity for diseased cells or specific tissues is difficult to achieve. In this article, we have presented evidence that this limitation can be overcome by blocking the lipoprotein receptor by preferential alkylation of active Lys side chains at the receptor-binding site. Alternatively, receptor binding might be capped or blocked by coordination to a suitable inhibitor, like heparin in the case of LDL or apoE (37), although such capping might only provide transient blockage of normal receptor binding. The particles can be rerouted to alternate receptors by attachment of appropriate targeting molecules. We have demonstrated this principle with folate, but examples of targeting somatostatin (25), Her2/neu (29, 52), ΔEGF (24), αvβ3 integrins (27), etc. have been reported in the literature and should be applicable to lipoproteins.
As noted above, the LBNPs have been used for delivery of near-infrared fluorophores (35, 53, 54), photodynamic therapy agents (32, 34), MRI agents (ref. 55; I. Corbin, H.L., J.C., J.D.G., and G.Z., unpublished data), and PET/SPECT agents (18, 21) to tumor cells that overexpress LDLR, and their binding was monitored by noninvasive imaging methods. This approach could be extended to ultrasound and bioluminescent probes as well as quantum dots. To date, MRI detection of these particles has been accomplished only with Gd chelates with lipophilic appendages that intercalate into the phospholipid monolayer (ref. 55; I. Corbin, H.L., J.C., J.D.G., and G.Z., unpublished data). Bifunctional Gd chelates containing fluorescent dyes that facilitate in vitro tracking by confocal microscopy have also been developed (55). A major limitation to such agents is the limited number of Gd chelates that can be attached to the phospholipid monolayer; thus, the relaxation enhancement provided by these agents is limited. Incorporation of ferromagnetic particles into lipoproteins might be advantageous and needs to be explored.
Potential Problems with Rerouted LBNPs. During this study, rerouting of LDL to FR has been achieved by alkylation of ≈50% of the exposed Lys side chains. Nevertheless, such high levels of FA labeling could compromise one of the key advantages of the LBNPs, their immunocompatibility. If immunogenicity is encountered, it could be minimized by lowering the extent of FA conjugation and/or by PEGylating the particle. Higher levels of alkylation of apoB-100 could also interfere with receptor binding, which might be recovered through the use of appropriate linkers. Future studies must be directed toward determining the effect of Lys side-chain alkylation on lipoprotein immunogenicity, delineation of the minimal degree of alkylation required for rerouting of the particles, and the effect of multiple targeting ligands on the affinity of LBNPs for cell surface receptors.
The amount of folate required to achieve a desired fraction of alkylated Lys side chains can be determined empirically by measuring LDL-FA absorbance and performing a protein assay by Lowry's method (39), but the relationship between the extent of folate conjugation and receptor affinity and the rate of endocytosis has yet to be determined. This relationship will have to be determined for each potential targeting group for various receptors.
Modified LDL may bind to the remnant receptor in the liver (56). The extent of such binding will have to be determined because it will compete with cell-surface receptors (e.g., FR) for binding of modified LDL particles.
Extension of Platform to Other Lipoproteins. There are a number of reasons why other lipoproteins, particularly HDL may be preferable to LDL for targeted delivery of diagnostic and therapeutic agents. The smaller size of HDL (7-12 nm) compared with LDL (≈22 nm) should make it more permeable to vascular membranes. In addition, HDL can be prepared from recombinant proteins and synthetic lipids (12) and is well tolerated by humans (57). The principal apolipoprotein of human HDL, ApoA-I, contains 243 amino acid residues in a single polypeptide chain that is ≈55% and 75% α-helical in the lipid-free and lipid-associated states, respectively. There are 21 Lys residues with one additional N-terminal Lys. The majority (86%, 18 of 21) of Lys residues in lipid-free apoA-I seem highly exposed to the solvent and, as such, exhibit a normal pKa value of 9.9-10.4. Once apoA-I is bound to the lipid, however, only 57% (12 of 21) to 67% (14 of 21) of Lys residues have a normal pKa value; the rest are active Lys residues. Thus, selective alkylation of Lys ε-amino groups on ApoA-I is possible and may be used to reroute HDL to selected disease-specific receptors. Also, HDL does not undergo endocytosis as LDL does but transfers cholesterol and lipids to the target cell after attachment to cell surface receptors (58, 59).
In summary, LBNPs provide a highly versatile biocompatible natural platform for diagnosis and therapy of cancer and various diseases. The feasibility of rerouting these particles to receptors other than their normal receptors has been demonstrated with LDL by targeting this lipoprotein to FRs instead of to LDLR. This principle should be applicable to other lipoproteins, particularly HDL-based particles. Potential applications of these agents include using them for delivering drugs to specific tumors and combining them with various imaging modalities for cancer diagnosis, for monitoring gene expression, and for stem cell tracking.
Acknowledgments
We thank Dr. Britton Chance (University of Pennsylvania) and Dr. Sissel Lund-Katz (Children's Hospital of Philadelphia) for insightful discussions. This work was supported by National Institutes of Health Grants R21-CA114463 (to G.Z.), N01-CO37119 (to G.Z.), and R24-CA83105 (to J.D.G.).
Author contributions: G.Z. and J.D.G. designed research; J.C. and H.L. performed research; J.C. and H.L. contributed new reagents/analytic tools; G.Z., J.C., and H.L. analyzed data; and G.Z. and J.D.G. wrote the paper.
Conflict of interest statement: While the manuscript was under review, the technology described in this study was licensed to Marillion Pharmaceuticals.
Abbreviations: LDL, low-density lipoprotein; HDL, high-density lipoprotein; LDLR, LDL receptor; apoB, apolipoprotein B; FA, folic acid; LBNP, lipoprotein-based nanoplatform; FR, folate receptor; SiPc-BOA, tetra-t-butyl-silicon phthalocyanine bisoleate; DiI, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine; NHS, N-hydroxysuccinimide; LDL-FA, folate-conjugated LDL; FA-NHS, folate-N-hydroxysuccinimide ester; r-Pc-LDL, LDL core-loading of SiPc-BOA; DiI-LDL, LDL surface-loading of DiI; DiI-LDL-FA, folate-conjugated DiI-LDL; r-Pc-LDL-FA, folate-conjugated r-Pc-LDL.
References
- 1.Quintana, A., Raczka, E., Piehler, L., Lee, I., Myc, A., Majoros, I., Patri, A. K., Thomas, T., Mule, J. & Baker, J. R., Jr. (2002) Pharm. Res. 19, 1310-1316. [DOI] [PubMed] [Google Scholar]
- 2.Roy, I., Ohulchanskyy, T. Y., Pudavar, H. E., Bergey, E. J., Oseroff, A. R., Morgan, J., Dougherty, T. J. & Prasad, P. N. (2003) J. Am. Chem. Soc. 125, 7860-7865. [DOI] [PubMed] [Google Scholar]
- 3.Xu, H., Aylott J. W. & Kopelman R. (2002) Analyst 127, 1471-1477. [DOI] [PubMed] [Google Scholar]
- 4.Pan, D., Turner, J. L. & Wooley, K. L. (2003) Chem. Commun., 2400-2401. [DOI] [PubMed]
- 5.Kasili, P. M., Song, J. M. & Vo-Dinh, T. (2004) J. Am. Chem. Soc. 126, 2799-2806. [DOI] [PubMed] [Google Scholar]
- 6.Ruoslahti, E. (2002) Cancer Cell 2, 97-98. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, S. A., Rader, R. K., Westlin, W. F., Null, C., Jackson, D., Lanza, G. M., Wickline, S. A. & Kotyk, J. J. (2000) Magn. Reson. Med. 44, 433-439. [DOI] [PubMed] [Google Scholar]
- 8.Hood, J. D., Bednarski, M., Frausto, R., Guccione, S., Reisfeld, R. A., Xiang, R. & Cheresh, D. A. (2002) Science 296, 2404-2407. [DOI] [PubMed] [Google Scholar]
- 9.Li, L., Wartchow, C. A., Danthi, S. N., Shen, Z., Dechene, N., Pease, J., Choi, H. S., Doede, T., Chu, P., Ning, S., et al. (2004) Int. J. Radiat. Oncol. Biol. Phys. 58, 1215-1227. [DOI] [PubMed] [Google Scholar]
- 10.Weissleder, R., Moore, A., Mahmood, U., Bhorade, R., Benveniste, H., Chiocca, E. A. & Basilion, J. P. (2000) Nat. Med. 6, 351-355. [DOI] [PubMed] [Google Scholar]
- 11.Gao, X., Cui, Y., Levenson, R. M., Chung, L. W. & Nie, S. (2004) Nat. Biotechnol. 22, 969-976. [DOI] [PubMed] [Google Scholar]
- 12.Rensen, P. C., de Vrueh, R. L., Kuiper, J., Bijsterbosch, M. K., Biessen, E. A. & van Berkel, T. J. (2001) Adv. Drug Deliv. Rev. 47, 251-276. [DOI] [PubMed] [Google Scholar]
- 13.Shaw, J. M., Shaw, K. V., Yanovich, S., Iwanik, M., Futch, W. S., Rosowsky, A. & Schook, L. B. (1987) Ann. N.Y. Acad. Sci. 507, 252-271. [DOI] [PubMed] [Google Scholar]
- 14.Versluis, A. J., van Geel, P. J., Oppelaar, H., van Berkel, T. J. & Bijsterbosch, M. K. (1996) Br. J. Cancer 74, 525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masquelier, M., Vitols, S. & Peterson, C. (1986) Cancer Res. 46, 3842-3847. [PubMed] [Google Scholar]
- 16.Firestone, R. A., Pisano, J. M., Falck, J. R., McPhaul, M. M. & Krieger M. (1984) J. Med. Chem. 27, 1037-1043. [DOI] [PubMed] [Google Scholar]
- 17.Samadi-Baboli, M., Favre, G., Canal, P. & Soula, G. (1993) Br. J. Cancer 68, 319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urizzi, P., Souchard, J. P., Palevody, C., Ratovo, G., Hollande, E. & Nepveu, F. (1997) Int. J. Cancer 70, 315-322. [DOI] [PubMed] [Google Scholar]
- 19.Krieger, M. (1986) Methods Enzymol. 128, 608-613. [DOI] [PubMed] [Google Scholar]
- 20.Krieger, M., Goldstein, J. L. & Brown, M. S. (1978) Proc. Natl. Acad. Sci. USA 75, 5052-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moerlein, S. M., Daugherty, A., Sobel, B. E. & Welch, M. J. (1991) J. Nucl. Med. 32, 300-307. [PubMed] [Google Scholar]
- 22.Bijsterbosch, M. K. & Van Berkel, T. J. (1990) Adv. Drug Deliv. Rev. 5, 231-251. [DOI] [PubMed] [Google Scholar]
- 23.Slamon, D. J., Godolphin, W., Jones, L. A., Holt, J. A., Wong, S. G., Keith, D. E., Levin, W. J., Stuart, S. G., Udove, J., Ullrich, A., et al. (1989) Science 244, 707-712. [DOI] [PubMed] [Google Scholar]
- 24.Wong, A. J., Ruppert, J. M., Bigner, S. H., Grzeschik, C. H., Humphrey, P. A., Bigner, D. S. & Vogelstein, B. (1992) Proc. Natl. Acad. Sci. USA 89, 2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker, A., Hessenius, C., Licha, K., Ebert, B., Sukowski, U., Semmler, W., Wiedenmann, B. & Grotzinger, C. (2001) Nat. Biotechnol. 19, 327-331. [DOI] [PubMed] [Google Scholar]
- 26.Leamon, C. P. & Low, P. S. (1991) Proc. Natl. Acad. Sci. USA 88, 5572-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruoslahti, E. (2002) Nat. Rev. Cancer 2, 83-90. [DOI] [PubMed] [Google Scholar]
- 28.Lund-Katz, S., Ibdah, J. A., Letizia, J. Y., Thomas, M. T. & Phillips, M. C. (1988) J. Biol. Chem. 263, 13831-13838. [PubMed] [Google Scholar]
- 29.Park, J. W., Kirpotin, D. B., Hong, K., Shalaby, R., Shao, Y., Nielsen, U. B., Marks, J. D., Papahadjopoulos, D. & Benz, C. C. (2001) J. Controlled Release 74, 95-113. [DOI] [PubMed] [Google Scholar]
- 30.Stevens, P. J., Sekido, M. & Lee, R. J. (2004) Pharm. Res. 21, 2153-2157. [DOI] [PubMed] [Google Scholar]
- 31.Moon, W. K., Lin, Y., O'Loughlin, T., Tang, Y., Kim, D. E., Weissleder, R. & Tung, C. H. (2003) Bioconjugate Chem. 14, 539-545. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, G., Li, H., Zhang, M., Lund-Katz, S., Chance, B. & Glickson, J. D. (2002) Bioconjugate Chem. 13, 392-396. [DOI] [PubMed] [Google Scholar]
- 33.Lee, R. J. & Low, P. S. (1994) J. Biol. Chem. 269, 3198-3204. [PubMed] [Google Scholar]
- 34.Li, H., Marotta, D., Kim, S. K., Busch, T. M., Wileyto, E. P. & Zheng, G. (2005) J. Biomed. Opt 10, 41203. [DOI] [PubMed] [Google Scholar]
- 35.Li, H., Zhang, Z., Blessington, D., Nelson, D. S., Zhou, R., Lund-Katz, S., Chance, B., Glickson, J. D. & Zheng, G. (2004) Acad. Radiol. 11, 669-677. [DOI] [PubMed] [Google Scholar]
- 36.Egorin, M. J., Zuhowski, E. G., Sentz, D. L., Dobson, J. M., Callery, P. S. & Eiseman, J. L. (1999) Cancer Chemother. Pharmacol. 44, 283-294. [DOI] [PubMed] [Google Scholar]
- 37.Lund-Katz, S., Laplaud, P. M., Phillips, M. C. & Chapman, M. J. (1998) Biochemistry 37, 12867-12874. [DOI] [PubMed] [Google Scholar]
- 38.Pitas, R. E., Boyles, J., Mahley, R. W. & Bissell, D. M. (1985) J. Cell Biol. 100, 103-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. (1951) J. Biol. Chem. 193, 265-275. [PubMed] [Google Scholar]
- 40.Innerarity, T. L., Weisgraber, K. H., Rall, S. C., Jr., & Mahley, R. W. (1987) Acta Med. Scand. Suppl. 715, 51-59. [DOI] [PubMed] [Google Scholar]
- 41.Law, A. & Scott, J. (1990) J. Lipid Res. 31, 1109-1120. [PubMed] [Google Scholar]
- 42.Raussens, V., Slupsky, C. M., Ryan, R. O. & Sykes, B. D. (2002) J. Biol. Chem. 277, 29172-29180. [DOI] [PubMed] [Google Scholar]
- 43.Ryan, R. O. (1996) Biochem. Cell Biol. 74, 155-164. [DOI] [PubMed] [Google Scholar]
- 44.Fan, D., Korando, L. A., Dothager, R. S., Li, Q. & Wang, J. (2004) J. Biomol. NMR 29, 419-420 (lett.). [DOI] [PubMed] [Google Scholar]
- 45.Raussens, V., Fisher, C. A., Goormaghtigh, E., Ryan, R. O. & Ruysschaert, J. M. (1998) J. Biol. Chem. 273, 25825-25830. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, C., Wardell, M. R., Weisgraber, K. H., Mahley, R. W. & Agard, D. A. (1991) Science 252, 1817-1822. [DOI] [PubMed] [Google Scholar]
- 47.Boren, J., Lee, I., Zhu, W., Arnold, K., Taylor, S. & Innerarity, T. L. (1998) J. Clin. Invest. 101, 1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstein, J. L., Brown, M. S., Anderson, R. G., Russell, D. W. & Schneider, W. J. (1985) Annu. Rev. Cell Biol. 1, 1-39. [DOI] [PubMed] [Google Scholar]
- 49.Knott, T. J., Rall, S. C., Jr., Innerarity, T. L., Jacobson, S. F., Urdea, M. S., Levy-Wilson, B., Powell, L. M., Pease, R. J., Eddy, R., Nakai, H., et al. (1985) Science 230, 37-43. [DOI] [PubMed] [Google Scholar]
- 50.Innerarity, T. L., Weisgraber, K. H., Arnold, K. S., Mahley, R. W., Krauss, R. M., Vega, G. L. & Grundy, S. M. (1987) Proc. Natl. Acad. Sci. USA 84, 6919-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boren, J., Ekstrom, U., Agren, B., Nilsson-Ehle, P. & Innerarity, T. L. (2001) J. Biol. Chem. 276, 9214-9218. [DOI] [PubMed] [Google Scholar]
- 52.Park, B. W., Zhang, H. T., Wu, C., Berezov, A., Zhang, X., Dua, R., Wang, Q., Kao, G., O'Rourke, D. M., Greene, M. I. & Murali, R. (2000) Nat. Biotechnol. 18, 194-198. [DOI] [PubMed] [Google Scholar]
- 53.Zheng, G., Li, H., Yang, K., Blessington, D., Licha, K., Lund-Katz, S., Chance, B. & Glickson, J. D. (2002) Bioorg. Med. Chem. Lett. 12, 1485-1488. [DOI] [PubMed] [Google Scholar]
- 54.Wu, S. P., Lee, I., Ghoroghchian, P. P., Frail, P. R., Zheng, G., Glickson, J. D. & Therien, M. J. (2005) Bioconjugate Chem. 16, 542-550. [DOI] [PubMed] [Google Scholar]
- 55.Li, H., Gray, B. D., Corbin, I., Lebherz, C., Choi, H., Lund-Katz, S., Wilson, J. M., Glickson, J. D. & Zhou, R. (2004) Acad. Radiol. 11, 1251-1259. [DOI] [PubMed] [Google Scholar]
- 56.Brown, M. S., Basu, S. K., Falck, J. R., Ho, Y. K. & Goldstein, J. L. (1980) J. Supramol. Struct. 13, 67-81. [DOI] [PubMed] [Google Scholar]
- 57.Pajkrt, D., Doran, J. E., Koster, F., Lerch, P. G., Arnet, B., van der Poll, T., ten Cate, J. W. & van Deventer, S. J. (1996) J. Exp. Med. 184, 1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lund-Katz, S., Liu, L., Thuahnai, S. T. & Phillips, M. C. (2003) Front. Biosci. 8, d1044-d1054. [DOI] [PubMed] [Google Scholar]
- 59.Acton, S., Rigotti, A., Landschulz, K. T., Xu, S., Hobbs, H. H. & Krieger, M. (1996) Science 271, 518-520. [DOI] [PubMed] [Google Scholar]