Abstract
Nuclear transfer (NT) has potential applications in agriculture and biomedicine, but the technology is hindered by low efficiency. Global gene expression analysis of clones is important for the comprehensive study of nuclear reprogramming. Here, we compared global gene expression profiles of individual bovine NT blastocysts with their somatic donor cells and fertilized control embryos using cDNA microarray technology. The NT embryos' gene expression profiles were drastically different from those of their donor cells and closely resembled those of the naturally fertilized embryos. Our findings demonstrate that the NT embryos have undergone significant nuclear reprogramming by the blastocyst stage; however, problems may occur during redifferentiation for tissue genesis and organogenesis, and small reprogramming errors may be magnified downstream in development.
Keywords: bovine, embryo, microarray, nuclear transfer
Since its advent in 1997 (1), nuclear transfer (NT), or cloning using differentiated somatic cells from adult mammals, has been achieved for a number of species. However, the technology is extremely inefficient, with many abnormalities leading to high pregnancy loss and neonatal death (2). These problems are hypothesized to result from incomplete nuclear reprogramming, the process of reversing a differentiated somatic nucleus to a totipotent embryonic state after NT. In support of this hypothesis, several studies have shown abnormally high levels of DNA methylation, as well as aberrant gene expression, in bovine NT embryos (3–8). Previous gene expression studies used methods that analyzed only a handful of genes from a single NT embryo; therefore, the extent of global nuclear reprogramming at early embryonic stages has yet to be ascertained (6–8). Here, we used microarray technology in conjunction with linear amplification to analyze the global gene expression of individual NT embryos. We examined the gene expression profiles of NT embryos and compared them with their donor fibroblast cells and expression profiles of preimplantation control embryos created by natural fertilization in vivo by artificial insemination (AI). Additionally, in vitro fertilized (IVF) embryos were included as in vitro controls and because IVF embryos are commonly used in the literature.
Methods
Detailed methods are shown in Supporting Text, which is published as supporting information on the PNAS web site.
Microarray Design and Annotation. Development of the 7,872 cattle cDNA microarray was done at the University of Illinois at Urbana–Champaign as described by Everts et al. (9). The 7,872 cDNA microarray consisted of the original 3,800 cDNA microarray (10) and was supplemented with sequences selected from normalized and subtracted placenta and spleen cDNA libraries. In total, the 7,872 cattle cDNA microarray potentially contains 6,298 unique sequences (5,325 unique UniGene hits and 973 putative novel genes or divergent orthologs).
Generation of NT, IVF, and AI Embryos. IVF and NT were based on Kubota et al. (11, 12). In this study, cultured skin fibroblast cells from an adult Holstein cow, which have led to clone term development in our laboratory (13), were used for NT. Fifteen NT embryos (35% blastocyst rate) from two different NT batches generated from the same fibroblast cell line were used for microarray analysis. Likewise, 15 IVF embryos (37% blastocyst rate) were produced under the same culture conditions (11) for comparison. For the NT and IVF embryos, after development to blastocyst stage (day 7), International Embryo Transfer Society quality grade 1 embryos were frozen and thawed according to a previously described vitrification protocol (14). Additionally, cryopreserved day 7 AI embryos from two donor cows/sires, 11 and 3 embryos, respectively, served as in vivo controls. Only Holstein embryos were used.
Linear Amplification. The linear amplification of the individual embryos and the donor cells was done as described by Baugh et al. (15) for 2 ng of total RNA.
Labeling and Microarray Hybridizations. The aminoallyl labeling and hybridization protocol was based on one developed by The Institute for Genomic Research (16). One microgram of amplified RNA (aRNA) was reverse-transcribed, labeled, and hybridized to each microarray. In total, 96 microarrays, all embryos and nuclear donor cells with dye-swap, were analyzed. On average across all of the embryos, the correlation coefficient between the Cy3 and Cy5 replicates was >0.90. All embryos were hybridized versus a standard reference, comprised of total RNA isolated from BL3°, MDBK, and EBTR cell lines and brain tissue from an Angus heifer. This reference design compares embryo profiles based on the expression of each gene in the embryo versus the standard reference. The background and standard deviation were calculated for each raw data file after scanning (genepix pro 4.0, Axon Instruments, Union City, CA), and only those spots with intensities three standard deviations above background were considered “expressed” and loaded into genespring 6.1 (Agilent Technologies, Palo Alto, CA). Loess normalization was applied to all microarrays before statistical comparison of samples (17). Genes present in either the standard reference or sample on 90% of the microarrays underwent further analysis.
Data Analysis. Both genespring 6.1 and our independent analysis using sas (SAS Institute, Cary, NC) statistical software used one-way ANOVA to determine gene expression differences, and there was a high degree of overlap between the two methods. A two-stage modeling approach, as implemented by Wolfinger et al. (18), was used. The first-stage model, fitted to the log-transformed and normalized ratio of embryo and standard reference samples across all genes, included the effects of dye, array, and array x dye interaction. The residuals were analyzed by gene in a second-stage model that included the fixed effects of dye and condition and the random effects of array. The heterogeneity of variance model provided a significantly better fit than the homogeneity of variance model for a large number of genes and was used for the analyses. Probability (P) values were adjusted for multiple comparisons using the false discovery rate approach. In addition to these statistical criteria, those genes that differed by ≥2-fold were considered to be differentially expressed.
Quantitative Real-Time RT-PCR. primer express software (Applied Biosystems) was used for primer and 5′ FAM-3′ TAMRA-labeled probe design. aRNAs from eight randomly selected NT embryos, eight AI embryos (except for two genes, DNAJC10 and HRH1, where 14 embryos were analyzed), and the donor cells were used. The relative standard curve method was used for quantification (Applied Biosystems Prism SDS 7700 User Bulletin #2) and was comprised of pooled aRNA from each sample tested. The same standard reference aRNA that was used in the microarray analysis was used as the calibrator sample. After relative quantities were determined, the ratios of all samples and the standard references were calculated, and the mean for each group was determined and compared for an overall fold change.
Results and Discussion
Linear amplification enabled production of sufficient amounts of aRNA for the microarray experiments and has been shown to reliably amplify the initial mRNA population (15, 19–21). Approximately 5 ng of total RNA can be isolated from a blastocyst-stage embryo (22); therefore, it was necessary to carry out multiple rounds of amplification. To test the reproducibility and fidelity of the amplification procedure, a validation experiment was conducted by using RNA from the kidney tissue of a newborn calf. Two replicates of the bovine kidney RNA were amplified and hybridized to the microarray. Comparisons were made between aRNA (rounds 1, 2, and 3) and unamplified RNA. Genes whose expression was detected with Cy3 and/or Cy5 in 90% of the microarrays were used for analysis. The correlation coefficient between the replicates of the unamplifed RNA was 0.98. Similarly, the correlation coefficient after three rounds of amplification was 0.94. One-way ANOVA identified differences in gene expression between amplified and unamplified samples. False discovery rate-corrected P values <0.05 and ≥2-fold were used as criteria for differential expression, n = 2,611 genes. Only one gene of the 2,611 genes analyzed was identified as differentially expressed between unamplified and samples subjected to three rounds of amplification. Thus, the linear amplification protocol used for the study of single bovine embryos accurately reflected the abundance of RNA in native samples and was highly reproducible.
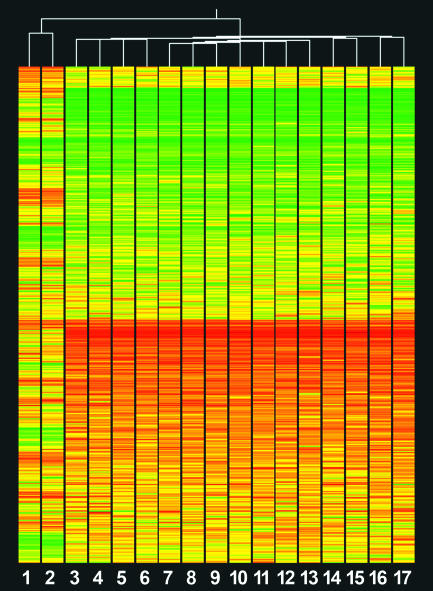
Gene Expression Profile Comparison of Donor Cells and NT Embryos. To examine the extent of nuclear reprogramming of the NT blastocyst-stage embryos, we compared their expression patterns with those of the donor somatic cells used for NT. Hierarchical clustering of 5,356 genes expressed in samples and/or the standard reference uncovered a difference of 84.2% between the expression profiles of the NT embryos and their donor cells (Fig. 1). A total of 1,546 genes were identified as differentially expressed (false discovery rate P value <0.05, ≥2-fold), representing 29% of all genes analyzed. Among these, 751 were up- and 795 down-regulated in the donor cells versus the NT embryos. The list of genes differentially expressed by at least 10-fold and the complete list of the 1,546 differentially expressed genes as well as their distribution in different Gene Ontology (GO) categories can be found in Tables 4 and 5 and Fig. 3, which are published as supporting information on the PNAS web site. Expression Analysis Systematic Explorer [National Institute of Allergy and Infectious Diseases (NIAID)/National Institutes of Health (NIH)] was used to find biological themes in the differentially expressed genes (23). GO categories that were the most significantly overrepresented (P < 10–5) in the genes up-regulated in the NT embryos were “mitochondrion,” “carrier activity,” “mitochondrial inner membrane,” “transporter activity,” “primary active transporter activity,” “RNA splicing” and “ion transporter activity.” Using keggcharts (24) (Database for Annotation, Visualization, and Integrated Discovery; NIAID/NIH), the following pathways were identified in the genes up-regulated in the NT embryos: oxidative phosphorylation, cell cycle, ATP synthesis, tricarboxylic acid cycle, purine metabolism, glycolysis/gluconeogenesis, pyruvate metabolism, and apoptosis.
Fig. 1.
Hierarchical cluster of 5,356 genes comparing donor cells (1–2) with NT embryos (3–17). Standard correlation reveals an 84.2% difference in gene expression profiles (top tree). Color indicates the normalized expression values (sample: standard reference ratios) for each gene examined. Red represents high expression in the sample compared with the standard reference; yellow, approximately equal expression; and green, low expression.
The inner cell mass of a blastocyst can generate totipotent embryonic stem (ES) cells that should display “stemness” characteristics. We analyzed the expression of 94 genes on the microarray known to be highly expressed in human and mouse ES cells as compared with other stem cell types (25–28). Twenty-three genes had significantly higher expression in the NT embryos than in the differentiated donor cells, including genes previously characterized as ES cell-specific: ODC1, PECAM1, and CCNE1 (29) (Table 1). Eight genes were significantly overrepresented by Expression Analysis Systematic Explorer analysis in GO categories “ATP binding” and/or “ATPase activity:” AK3, ATP5A1, CCT3, CCT8, EIF4A1, HSPA8, HSPA4, and HSPE1. Taken together with the dissimilarity of gene expression profiles of donor cells and NT embryos, our data indicate significant nuclear reprogramming of the donor cell nuclei after cloning at the blastocyst stage of embryo development.
Table 1. ES cell-enriched genes up-regulated in NT embryos compared with donor cells.
| Gene symbol | Sequence identifier | UniGene ID* | Fold Δ† |
|---|---|---|---|
| ODC1 | BF043697 | Hs.443409 | 20.1 |
| CCNE1 | BF045665 | Hs.244723 | 6.7 |
| HDAC1 | BF040519 | Hs.88556 | 5.7 |
| HSPE1 | BF039685 | Hs.1197 | 5.6 |
| AK3 | AW465803 | Hs.10862 | 4.6 |
| PRDX1 | BF046014 | Hs.180909 | 4.1 |
| SLC7A7 | AW463705 | Hs.194693 | 3.7 |
| PECAM1 | AW462978 | Hs.78146 | 3.5 |
| MGST2 | AW463844 | Hs.81874 | 3.4 |
| CCNC | BF045762 | Hs.435450 | 3.2 |
| ATP5A1 | BF039512 | Hs.298280 | 3.1 |
| DUSP16 | AW466062 | Hs.20281 | 3.1 |
| RPA3 | AW461938 | Hs.1608 | 2.9 |
| CCT3 | AW464746 | Hs.1708 | 2.9 |
| K-ALPHA-1 | AW463276 | Hs.446608 | 2.9 |
| TLE4 | BF042408 | Hs.494269 | 2.7 |
| HSPA8 | AW463634 | Hs.180414 | 2.5 |
| HSPA4 | BF040631 | Hs.90093 | 2.5 |
| HNRPA1 | AW464603 | Hs.356721 | 2.4 |
| ECT2 | AW461921 | Hs.293257 | 2.4 |
| CCT8 | BF040432 | Hs.416211 | 2.3 |
| EIF4A1 | AW464970 | Hs.129673 | 2.2 |
| HMGB1 | BF040515 | Hs.434102 | 2.2 |
Annotation using Human UniGene.
Fold change is expressed as the ratio of the normalized fluorescence intensity of the embryos:standard reference divided by the donor cells:standard reference.
Gene Expression Profile Comparison of NT and Fertilized Control Embryos. The gene expression profiles of the individual NT embryos (n = 15) were compared with those of naturally fertilized control embryos created by AI (n = 14) and IVF embryos (n = 15). Of the 5,174 genes used in this comparison, 3,490 were expressed at intensities of ≥100 above background in the AI, IVF, and NT embryos.
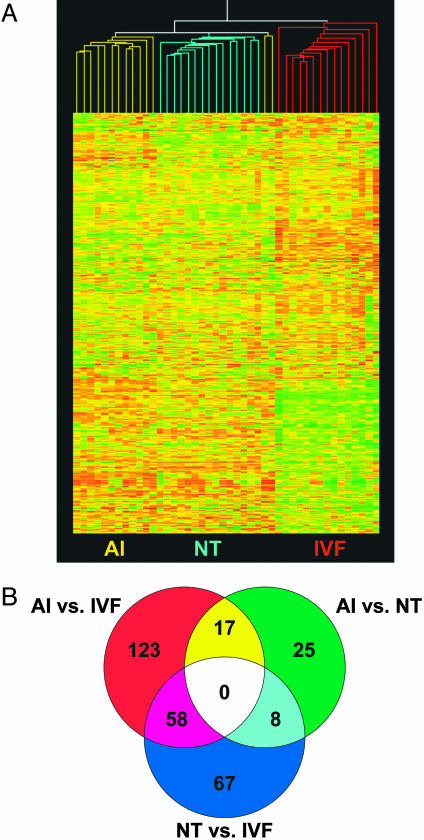
Hierarchical clustering of the three embryo types based on 5,174 analyzed genes revealed that the gene expression profiles of the NT and AI embryos were more similar than those of the IVF and AI embryos (Fig. 2A). The correlation coefficient between the NT and AI embryos was 0.808 but only 0.714 between the IVF and AI embryos. The correlation coefficient between NT and IVF embryos was 0.718, and the overall correlation coefficient across all three embryo types was 0.713. Interestingly, we observed appreciably less variation among the individual NT embryos, with the correlation coefficient being 0.838, as opposed to 0.733 observed among IVF embryos (correlation coefficient among the AI embryos was 0.812). The low variability exhibited among the NT embryos (similar to control AI embryos) suggests a relatively uniform reprogramming mechanism for the NT embryos by the blastocyst stage. It is unlikely that the variation observed among IVF embryos is due solely to in vitro culture, because the NT embryos were subjected to the same culture conditions yet did not display as variable expression profiles. The variation seen among the IVF embryos may be partially attributed to maternally inherited genetic variability, because the IVF embryos were derived from different cows (slaughterhouse ovaries).
Fig. 2.
Hierarchical cluster of AI, NT, and IVF embryos and characterization of differentially expressed genes. (A)(Inset) The 5,174-gene cluster arranged to highlight differential gene expression in IVF embryos (red) as compared with both AI (yellow) and NT embryos (blue). (B) Venn diagram characterizing differential gene expression between and specific to individual embryo types. Each circle represents the differential expression between two embryo types out of 5,174 analyzed genes. The circle (upper right) shows the 50 genes that are differentially expressed between AI and NT embryos; 25 genes (green) are uniquely expressed in AI vs. NT only, 17 genes (yellow) are specific to AI embryos, and 8 genes (aqua) are NT-specific.
Despite the similarity of expression profiles between the NT embryos and the naturally produced AI embryos, 50 of 5,174 analyzed genes were differentially expressed (versus 61 genes identified between the two groups of AI embryos). This value represents ≈1% of the total genes in this analysis. Of the 50 genes differentially expressed between the NT and AI embryos (Table 2), 25 were unique to this comparison. The remaining 25 genes also differed in AI-to-IVF or NT-to-IVF embryo comparisons. Conceivably, the differential expression of at least some of these genes could be credited to the fertilization in vivo of the naturally produced embryos. Among the remaining 25 genes, eight were uniquely expressed in the NT embryos; this could be a specific effect of nuclear reprogramming or the NT procedure. Likewise, 17 genes were uniquely expressed in the AI embryos. In summary, it is likely that many of the differentially expressed genes could be related to differential effects of the in vivo and in vitro environments (i.e., developmental timing, morphology, intracellular signaling, etc.) or genetic differences between the NT and AI embryos.
Table 2. Fifty genes differentially expressed between AI and NT embryos.
| Gene symbol | Sequence identifier | UniGene ID* | Fold Δ† |
|---|---|---|---|
| Unique genes up-regulated in AI embryos | |||
| FLJ11806 | AW466113 | Hs.323443 | 2.9 |
| CCL26 | BM365027 | Hs.131342 | 2.9 |
| MYO1D | BF039614 | Hs.39871 | 2.8 |
| GMPR | BF041701 | Hs.1435 | 2.8 |
| CPNE3 | AW464901 | Hs.14158 | 2.4 |
| TRIM38 | BM362887 | Hs.511746 | 2.0 |
| Down-regulated in AI embryos | |||
| AW464485 | Hs.240443 | 3.9 | |
| BF041274 | 3.7 | ||
| ATF3 | AW464633 | Hs.460 | 2.8 |
| ADRB2 | BF040456 | Hs.2551 | 2.5 |
| AKAP11 | BF045445 | Hs.414995 | 2.4 |
| CA12 | AW461641 | Hs.279916 | 2.2 |
| ATF4 | BF043185 | Hs.181243 | 2.1 |
| PPAP2A | BF041418 | Hs.482121 | 2.1 |
| ABCG2 | AW462521 | Hs.194720 | 2.1 |
| LOC339287 | AW464327 | Hs.350229 | 2.1 |
| KCNK1 | BF043113 | Hs.376874 | 2.0 |
| Unique genes up-regulated in NT embryos | |||
| PSPH | BF043883 | Hs.512656 | 5.1 |
| ZNRD1 | AW464801 | Hs.57813 | 2.2 |
| DDIT4 | AW462331 | Hs.111244 | 2.1 |
| HARSL | AW464854 | Hs.432560 | 2.1 |
| PIP5K1A | BF044038 | Hs.149255 | 2.1 |
| Down-regulated in NT embryos | |||
| DUSP6‡ | BF040804 | Hs.298654 | 2.8 |
| SNX10 | AW463500 | Hs.418132 | 2.2 |
| SCD | BF044532 | Hs.119597 | 2.1 |
| Unique genes up-regulated in AI embryos compared with NT embryos | |||
| ACOX1 | AW465214 | Hs.379991 | 2.9 |
| MEIS2 | BF040423 | Hs.362805 | 2.7 |
| SCAND1 | BF045856 | Hs.274411 | 2.7 |
| RNH | AW465255 | Hs.130958 | 2.6 |
| TFAP2A | BF039106 | Hs.210911 | 2.6 |
| AW463677 | Bt.9985 | 2.5 | |
| CD81 | AW465486 | Hs.54457 | 2.5 |
| FARP1 | BF043795 | Hs.207428 | 2.4 |
| FLJ23186 | BF046601 | Hs.434247 | 2.4 |
| MITF | BF040850 | Hs.166017 | 2.3 |
| IHPK2 | AW464276 | Hs.323432 | 2.1 |
| DUSP6 | BF041073 | Hs.298654 | 2.1 |
| PGA5 | AW465637 | Hs.432854 | 2.0 |
| KIAA0284 | AW462372 | Hs.182536 | 2.0 |
| CASP6 | BF040727 | Hs.3280 | 2.0 |
| COL4A1 | AW465560 | Hs.437173 | 2.0 |
| FOLR1 | BF044613 | Hs.73769 | 2.0 |
| CBR1 | AW461769 | Hs.88778 | 2.0 |
| Unique genes up-regulated in NT embryos compared with AI embryos | |||
| BF042216 | Hs.55987 | 2.9 | |
| FLJ20160 | BF045743 | Hs.418581 | 2.8 |
| LOC80298 | BF044934 | Hs.5009 | 2.6 |
| HRH1 | AW464562 | Hs.1570 | 2.5 |
| BF040178 | 2.5 | ||
| DNAJC10 | AW463952 | Hs.516632 | 2.3 |
| AW462843 | Bt.1173 | 2.2 | |
Some sequences do not have human UniGene IDs and are annotated with the Cattle UniGene names or are previously unannotated bovine sequences.
Fold change is expressed as the ratio of the normalized fluorescence intensity of the AI embryos:standard reference divided by the NT embryos: standard reference. The reciprocal values are presented if the ratio of the ratios was <0.5.
DUSP6 is present in two lists and could be a potential isoform or related gene.
The majority of the 50 differentially expressed genes were scattered on many different bovine chromosomes, but two of the four genes on chromosome 23 were linked. However, one (ZNRD1) was up-regulated in the NT embryos, whereas the other (TFAP2A) was down-regulated. Therefore, it is unlikely that the differential expression is due to aberrant regional chromatin remodeling.
Some of the 25 uniquely expressed genes between NT and AI embryos were identified in the literature as involved in development. For example, MITF is required for retina pigment epithelium specification in vertebrates (30), and TFAP2A is critical for neural tube, body wall, and cardiac development (31, 32). Likewise, MEIS2 and DUSP6 are involved in vertebrate limb development (33, 34). The homeobox gene MEIS2 serves as a determinant of proximo-distal limb identity (34). It is also found expressed during mouse placentation (35). Folate receptor 1 (FOLR1) is involved in maternal–fetal folate transport. A study using a mouse knockout of Folbp1 found that the embryos died in utero at embryonic day 10 and had neural tube defects (36). These results suggest that FOLR1 is essential for normal embryonic development. Collagen IV isoform α 1(COL4A1)-deficient mouse embryos exhibited growth retardation and developed up to embryonic day 9.5 (36). Additionally, COL4A1 was critical for basement membrane stability and integrity. Examination of deficient mouse placentas revealed aberrant development of the labyrinth layer and an abnormal deposition of cells forming a barrier between maternal and fetal blood pools. All of these genes were down-regulated in the NT embryos and could be involved in the abnormal development and mortality observed in NT fetuses.
There were a greater number of differentially expressed genes between AI and IVF embryos (198) and NT and IVF embryos (133) than between NT and AI embryos. Among these, 123 and 67 genes were uniquely expressed in the AI-to-IVF and NT-to-IVF embryo comparisons, respectively. Fifty-eight genes were uniquely expressed in the IVF embryos as compared with AI and NT embryos (Table 6, which is published as supporting information on the PNAS web site). The number of differentially expressed genes between and specific to the embryo types is depicted in Fig. 2B.
Many of the abnormalities in cloned animals suggest imprinting disruptions (38, 39). We examined the expression of 21 genes on the microarray that are imprinted in mice and/or humans (Imprinted Gene Catalog; ref. 40) and are putatively imprinted in cattle. Among these, 20 were similarly expressed in the AI, IVF, and NT embryos: CDKN1C, COPG2, CPA4, DCN, DLK1, GNAS, GRB10, H19, IGF2, IGF2R, L3MBTL, MEG3, MEST, NAP1L5, PEG10, PEG3, PLAGL1, SDHD, SGCE, and UBE3A. Only CD81, a gene imprinted in the mouse placenta (41), was differentially expressed between the NT and AI embryos. The aberrant expression of this imprinted gene in the NT embryos could be linked to the high incidence of large offspring syndrome observed in NT calves. The observation that there was only a single differentially expressed imprinted gene between the NT and AI embryos indicates that the other imprinted genes on the microarray were properly reprogrammed in the NT embryos. When the donor cells and NT embryos were compared, 10 of the imprinted genes studied, CD81, COPG2, DCN, GNAS, GRB10, IGF2R, MEST, PEG3, PLAGL1, and SGCE, were significantly up-regulated in the donor cells. Conversely, one imprinted gene, SDHD, known to be a component of the electron transport system, was up-regulated in the NT embryos. None of the differentially expressed imprinted genes were located on the same bovine chromosome.
Aberrant expression of X-linked genes was previously reported in bovine NT embryos and the tissues of deceased clones (13, 42). Of the 123 human X-linked genes examined in this study, none were differentially expressed between the NT and AI embryos. No systematic study of X-chromosome inactivation (XCI) has been conducted in cattle. However, De La Fuente et al. (43) determined that XCI is completely established by day 14–15 in in vivo bovine embryos. It is therefore likely that XCI has not occurred in the embryos used in the present study.
In the normal bovine preimplantation embryo, active and passive demethylation occurs and is followed by de novo methylation at the 8- to 16-cell stage (3). Bovine NT embryos have been shown to be abnormally hypermethylated (3–5). We examined genes on the microarray that are involved in methylation regulation: ATF7IP, DMAP1, DNMT2, DNMT3A, DNMT3B, FOS, MBD4, MIZF, and p66alpha. These genes were not differentially expressed among the AI, IVF, and NT embryos. This was consistent with our observation of similar expression of imprinted genes between NT and normal AI embryos and illustrates that the methylation regulation involved in nuclear reprogramming is not impaired in the NT embryos. Further substantiation came from the observation that both the de novo methyltransferases, DNMT3A and DNMT3B, which act to methylate the genome after demethylation (44), were very highly and consistently expressed in all three types of embryos studied. This level of expression was not seen in the donor cells, indicating that these de novo methyltransferases were reprogrammed in the NT embryos.
Remodeling of chromatin is also involved in epigenetic reprogramming (45), and abnormally high histone H3-K9 methylation and acetylation have been observed in preimplantation NT embryos (46). Twenty-six genes associated with chromatin modification and epigenetic regulation were examined: ARID1A; ASF1A; BAT8; BAZ1B; CHD4; CHRAC1; CPA4; CTCF; CUGBP1; HDAC1, -2, -3, and -7A; L3MBTL; MLL3; MSL3L1; MYST1 and -4; RBM14; RPS6KA5; SET07; SIRT5; SMARCA5; SMARCC1; SMARCD3; and TRIM28. None were differentially expressed among the AI, IVF, and NT embryos. However, ASF1A, BAZ1B, HDAC1, MLL3, RPS6KA5, and TRIM28 were up-regulated in the NT embryos, and HDAC7A and SMARCD3 were up-regulated in the donor cells. These findings point to proper function, modification, and remodeling of chromatin in concordance with reprogramming.
We examined 434 genes that were categorized as involved in developmental processes (Table 7, which is published as supporting information on the PNAS web site). Only five were differentially expressed between the NT and AI embryos: AKAP11, CCL26, PIP5K1A, PPAP2A, and TFAP2A. In contrast, 14 genes between the NT and IVF embryos and 24 genes between AI and IVF embryos were differentially expressed. Eight genes, CD59, IQGAP1, NASP, PLAU, PTPRF, SFN, TM4SF12, and TXNRD1, were uniquely expressed in IVF embryos and were either up- or down-regulated compared with AI and NT embryos. These results support a specific effect of IVF on gene expression patterns. Likewise, three genes were specific to AI embryos (AKAP11, CCL26, and PPAP2A), suggesting that their expression is associated with better survival in later embryonic development or a response to the in vivo environment. The gene PIP5K1A was up-regulated exclusively in the NT embryos and is involved in signal transduction.
In addition to examining developmentally important genes, we also identified similarly expressed genes across all embryo types. These genes may be essential for embryonic development, because they were consistently expressed. In the AI, NT, and IVF embryos, 339 genes were expressed with ≥90% similarity (Table 8, which is published as supporting information on the PNAS web site). Also, there were 1,576, 1,001, and 887 genes expressed with 90% similarity in AI and NT, IVF and NT, and AI and IVF embryo comparisons, respectively. This further illustrates that the AI and NT embryos have the most similar expression profiles of all embryo types. Expression Analysis Systematic Explorer analysis determined that “cellular catabolism,” “intracellular,” “Arp2/3 protein complex,” “cytoplasm,” “RNA metabolism,” “RNA processing,” “Golgi apparatus,” and “intracellular organelle” GO categories were significantly (P < 0.05) overrepresented among the 339 genes. Additionally, cell cycle, tricarboxylic acid cycle, pyruvate metabolism, and ribosome pathways were identified by keggcharts.
To confirm the microarray results, we conducted quantitative real-time RT-PCR. We selected 12 genes for confirmation. Six genes from the donor cell and NT embryo comparison (CITED1, COL5A2, DNMT3B, HDAC1, PECAM1, and VIM) were selected on the basis of cell-type specificity and role in nuclear reprogramming. The other six genes from the AI and NT embryo comparison (ACOX1, CD81, DNAJC10, HRH1, IHPK2, and TFAP2A) were chosen because they are involved in metabolism, transcription and signal transduction. The β-actin (ACTB) gene was selected as the endogenous control, and the analysis was done by using the relative standard curve method with the standard reference as the calibrator. The primers and probes used for the real-time PCR are in Table 9, which is published as supporting information on the PNAS web site. Eleven of the 12 genes studied were validated by real-time PCR (Table 3). Real-time PCR of IHKP2 transcripts did not correspond with the microarray data that showed a 2.1-fold increase in expression in the AI embryos as compared with the NT embryos. Instead, the real-time PCR fold change was a 1.2-fold increase in the NT embryos. A one-way ANOVA was performed on the ratios of each embryo to the standard reference, and there was no significant difference between the AI and NT embryos. With this one exception, the real-time PCR results firmly substantiate the differential gene expression obtained by using microarrays.
Table 3. Quantitative real-time RT-PCR results for selected genes.
| Comparison | Gene symbol | Microarray fold Δ* | Expression | Real-time fold Δ* |
|---|---|---|---|---|
| COL5A2 | 9.8 | ↑ Cells | 20.3 | |
| VIM | 17.2 | ↑ Cells | 8.0 | |
| Donor cells vs. | HDAC1 | 5.7 | ↑ NT | 4.6 |
| NT embryos | PECAM1 | 3.0 | ↑ NT | 9.8 |
| DNMT3B | 16.6 | ↑ NT | 9.6 | |
| CITED1 | 3.9 | ↑ NT | 10.2 | |
| ACOX1 | 2.9 | ↑ AI | 2.7 | |
| TFAP2A | 2.6 | ↑ AI | 5.0 | |
| AI vs. NT embryos | CD81 | 2.5 | ↑ AI | 2.4 |
| IHKP2† | 2.1 | ↑ AI, ↑ NT | 1.2 | |
| DNAJC10 | 2.3 | ↑ NT | 3.8 | |
| HRH1 | 2.6 | ↑ NT | 8.8 |
Fold change is expressed as the ratio of the value of the NT embryos:standard reference divided by the donor cells:standard reference or the AI embryos:standard reference divided by the NT embryos:standard reference. The reciprocal values are presented if the ratio of the ratios was <0.5.
IHKP2 gene did not correspond with results obtained by microarray analysis.
Conclusion
Our data documented that the NT embryos' gene expression profiles were very different from those of their donor cells and very closely resembled those of naturally fertilized AI embryos, more so than IVF embryos. We conclude that the NT embryos have undergone significant nuclear reprogramming by the blastocyst stage. Our finding of more similarity between AI and NT embryos was unexpected. The developmental competence of IVF embryos is vastly superior to that of NT embryos, because they have much higher pregnancy (40–45%) and calving rates (≈35–40%) than NT embryos (20–30% and 5–10%, respectively). Thus, our original expectations were that the gene expression profiles of the NT embryos would be more different than the IVF embryos when compared with the naturally fertilized control AI embryos, even though IVF embryos (compared with AI embryos) have lower pregnancy and higher abortion rates and produce calves with large offspring syndrome. Additionally, because various abnormalities occur in NT embryos at various stages of postimplantation development, we originally expected that the gene expression profiles among NT embryos would be more variable. Surprisingly, we observed appreciably less variation among the individual NT embryos than that observed among individual IVF embryos. The significant gene expression differences between NT and IVF embryos are expected, as reported by Pfister-Genskow et al. (45). However, the explanation of this observation differed in this study, because our data showed that the NT embryos had undergone significant nuclear reprogramming, with similar gene expression profiles by the blastocyst stage to those of normally fertilized control embryos. The gene expression differences between the IVF and NT embryos may be partially attributed to developmental competence variability (compared with AI embryos), as well as the maternally inherited genetic variability among IVF embryos.
Because the gene expression profiles of the NT embryos closely resembled those of AI embryos, and there was lower variability in global gene expression profiles among the NT embryos, the NT embryos examined had significant and uniform nuclear reprogramming at the blastocyst stage of embryo development. This is supported by the high developmental rates of NT embryos to the blastocyst stage and high efficiency of nuclear transfer ES (ntES) cell line derivation from NT embryos. Recently, Hwang et al. (48) reported that human embryos derived from NT can be used successfully for generating ntES cell lines (35% efficiency), an efficiency similar to that from normally fertilized embryos (49, 50). Similar findings for ntES cell line derivation (20–30%) were reported from mouse NT embryos (51, 52), which are commonly known to have poor fetal/term development (1–2%). Furthermore, our recent report revealed that bovine NT embryos are more efficient than IVF embryos for generating bovine ES cell lines (53).
We used a 7,872 cDNA bovine microarray to compare gene expression profiles between cloned and fertilized control embryos. Although this microarray was primarily derived from the bovine placental and spleen cDNA libraries, we identified ≈3,500 genes expressed in the embryos. Most importantly, the microarray contains sequences from all categories of interest for studying nuclear reprogramming, including imprinted genes; X-linked genes; genes involved in development; regulators of methylation; and chromatin modification and other epigenetic regulators, in addition to hundreds of GO categories. This microarray, however, is deficient for certain embryonic-specific genes. Thus, future studies are planned using microarrays containing genes from this microarray, as well as sequences from embryonic/fetal or uterine cDNA libraries that we are currently developing.
Our results suggest that the commonly observed low developmental efficiency of NT embryos may not be largely due to nuclear reprogramming during early embryo development (reprogramming of the somatic donor cell genome from a differentiated to a totipotent status, i.e., gene dedifferentiation) but may be potentially caused by abnormal gene reprogramming during postimplantation fetal/placental development. Additional work is needed to determine whether small early-stage reprogramming errors (<1% of genes examined) are magnified downstream in development. The success of NT may depend upon nuclear reprogramming of gene expression for dedifferentiation of the donor somatic cell nuclei during early embryo development and reprogramming of gene expression for redifferentiation of NT embryos during tissue genesis and organogenesis in later development. Humpherys et al. (54) revealed abnormal gene expression in the livers and placentas of murine NT neonates. Additional research is needed to address whether the low rate and aberrant development of NT embryos are caused by abnormal gene reprogramming at later stages of development in the bovine.
Supplementary Material
Acknowledgments
We thank B. Pedersen, DVM (Delaware Valley Veterinary Service, P.C. Delhi, NY) and Cyagra, Inc. (Elizabethtown, PA) for the AI embryos, S. Plummer for careful editing of the manuscript, and V. Kask for help with the figures. This project was supported in part by National Research Initiative Grant 2002-35205-11548 from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service and U.S. Department of Agriculture Agricultural Research Service Contracts AG 58-1265-2-018 and 58-1265-2-020.
Author contributions: S.L.S., R.E.E., X.C.T., J.-P.R., H.A.L., and X.Y. designed research; S.L.S., F.D., L.-Y.S., and B.-S.J. performed research; R.E.E. and H.A.L. contributed new reagents/analytic tools; S.L.S. and S.L.R.-Z. analyzed data; and S.L.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: NT, nuclear transfer; AI, artificial insemination; IVF, in vitro fertilized/fertilization; aRNA, amplified RNA; GO, Gene Ontology; ES, embryonic stem.
Data deposition: The microarray data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE3568 and GSE3569).
References
- 1.Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A. J. & Campbell, K. H. (1997) Nature 385, 810–813. [DOI] [PubMed] [Google Scholar]
- 2.Hill, J. R., Roussel, A. J., Cibelli, J. B., Edwards, J. F., Hooper, N. L., Miller, M. W., Thompson, J. A., Looney, C. R., Westhusin, M. E., Robl, J. M., et al. (1999) Theriogenology 51, 1451–1465. [DOI] [PubMed] [Google Scholar]
- 3.Dean, W., Santos, F., Stojkovic, M., Zakhartchenko, V., Walter, J., Wolf, E. & Reik, W. (2001) Proc. Natl. Acad. Sci. USA 98, 13734–13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang, Y. K., Koo, D. B., Park, J. S., Choi, Y. H., Chung, A. S., Lee, K. K. & Han, Y. M. (2001) Nat. Genet. 28, 173–177. [DOI] [PubMed] [Google Scholar]
- 5.Bourc'his, D., Le Bourhis, D., Patin, D., Niveleau, A., Comizzoli, P., Renard, J. P. & Viegas-Pequignot, E. (2001) Curr. Biol. 11, 1542–1546. [DOI] [PubMed] [Google Scholar]
- 6.Daniels, R., Hall, V. & Trounson, A. O. (2000) Biol. Reprod. 63, 1034–1040. [DOI] [PubMed] [Google Scholar]
- 7.Wrenzycki, C., Wells, D., Herrmann, D., Miller, A., Oliver, J., Tervit, R. & Niemann, H. (2001) Biol. Reprod. 65, 309–317. [DOI] [PubMed] [Google Scholar]
- 8.Han, D. W., Song, S. J., Uhum, S. J., Do, J. T., Kim, N. H., Chung, K. S. & Lee, H. T. (2003) Zygote 11, 245–252. [DOI] [PubMed] [Google Scholar]
- 9.Everts, R. E., Band, M. R., Liu, Z. L., Kumar, C. G., Liu, L., Loor, J. J., Oliveira, R. & Lewin, H. A. (2005) Vet. Immunol. Immunopathol. 105, 235–245. [DOI] [PubMed] [Google Scholar]
- 10.Band, M. R., Olmstead, C., Everts, R. E., Liu, Z. L. & Lewin, H. A. (2002) Anim. Biotechnol. 13, 163–172. [DOI] [PubMed] [Google Scholar]
- 11.Kubota, C., Yang, X., Dinnyes, A., Todoroki, J., Yamakuchi, H., Mizoshita, K., Inohae, S. & Tabara, N. (1998) Mol. Reprod. Dev. 51, 281–286. [DOI] [PubMed] [Google Scholar]
- 12.Kubota, C., Yamakuchi, H., Todoroki, J., Mizoshita, K., Tabara, N., Barber, M. & Yang, X. (2000) Proc. Natl. Acad. Sci. USA 97, 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue, F., Tian, X. C., Du, F., Kubota, C., Taneja, M., Dinnyes, A., Dai, Y., Levine, H., Pereira, L. V. & Yang, X. (2002) Nat. Genet. 31, 216–220. [DOI] [PubMed] [Google Scholar]
- 14.Nedambale, T. L., Dinnyes, A., Yang, X. & Tian, X. C. (2004) Biol. Reprod. 71, 1671–1676. [DOI] [PubMed] [Google Scholar]
- 15.Baugh, L. R., Hill, A. A., Brown, E. L. & Hunter, C. P. (2001) Nucleic Acids Res. 29, E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegda, P., Qi, R., Abernathy, R., Gay, C., Dharap, S., Gaspard, R., Earle-Hughes, J., Snesrud, E., Lee, N. H. & Quackenbush, J. (2000) BioTechniques 29, 548–562. [DOI] [PubMed] [Google Scholar]
- 17.Clevel, W. S. & Devlin, S. J. (1988) J. Am. Stat. Assoc. 83, 596–610. [Google Scholar]
- 18.Wolfinger, R. D., Gibson, G., Wolfinger, E. D., Bennett, L., Hamadeh, H., Bushel, P., Afshari, C. & Paules, R. S. (2001) J. Comput. Biol. 8, 625–637. [DOI] [PubMed] [Google Scholar]
- 19.Wang, E., Miller, L. D., Ohnmacht, G. A., Liu, E. T. & Marincola, F. M. (2000) Nat. Biotechnol. 18, 457–459. [DOI] [PubMed] [Google Scholar]
- 20.Scheidl, S. J., Nilsson, S., Kalen, M., Hellstrom, M., Takemoto, M., Hakansson, J. & Lindahl, P. (2002) Am. J. Pathol. 160, 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenson, S. D., Robetorye, R. S., Bohling, S. D., Schumacher, J. A., Morgan, J. W., Lim, M. S. & Elenitoba-Johnson, K. S. (2003) Mol. Pathol. 56, 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bilodeau-Goeseels, S. & Schultz, G. A. (1997) Biol. Reprod. 56, 1323–1329. [DOI] [PubMed] [Google Scholar]
- 23.Hosack, D. A., Dennis, G., Jr., Sherman, B. T., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 25.Abeyta, M. J., Clark, A. T., Rodriguez, R. T., Bodnar, M. S., Pera, R. A. & Firpo, M. T. (2004) Hum. Mol. Genet. 13, 601–608. [DOI] [PubMed] [Google Scholar]
- 26.Ivanova, N. B., Dimos, J. T., Schaniel, C., Hackney, J. A., Moore, K. A. & Lemischka, I. R. (2002) Science 298, 601–604. [DOI] [PubMed] [Google Scholar]
- 27.Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. (2002) Science 298, 597–600. [DOI] [PubMed] [Google Scholar]
- 28.Sato, N., Sanjuan, I. M., Heke, M., Uchida, M., Naef, F. & Brivanlou, A. H. (2003) Dev. Biol. 260, 404–413. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, D. L. & Rizzino, A. (2000) Mol. Reprod. Dev. 56, 113–123. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Morales, J. R., Rodrigo, I. & Bovolenta, P. (2004) BioEssays 26, 766–777. [DOI] [PubMed] [Google Scholar]
- 31.Brewer, S., Jiang, X., Donaldson, S., Williams, T. & Sucov, H. M. (2002) Mech. Dev. 110, 139–149. [DOI] [PubMed] [Google Scholar]
- 32.Brewer, S. & Williams, T. (2004) Dev. Biol. 267, 399–417. [DOI] [PubMed] [Google Scholar]
- 33.Kawakami, Y., Rodriguez-Leon, J., Koth, C. M., Buscher, D., Itoh, T., Raya, A., Ng, J. K., Esteban, C. R., Takahashi, S., Henrique, D., et al. (2003) Nat. Cell Biol. 5, 513–519. [DOI] [PubMed] [Google Scholar]
- 34.Mercader, N., Leonardo, E., Azpiazu, N., Serrano, A., Morata, G., Martinez, C. & Torres, M. (1999) Nature 402, 425–429. [DOI] [PubMed] [Google Scholar]
- 35.Sapin, V., Bouillet, P., Oulad-Abdelghani, M., Dastugue, B., Chambon, P. & Dolle, P. (2000) Mech. Dev. 92, 295–299. [DOI] [PubMed] [Google Scholar]
- 36.Piedrahita, J. A., Oetama, B., Bennett, G. D., van Waes, J., Kamen, B. A., Richardson, J., Lacey, S. W., Anderson, R. G. & Finnell, R. H. (1999) Nat. Genet. 23, 228–232. [DOI] [PubMed] [Google Scholar]
- 37.Poschl, E., Schlotzer-Schrehardt, U., Brachvogel, B., Saito, K., Ninomiya, Y. & Mayer, U. (2004) Development (Cambridge, U.K.) 131, 1619–1628. [DOI] [PubMed] [Google Scholar]
- 38.Mann, M. R., Chung, Y. G., Nolen, L. D., Verona, R. I., Latham, K. E. & Bartolomei, M. S. (2003) Biol. Reprod. 69, 902–914. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa, H., Ono, Y., Shimozawa, N., Sotomaru, Y., Katsuzawa, Y., Hiura, H., Ito, M. & Kono, T. (2003) Reproduction 126, 549–557. [DOI] [PubMed] [Google Scholar]
- 40.Morison, I. M., Paton, C. J. & Cleverley, S. D. (2001) Nucleic Acids Res. 29, 275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis, A., Mitsuya, K., Umlauf, D., Smith, P., Dean, W., Walter, J., Higgins, M., Feil, R. & Reik, W. (2004) Nat. Genet. 36, 1291–1295. [DOI] [PubMed] [Google Scholar]
- 42.Wrenzycki, C., Lucas-Hahn, A., Herrmann, D., Lemme, E., Korsawe, K. & Niemann, H. (2002) Biol. Reprod. 66, 127–134. [DOI] [PubMed] [Google Scholar]
- 43.De La Fuente, R., Hahnel, A., Basrur, P. K. & King, W. A. (1999) Biol. Reprod. 60, 769–775. [DOI] [PubMed] [Google Scholar]
- 44.Okano, M., Bell, D. W., Haber, D. A. & Li, E. (1999) Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 45.Li, E. (2002) Nat. Rev. Genet. 3, 662–673. [DOI] [PubMed] [Google Scholar]
- 46.Santos, F., Zakhartchenko, V., Stojkovic, M., Peters, A., Jenuwein, T., Wolf, E., Reik, W. & Dean, W. (2003) Curr. Biol. 13, 1116–1121. [DOI] [PubMed] [Google Scholar]
- 47.Pfister-Genskow, M., Myers, C., Childs, L. A., Lacson, J. C., Patterson, T., Betthauser, J. M., Goueleke, P. J., Koppang, R. W., Lange, G., Fisher, P., et al. (2005) Biol. Reprod. 72, 546–555. [DOI] [PubMed] [Google Scholar]
- 48.Hwang, W. S., Roh, S. I., Lee, B. C., Kang, S. K., Kwon, D. K., Kim, S., Kim, S. J., Park, S. W., Kwon, H. S., Lee, C. K., et al. (2005) Science 308, 1777–1783. [DOI] [PubMed] [Google Scholar]
- 49.Hoffman, L. M. & Carpenter, M. K. (2005) Nat. Biotechnol. 23, 699–708. [DOI] [PubMed] [Google Scholar]
- 50.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S. & Jones, J. M. (1998) Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- 51.Li, J., Ishii, T., Feinstein, P. & Mombaerts, P. (2004) Nature 428, 393–399. [DOI] [PubMed] [Google Scholar]
- 52.Wakayama, S., Ohta, H., Kishigami, S., Thuan, N. V., Hikichi, T., Mizutani, E., Miyake, M. & Wakayama, T. (2005) Biol. Reprod. 72, 932–936. [DOI] [PubMed] [Google Scholar]
- 53.Wang, L., Duan, E., Sung, L. Y., Jeong, B. S., Yang, X. & Tian, X. C. (2005) Biol. Reprod. 73, 149–155. [DOI] [PubMed] [Google Scholar]
- 54.Humpherys, D., Eggan, K., Akutsu, H., Friedman, A., Hochedlinger, K., Yanagimachi, R., Lander, E. S., Golub, T. R. & Jaenisch, R. (2002) Proc. Natl. Acad. Sci. USA 99, 12889–12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.