Abstract
Snf5 (Ini1/Baf47/Smarcb1), a core member of the Swi/Snf chromatin remodeling complex, is a potent tumor suppressor whose mechanism of action is largely unknown. Biallelic loss of Snf5 leads to the onset of aggressive cancers in both humans and mice. We have developed an innovative and widely applicable analytical technique for cross-species validation of cancer models and show that the gene expression profiles of our Snf5 murine models closely resemble those of human Snf5-deficient rhabdoid tumors. We exploit this system to produce what we believe to be the first report documenting the effects on gene expression of inactivating a Swi/Snf subunit in normal mammalian cells and to identify the transcriptional pathways regulated by Snf5. We demonstrate that the tumor suppressor activity of Snf5 depends on its regulation of cell cycle progression; Snf5 inactivation leads to aberrant up-regulation of E2F targets and increased levels of p53 that are accompanied by apoptosis, polyploidy, and growth arrest. Further, conditional mouse models demonstrate that inactivation of p16Ink4a or Rb (retinoblastoma) does not accelerate tumor formation in Snf5 conditional mice, whereas mutation of p53 leads to a dramatic acceleration of tumor formation.
Keywords: chromatin remodeling, gene expression analysis, mouse models, Swi/Snf
The Swi/Snf ATPase chromatin remodeling complex has recently been linked to growth control and cancer development (reviewed in ref. 1). Specific inactivating mutations in the core Snf5 subunit are a hallmark of rhabdoid tumor (RT), an aggressive pediatric cancer, and also lead to a familial cancer predisposition syndrome (2-5). In mice, 15-30% of heterozygotes develop RTs, all of which have lost the functional Snf5 allele (6-8). Induced inactivation of conditional Snf5 leads to 100% of mice developing lymphomas or RTs with a median onset of only 11 weeks (9). This rate of cancer onset is extremely rapid for inactivation of a single gene and indicates a critical role for Snf5 in preventing cancer. However, the mechanistic basis for this activity is largely unknown. Because Snf5 is present in all variants of the Swi/Snf complex and is itself a bona fide tumor suppressor, we sought to use our Snf5-targeted mouse models to identify genes whose transcription is regulated by Snf5 and determine whether altered expression of these targets contributes to oncogenesis.
Materials and Methods
Cell Culture. Murine embryonic fibroblasts (MEFs) were harvested at embryonic day 13.5. An adenoviral vector containing the CMV promoter driving expression of green fluorescent protein (Ad5CMV-GFP, University of Iowa Gene Transfer Vector Core, Iowa City) was used to determine the most efficient multiplicity of infection (moi). Excision of Snf5 was achieved with Ad5CMVcre (University of Iowa Gene Transfer Vector Core) at an moi of 50 to 2 × 106 MEFs on 10-cm plates. To rule out contamination due to overgrowth of nondeleted cells, findings at later time points were confirmed in parallel experiments with transduction of a selectable pBabe-Cre-puromycinr vector.
Western Blots. SNF5/BAF-47 and β-tubulin antibodies were from BD Biosciences, p16Ink4a and p21 antibodies were from Santa Cruz Biotechnology, p19Arf antibody was from Abcam, Inc. (Cambridge, MA), and p53 antibody was from NovoCastra (Newcastle, U.K.).
Southern Analysis. Southern analysis was performed as described in ref. 9.
RNA Isolation and Microarray Analysis. cRNA was prepared for the Affymetrix GeneChip Mouse Genome 430A 2.0 Array as described in ref. 10. MEF expression data were analyzed for fold changes after the deletion of Snf5 by using the dna-chip analyzer (dchip) software following standard procedures for normalization and modeled-based expression using default settings (11). Probes were excluded if the presence call for either the conditional or control samples was <20%. The remaining probes were then compared with each other based on a criterion of at least 1.5-fold change between the mean value of Snf5+/- and Snf5flox/- samples. Real-time PCR was used to confirm microarray data (Figs. 7 and 8, which are published as supporting information on the PNAS web site). Primers used for this analysis are listed in Table 1, which is published as supporting information on the PNAS web site.
Patient Samples. All patient samples have been previously published with the exception of the 10 new human RTs. Studies were done with the approval of the Office for Protection of Research Subjects at the Dana-Farber Cancer Institute, the Committee for Clinical Investigation at Children's Hospital Boston, and the Institutional Review Board of The Children's Hospital of Philadelphia.
Matching of Human and Mouse Probes, Preprocessing, and Normalization. The orthologous UniGene symbols were identified for each mouse or human probe by using the Affymetrix software, and the corresponding species-specific probe was then determined. For the nonnegative matrix factorization (NMF) analysis, gene expression values were thresholded (to 50). Genes with either a <5-fold change or <500 units of absolute change across the collection of samples were eliminated from further analysis. There was a final common set of 2,653 gene symbols to proceed with the analysis. Because the data sets correspond to different types of human and mouse microarrays (e.g., Hu6800, U95, and U133), we normalized the data before decomposing or projecting it. We replaced the expression value for each gene in each data set by its rank in its column.
NMF Dimension Reduction and Projection. We constructed an expression matrix A of size 2,653 × 34 whose rows contain the expression levels of the 2,653 genes in the 34 samples of human pediatric brain tumors (12). We then used NMF to factor matrix A into two matrices with positive entries, A ≈ WH. The matrix W has size 2,653 × 4, with each of the four columns defining a metagene; entry wij is the coefficient of gene i in metagene j. The matrix H has size 4 × 34, with each of the 34 columns representing the metagene expression pattern of the corresponding sample; entry hij represents the “expression level” of metagene i in sample j. We chose four as the middle dimension because the corresponding metagene expression levels can be used to yield an accurate clustering of the data into the four represented subtypes (13). To project other signatures into the 4D metagene set obtained from the Pomeroy et al. (12) data, we computed the Moore-Penrose generalized pseudoinverse of W and applied this value to all of the data, including the original. This computation yielded Ap, a 4 × 58 metagene expression matrix for all of the signatures under consideration, which could be viewed as a heat map (Fig. 2a).
Fig. 2.
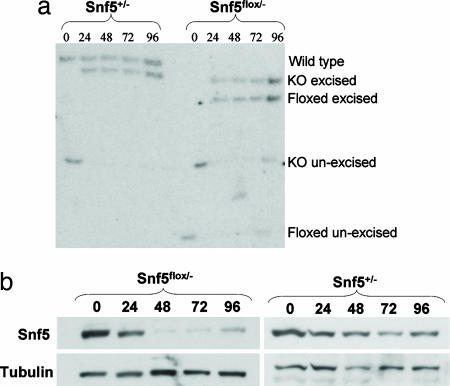
Inactivation of Snf5 in MEFs. (a) Southern analysis of Cre-mediated recombination at the Snf5 locus in MEFs at 0, 24, 48, 72, and 96 h. At time 0, before transduction, the knockout and floxed alleles are unexcised. Cre-mediated recombination results in excision of the neomycin resistance cassette in the knockout allele and excision of exon 1 in the floxed allele. (b) Anti-Snf5 Western blot of Snf5flox/- and Snf5+/- cells at 0, 24, 48, 72, and 96 h after adenovirus-Cre transduction.
Gene Set Enrichment Analysis (GSEA). GSEA was performed as described in ref. 14. The cumulative distribution function was constructed by performing 10,000 random gene set membership assignments; thus, the smallest obtainable P value was P = 1 × 10-4.
Biplot. Ap was visualized by using 2D and 3D biplots (15), which use metric scaling to map variables and observation in a single plot. The biplots were implemented in r by calling the “princomp” routine and rescaling to match the observations.
Software. The program to compute the NMF decomposition is our own (13). The pseudoinverse, biplot map, heat map, hierarchical clustering, and scatter plots were implemented by using functions in the r environment (i.e., ginv, biplot, image, hclust, plot, and scatterplot3d).
Results
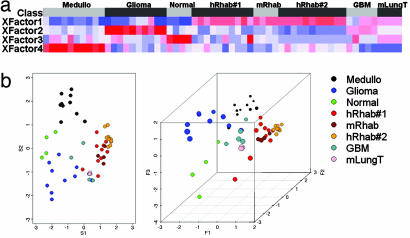
The Gene Expression Profile of Murine RTs Is Highly Related to That of Human RTs. We first sought to use cross-species conservation of gene expression to validate the Snf5 murine cancer model system. Of note, the histological appearance of the murine tumors is essentially indistinguishable from human RT, including the presence of classic rhabdoid cells (6). We developed a unique analytical projection approach partially based on our previous work using NMF (13) and principal component analysis projection of tumor samples (12). NMF is an algorithm based on decomposition by parts that can reduce the dimension of expression data from thousands of genes to a handful of metagenes. Each metagene represents a weighted average of several genes that tend to be coexpressed in a particular sample class. NMF is an efficient method for identification of distinguishing molecular patterns and provides a powerful method for class discovery. We used NMF on a data set containing our previously published gene expression signatures derived from four classes of human pediatric brain samples: RTs, medulloblastomas, gliomas, and normal cerebellum (12). We identified a small number of gene combinations (metagenes) whose profiles best represented the most distinguishing features of the expression patterns of this four-class data set (Fig. 1a). We then independently evaluated the expression levels of these metagenes in four murine RTs. As additional controls, we included signatures from four murine lung tumors, an additional 10 human RTs, and four glioblastomas derived from a previously published CNS tumor data set (17). These results revealed that murine Snf5 tumor signatures are closely related to those of both sets of human RT samples and are distinct from the controls (Fig. 1a). We then used a biplot transformation to project a representation of each signature into 2D and 3D space. The murine RTs cluster within the human RTs, indicating that the NMF expression signatures of these tumors are more closely related to each other than to the human and murine control samples (Fig. 1b).
Fig. 1.
The expression signature of murine RT is highly related to that of human RT. (a) NMF was performed on the human pediatric CNS samples, and four metagene factors whose profiles best represented the most salient features of the expression patterns were identified. Expression values for these metagenes were then independently calculated for the four murine RT samples. A heat map reveals that expression of the metagenes is highly correlated between both sets of human RT sample and the murine RT samples. (b) A biplot transformation was performed on the NMF results, and a representation of each sample was projected into 2D (Left) and 3D (Right) space. Note that the murine RTs cluster with the human RTs and are distinct from all other controls. Samples originally published in Pomeroy et al. (12) are medulloblastomas (Medullo), gliomas, normal cerebellum (normal), and RTs (hRab#1). mRhab, murine RTs; hRhab#2, the 10 previously unpublished human RTs; GBM, published glioblastoma multiformae tumors (17); mLungT, murine KRAS2 mutant lung tumors (21).
Snf5 Loss Leads to Transcriptional Activation. Having established the validity of the murine model, we sought to investigate the role of Snf5 in transcriptional regulation. We wanted to use a highly controlled system that enabled complete inactivation of Snf5, avoided off-target effects on gene expression, could be performed in normal nontransformed cells, and could be readily validated. MEFs from mice carrying conditional Snf5 alleles met these criteria. We thus isolated conditional (Snf5flox/-) and control (Snf5+/-) MEFs. We focused on early changes in gene expression after Snf5 inactivation to avoid secondary effects. An adenoviral-Cre vector was used to inactivate Snf5 because it provided highly efficient and rapid recombination in MEFs (Fig. 9, which is published as supporting information on the PNAS web site). Excision of the floxed allele was complete by 24 h, and the level of Snf5 protein had decreased significantly by 24 h and was essentially absent by 48 h (Fig. 2). Pilot data revealed minimal changes in gene expression at 24 h, and we therefore isolated RNA at 48, 72, and 96 h in triplicate from independent experiments. There was no difference in the growth rate of the two genotypes at these early times. Gene expression results from the nine Snf5flox/- arrays were compared with results from the nine Snf5+/- arrays.
Deletion of Snf5 resulted in 1,457 probes corresponding to 1,300 genes having a >1.5-fold change in expression level (Table 2, which is published as supporting information on the PNAS web site). Considerably more probes were up-regulated (1,097) than were down-regulated (322). We defined the up-regulated probes (corresponding to 978 genes) as the MEF Snf5 up-regulated gene set and the 360 down-regulated probes (corresponding to 322 genes) as the MEF Snf5 down-regulated gene set.
A Subset of the MEF Snf5 Gene Sets Are Differentially Expressed in Human RT. To both validate findings from our MEF model system and determine whether genes in the MEF Snf5 gene signature were differentially expressed in human RT, we compared the MEF Snf5 up- and down-regulated gene sets to the data set containing our previously published gene expression signatures derived from RT and other human pediatric brain tumors (12). Many of the transcriptional pathways related to the general phenotype of cancer are undoubtedly shared among RT and other cancers. Additionally, many of the genes displaying altered transcription after Snf5 loss likely have their expression modified by subsequent genetic mutations that contribute to cancer formation. Nonetheless, our hypothesis was that there would be a conserved subset of genes that meet the following three criteria: (i) they are affected by Snf5 inactivation in normal mouse cells, (ii) they are similarly affected by Snf5 loss in human cancer cells, and (iii) they remain a distinguishing feature of RT when compared with other cancers. We compared expression data from 10 human RTs to expression data from 10 medulloblastomas, 10 glioblastomas, and four normal cerebellum samples to generate the human RT data set. GSEA was then used to independently evaluate the distribution of the MEF Snf5 up- and down-regulated gene sets in the human RT data set. GSEA is a computational method that determines whether a defined set of genes (e.g., members of the MEF Snf5 up-regulated gene set) shows statistically significant, concordant differences between two biological states (e.g., human RT vs. other pediatric tumors) (14). GSEA provides an enrichment score (ES) that measures the degree of enrichment of the gene set at the top (up-regulated in RT vs. other pediatric CNS tumors) or bottom (down-regulated in RT vs. other pediatric CNS tumors) of a rank-ordered gene list derived from the data set. A nominal P value is used to assess the significance of the ES.
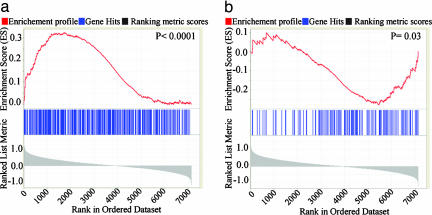
We found significant enrichment of the MEF Snf5 up-regulated gene set in genes that are up-regulated in the human RT data set (P < 1 × 10-4) (Fig. 3a; see also Table 3, which is published as supporting information on the PNAS web site). Similarly, there was significant enrichment of the MEF Snf5 down-regulated gene set in genes that are down-regulated in the human RT data set (P = 0.03) (Fig. 3b and Table 3). These results validate the MEF data, reveal that at least a subset of Snf5 target genes remain distinguishing features of RT, and demonstrate shared biological pathways of gene expression between Snf5-deficient MEFs and Snf5-deficient human RT.
Fig. 3.
A subset of the MEF Snf5 gene sets is differentially expressed in human RT. (a) Each vertical blue line represents the human ortholog of a probe in the MEF Snf5 up-regulated gene set. The left-to-right position of each line indicates the relative position of the probe within the rank ordering of the 7,129 probes present on the affyHu6800 human microarray from the probe most up-regulated in RT compared with other human tumors (position 1 on the left) to the most down-regulated (position 7129 on the right). The probes near the middle are not differentially expressed. The MEF Snf5 up-regulated gene set is enriched among genes up-regulated in the human RT data set, as evidenced by the increased number of blue marks on the left side and the positive enrichment score marked by the red line (P < 1 × 10-4). (b) The MEF Snf5 down-regulated gene set is enriched among the down-regulated genes in the human RT data set, as indicated by the increased number of blue marks on the right side and negative enrichment score (P = 0.03).
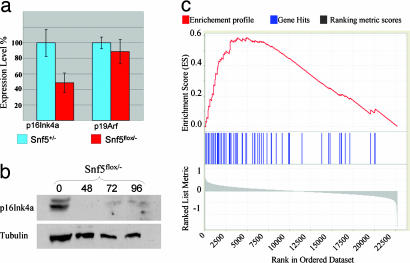
Inactivation of Snf5 Leads to Activation of E2F Targets but Decreased Growth of MEFs Due to Apoptosis, Growth Arrest, and Polyploidy. We sought to use the MEF model system to investigate the normal function of Snf5 and identify transcriptional pathways that may underlie its tumor suppressor activity. We therefore rank-ordered the murine probes from the most up-regulated after Snf5 inactivation in MEFs (probe 1) to the most down-regulated (probe 22690) and used GSEA to compare this list to an annotated functional database of 526 metabolic and signaling pathway gene sets (available with our GSEA software (14). Two of the gene sets whose members were clustered high on the rank-ordered list were “cell cycle,” which contained 130 probes (P < 0.001) and “cell cycle checkpoint,” which contained 40 probes (P < 0.001). Inspection of the lists revealed that many of the enriched genes were targets of the E2F pathway. We thus asked whether targets of the p16Ink4a/Rb (retinoblastoma)/E2F pathway are regulated by Snf5 within normal cells. Because the probe for p16Ink4a on the Affymetrix chips cannot distinguish between the overlapping p16Ink4a and p19Arf genes, we used quantitative real-time PCR and Western blotting to evaluate these genes. After inactivation of Snf5, p16Ink4a mRNA is down-regulated 2-fold and the protein becomes essentially undetectable, whereas the expression of p19Arf is unchanged (Figs. 4 a and b and 5c). The decreased levels of transcript are not due to methylation at the p16Ink4a promoter (not shown). Evaluation of microarray data can be complicated by subjective identification bias. Hence, we compared our MEF Snf5 rank-ordered list to an independently derived set of E2F targets derived from cells expressing mutant Rb or p16Ink4a (18). There was indeed marked enrichment of this independent E2F gene set within genes up-regulated in the MEF Snf5 rank-ordered data set (P < 1 × 10-4) (Fig. 4c; see also Tables 4 and 5, which are published as supporting information on the PNAS web site).
Fig. 4.
Loss of Snf5 leads to down-regulation of p16Ink4 and up-regulation of E2F target genes. (a) Quantitative real-time PCR analysis of p16Ink4a and p19Arf at 72 h after adenovirus-Cre transduction. (b) Anti-p16Ink4a Western blot of protein from Snf5flox/- cells at 0, 48, 72, and 96 h after adenovirus-Cre transduction. (c) E2F target genes are up-regulated after inactivation of Snf5. Each vertical blue line represents an E2F target as defined by the work of Vernell et al. (18). The left-to-right position of each line indicates the relative position of the probe within the rank ordering of the 22,690 probes present on the affy430A2.0 mouse microarray from the probe most up-regulated after Snf5 inactivation (position 1 on the left) to the most down-regulated (position 22690 on the right). The genes near the middle are unaffected by Snf5 loss. The E2F target gene set is clearly enriched among genes up-regulated in the MEF Snf5 data set, as evidenced by the increased number of blue marks on the left side of the distribution and the positive enrichment score marked by the red line (P < 1 × 10-4).
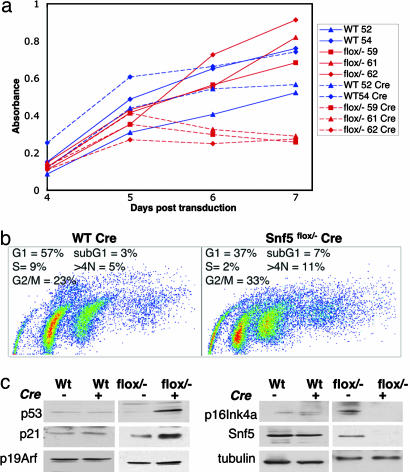
Fig. 5.
Loss of Snf5 leads to increased apoptosis, polyploidy, and poor cell growth. (a) MEFs from two wild-type embryos (blue) and three Snf5flox/- embryos (red) were transduced with pBabe-puror (dotted lines, control) or with pBabe Cre-puror (solid lines). Two days after transduction, cells were selected in puromycin, and at day 4, cells were plated for growth assay and monitored daily by absorbance of the Wst-1 reagent. (b) Cell cycle analysis by means of BrdUrd incorporation and propidium iodide staining reveals cell cycle arrest in Snf5flox/- Cre cells as evidenced by decreased S phase and increased G2/M; apoptosis evidenced by increased sub-G1 population and increased polyploidy seen as >4N population. (c) Western blot analysis demonstrating decreased levels of both Snf5 and p16Ink4a and increased levels of both p53 and p21 after deletion of Snf5.
Thus, the gene expression data were consistent with aberrant cell cycle regulation in Snf5-deficient MEFs. However, because levels of many cell cycle regulators are also affected by post-transcriptional processes, we sought to evaluate the phenotypic consequences of Snf5 inactivation in MEFs. Our microarray analysis was performed soon after Snf5 deletion at a time when there was no observable phenotypic difference between Snf5-deficient and control MEFs. To determine whether these transcriptional alterations would ultimately manifest in phenotypic changes, we evaluated Snf5-deficient MEFs at later times. Shortly after E2F target genes are up-regulated, Snf5-deficient MEFs no longer grow in culture (Fig. 5a). Cell cycle analysis revealed a significant increase in apoptosis and a cell cycle arrest with accumulation of cells in G2 (Fig. 5b). In addition, there was a significant increase in polyploidy in Snf5-deficient MEFs (Fig. 5b). No significant change in senescence was detected by senescence-associated-β-gal staining (not shown). Under normal growth conditions, expression of E2F targets is concomitant with cell cycle progression and growth. However, forced expression of E2F in normal primary cells typically triggers a p53-dependent checkpoint that leads to apoptosis and cell cycle arrest (19). Given the apoptosis and growth arrest present in Snf5-deficient MEFs, we examined levels of p53 and p21 by Western blot. Inactivation of Snf5 leads to marked up-regulation of both p53 and p21 (Fig. 5c).
Inactivation of p53 Synergizes with Snf5 Inactivation in Oncogenic Transformation. These findings were consistent with the possibility that the tumor suppressor effects of Snf5 could be mediated through stimulation of cell cycle progression via the E2F pathway. Further, it seems possible that up-regulation of p53 serves to combat the prooncogenic effects of Snf5 loss. We therefore used mouse models to evaluate this. We have previously reported that in vivo induced inactivation of a conditional inverting allele of Snf5 leads to apoptosis of most Snf5-deficient cells but also to the rapid onset of fully penetrant cancer at a median of 11 weeks (9). It should be noted that expression of Cre from the IFN-inducible Mx promoter is slightly leaky. Consequently, even when not treated with polyI/polyC, Mx-Cre+ Snf5inv/- mice succumb to CD8+ mature T cell lymphomas, although at a median of 29 weeks rather than the 11-week median when polyI/polyC is given.
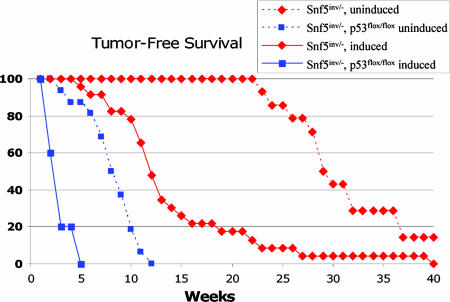
To evaluate the significance of increased expression of E2F targets and to determine whether p53 serves a functional role in suppressing oncogenic transformation after Snf5 loss, we bred Snf5 inverting conditional mice to p16Ink4a-knockout, Rb conditional, and p53 conditional mice in the presence of the Mx-Cre transgene. Coinactivation of either p16Ink4a or Rb with Snf5 had no significant effect on the rate of tumor formation, suggesting that these mutations may be redundant to Snf5, possibly because Snf5 loss has already led to E2F activation. In contrast, coinactivation of p53 led to a dramatic acceleration of tumor onset in Snf5 conditional mice. The median latency of tumor formation in noninduced mice decreased from 29 weeks to 7 weeks, whereas the latency in induced mice further decreased from 11 weeks to a remarkably rapid onset of <3 weeks (Fig. 6).
Fig. 6.
Coinactivation of Snf5 and p53 leads to a dramatic acceleration of tumor formation. Mx-Cre+ Snf5inv/- mice that are not treated with polyI/polyC (uninduced) develop tumors at a median of 29 weeks (red dotted line). Administration of polyI/polyC to Mx-Cre+ Snf5inv/- mice leads to tumor formation at a median of 11 weeks (red solid line). Mx-Cre+ Snf5inv/- p53flox/flox uninduced mice develop tumors with a median onset of 7 weeks (blue dotted line), and treatment with polyI/polyC further accelerates tumor onset to a median of 3 weeks (blue solid line).
Discussion
Validating Human Cancer Models. Genetically modified mice are often used to model human cancers, but there is active debate on the extent to which such models represent human disease. Thus, a major goal in the field has been to develop analytical tools for evaluating and validating mouse models. The advent of microarray-based gene expression profiling combined with whole-genome sequencing and annotation has recently enabled cross-species comparison of murine and human tumors (20, 21). Here, we report development of an innovative analytical projection approach for validating mouse models that extends our previous work using NMF clustering (13) and unites it with a projection algorithm. There are unique advantages to our approach. The method-specific preprocessing helps to normalize the data sets in a way that diminishes platform idiosyncrasies and reduces noise. The NMF algorithm then reduces expression data from thousands of genes to a small number of metagenes, each of which best captures the distinguishing features of one of the classes being modeled. In our case, four metagenes were identified that best differentiated the four classes of samples in the human CNS data set from each other. By subsequently using the metagene matrix, which has been optimized for human tumors, on individual mouse models and projecting the results, it is possible to evaluate how well a murine model matches the human condition. Unlike hierarchical clustering, our method is not dependent on a strict tree structure and instead provides projections that better expose the overall biological similarities and differences in the samples. In our case, it is noteworthy that the murine RTs cluster with the human RTs because it suggests that the transcriptional changes due to Snf5 loss and the resultant distinct mesodermal appearance of the tumors readily distinguish them from the human and murine controls. Our unique NMF-based projection method for cross-species validation should have wide applicability in assessing the fit of other mouse models for human tumors.
Transcriptional Regulation by Snf5. Members of the Swi/Snf complex were originally identified in yeast during screens for transcriptional activators. Subsequently, whole-genome expression analysis in yeast demonstrated a role for the Swi/Snf complex in transcriptional repression as well as activation. Intriguingly, studies examining wing vein development in Drosophila suggest that SNR1, the Snf5 Drosophila ortholog, may repress transcriptional activation by the ATPase Brm, a core Swi/Snf subunit (22). We found that three times as many genes were activated (978) as repressed (322) after Snf5 inactivation in MEFs, suggesting that mammalian Snf5 may in fact act to repress transcriptional activation by the Swi/Snf complex.
Studies of mammalian Swi/Snf regulated gene expression have, to date, been performed on cancer cell lines that, in addition to other oncogenic mutations, possess a mutant Swi/Snf subunit. Restoration of the deficient Swi/Snf subunit in these cell lines leads to growth arrest. This approach has significant limitations because both phenotypic states being compared are abnormal: a cancer cell line with a deficient subunit compared with a cell line with normal Swi/Snf expression but an abnormal growth arrest phenotype due to other genetic mutations. We took the approach of using normal embryonic fibroblasts in which both the Snf5 conditional and control cells were treated in an identical manner. The Snf5+/- and Snf5flox/- genotypes for MEFs were chosen to closely match control cells to experimental cells. Both the experimental and control cells started with one active allele of Snf5 and, as the neomycin resistance cassette that has replaced exon 1 in the knockout allele (Snf5-) is flanked by lox P sites, both cells underwent Cre-mediated recombination at the Snf5 locus.
We used cross-species comparison of gene expression to validate the MEF model system. We identified a statistically significant subset of genes that are both rapidly affected by Snf5 loss in MEFs and also differentially expressed in RT compared with other brain tumors. It is likely that these genes are required for development of RT and may thus constitute useful therapeutic targets for RT and other Swi/Snf mutated cancers.
Stimulation of Cell Cycle Progression and Synergy with p53 Loss. The Swi/Snf complex has been shown to bind directly to Rb (23, 24). The Brg1 and Brm ATPase subunits of the Swi/Snf complex are required for Rb-mediated cell cycle arrest, whereas Snf5 is dispensable for this activity (25). Reexpression of Snf5 in RT cell lines leads to cell cycle arrest that is associated with induction of p16Ink4a and repression of cyclin D1 (25-28). The significance of these findings with respect to tumor suppressor activity has been unclear because the spectrum and kinetics of tumor formation is distinct in Rb, p16Ink4a, and Snf5 mutant mice. Recently, however, cyclin D1 was shown to be a critical down-stream regulator of tumor formation in the absence of Snf5; inactivation of cyclin D1 prevents formation of tumors in Snf5+/- mice (29).
By inactivating Snf5 in normal cells, we demonstrate that primary loss of Snf5 leads to perturbation of cell cycle control pathways, including up-regulation of E2F targets and increased levels of p53, apoptosis, polyploidy, and cell cycle arrest. Interestingly, in the absence of cooperating hits, the same growth-promoting mutations found in cancers typically trigger these events in primary cells (30). For example, overexpression of E2Fs in primary cells also leads to p53-dependent apoptosis, polyploidy, and growth arrest (19, 31, 32).
To evaluate whether perturbation of the E2F pathway may be the basis for tumor suppression by Snf5, we crossed Snf5 conditional mice to mice carrying targeted mutations in Rb, p16Ink4a, and p53. There was a lack of synergy between induced inactivation of Snf5 and either Rb or p16Ink4a. Of note, we cannot entirely rule out interactions, because there is some redundancy between the Rb and p16Ink4a tumor suppressors. Longer-term studies using the uninduced strains may help determine whether subtle interactions exist. In contrast to the case with Rb or p16Ink4a, there is marked synergy between inactivation of Snf5 and p53 that leads to tumor onset at 3 weeks in induced mice. Intriguingly, these tumors were exclusively of the phenotypic profile seen in Snf5 conditional mice (i.e., peripheral CD8+/CD4- mature T cell lymphomas), suggesting that the developmental phenotype of the cancers is determined by Snf5 loss and that p53 deletion simply accelerates tumor onset rather than affecting tumor spectrum. It is also worth noting that, despite their rapid onset, these tumors were clonal, indicating that an additional mutation is required for full oncogenic transformation. Indeed, we have preliminary data that inactivation of p53 is not sufficient to fully reverse the negative growth effects of Snf5 loss.
Collectively, our findings generate insight into the mechanism by which Snf5 acts as a potent tumor suppressor. We speculate that the Swi/Snf complex regulates overall signaling through p16Ink4a/Rb/E2F at multiple points in this pathway, including regulation of p16Ink4a expression and direct interaction with Rb. In itself, decreased p16Ink4a activity is unlikely to explain the aberrant effects we observed, because p16Ink4a is not required for appropriate E2F regulation in primary MEFs (16). As a consequence of Snf5 loss, the role of p16Ink4a and Rb in repressing cell cycle progression may be compromised, which could lead to increased expression of E2F target genes and aberrant cell cycle control, causing up-regulation of p53 accompanied by a G2 arrest, polyploidy, and apoptosis. We have identified the conserved transcriptional pathways regulated by Snf5 as well as the subset of these genes with differential expression in RT, and these data can now be used to further elucidate the role of the Swi/Snf complex in transcriptional regulation and also to identify and evaluate potential targets for therapeutic intervention in RT and other Swi/Snf mutated cancers.
Supplementary Material
Acknowledgments
We thank H. Hock, A. Sweet-Cordero, and A. Kung for critical reading of the manuscript. J.A.B. was supported by National Institutes of Health Grant CA 46274. C.W.M.R. was supported in part by the Charles H. Hood Foundation, the Garrett B. Smith Foundation, the Claudia Adams Barr Foundation, and a National Cancer Institute mentored clinical scientist grant.
Author contributions: M.S.I. and C.W.M.R. designed research; M.S.I., C.G.S., J.A.E., C.M.F., X.W., and C.W.M.R. performed research; P.T., A.S., J.A.B., S.L.P., and J.P.M. contributed new reagents/analytic tools; P.T., A.S., and J.P.M. analyzed data; and C.W.M.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: RT, rhabdoid tumor; NMF, nonnegative matrix factorization; MEF, murine embryonic fibroblast; GSEA, Gene Set Enrichment Analysis; Rb, retinoblastoma.
References
- 1.Roberts, C. W. & Orkin, S. H. (2004) Nat. Rev. Cancer 4, 133-142. [DOI] [PubMed] [Google Scholar]
- 2.Biegel, J. A., Zhou, J. Y., Rorke, L. B., Stenstrom, C., Wainwright, L. M. & Fogelgren, B. (1999) Cancer Res. 59, 74-79. [PubMed] [Google Scholar]
- 3.Sevenet, N., Sheridan, E., Amram, D., Schneider, P., Handgretinger, R. & Delattre, O. (1999) Am. J. Hum. Genet. 65, 1342-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Versteege, I., Sevenet, N., Lange, J., Rousseau-Merck, M. F., Ambros, P., Handgretinger, R., Aurias, A. & Delattre, O. (1998) Nature 394, 203-206. [DOI] [PubMed] [Google Scholar]
- 5.Biegel, J. A., Fogelgren, B., Wainwright, L. M., Zhou, J. Y., Bevan, H. & Rorke, L. B. (2000) Genes Chromosomes Cancer 28, 31-37. [DOI] [PubMed] [Google Scholar]
- 6.Roberts, C. W., Galusha, S. A., McMenamin, M. E., Fletcher, C. D. & Orkin, S. H. (2000) Proc. Natl. Acad. Sci. USA 97, 13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klochendler-Yeivin, A., Fiette, L., Barra, J., Muchardt, C., Babinet, C. & Yaniv, M. (2000) EMBO Rep. 1, 500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guidi, C. J., Sands, A. T., Zambrowicz, B. P., Turner, T. K., Demers, D. A., Webster, W., Smith, T. W., Imbalzano, A. N. & Jones, S. N. (2001) Mol. Cell. Biol. 21, 3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts, C. W., Leroux, M. M., Fleming, M. D. & Orkin, S. H. (2002) Cancer Cell 2, 415-425. [DOI] [PubMed] [Google Scholar]
- 10.Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286, 531-537. [DOI] [PubMed] [Google Scholar]
- 11.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415, 436-442. [DOI] [PubMed] [Google Scholar]
- 13.Brunet, J. P., Tamayo, P., Golub, T. R. & Mesirov, J. P. (2004) Proc. Natl. Acad. Sci. USA 101, 4164-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S. & Mesirov, J. P. (2005) Proc. Natl. Acad. Sci. USA 102, 15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gower, J. C. & Hand, D. J. (1996) Biplots (Chapman & Hall, London).
- 16.Sharpless, N. E., Ramsey, M. R., Balasubramanian, P., Castrillon, D. H. & DePinho, R. A. (2004) Oncogene 23, 379-385. [DOI] [PubMed] [Google Scholar]
- 17.Nutt, C. L., Mani, D. R., Betensky, R. A., Tamayo, P., Cairncross, J. G., Ladd, C., Pohl, U., Hartmann, C., McLaughlin, M. E., Batchelor, T. T., et al. (2003) Cancer Res. 63, 1602-1607. [PubMed] [Google Scholar]
- 18.Vernell, R., Helin, K. & Muller, H. (2003) J. Biol. Chem. 278, 46124-46137. [DOI] [PubMed] [Google Scholar]
- 19.Wu, X. & Levine, A. J. (1994) Proc. Natl. Acad. Sci. USA 91, 3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. S., Chu, I. S., Mikaelyan, A., Calvisi, D. F., Heo, J., Reddy, J. K. & Thorgeirsson, S. S. (2004) Nat. Genet. 36, 1306-1311. [DOI] [PubMed] [Google Scholar]
- 21.Sweet-Cordero, A., Mukherjee, S., Subramanian, A., You, H., Roix, J. J., Ladd-Acosta, C., Mesirov, J., Golub, T. R. & Jacks, T. (2005) Nat. Genet. 37, 48-55. [DOI] [PubMed] [Google Scholar]
- 22.Marenda, D. R., Zraly, C. B. & Dingwall, A. K. (2004) Dev. Biol. 267, 279-293. [DOI] [PubMed] [Google Scholar]
- 23.Dunaief, J. L., Strober, B. E., Guha, S., Khavari, P. A., Alin, K., Luban, J., Begemann, M., Crabtree, G. R. & Goff, S. P. (1994) Cell 79, 119-130. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, H. S., Gavin, M., Dahiya, A., Postigo, A. A., Ma, D., Luo, R. X., Harbour, J. W. & Dean, D. C. (2000) Cell 101, 79-89. [DOI] [PubMed] [Google Scholar]
- 25.Betz, B. L., Strobeck, M. W., Reisman, D. N., Knudsen, E. S. & Weissman, B. E. (2002) Oncogene 21, 5193-5203. [DOI] [PubMed] [Google Scholar]
- 26.Oruetxebarria, I., Venturini, F., Kekarainen, T., Houweling, A., Zuijderduijn, L. M., Mohd-Sarip, A., Vries, R. G., Hoeben, R. C. & Verrijzer, C. P. (2003) J. Biol. Chem. 279, 3807-3816. [DOI] [PubMed] [Google Scholar]
- 27.Versteege, I., Medjkane, S., Rouillard, D. & Delattre, O. (2002) Oncogene 21, 6403-6412. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, Z. K., Davies, K. P., Allen, J., Zhu, L., Pestell, R. G., Zagzag, D. & Kalpana, G. V. (2002) Mol. Cell. Biol. 22, 5975-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsikitis, M., Zhang, Z., Edelman, W., Zagzag, D. & Kalpana, G. V. (2005) Proc. Natl. Acad. Sci. USA 102, 12129-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharpless, N. E. & DePinho, R. A. (2005) Nature 436, 636-637. [DOI] [PubMed] [Google Scholar]
- 31.Hernando, E., Nahle, Z., Juan, G., Diaz-Rodriguez, E., Alaminos, M., Hemann, M., Michel, L., Mittal, V., Gerald, W., Benezra, R., et al. (2004) Nature 430, 797-802. [DOI] [PubMed] [Google Scholar]
- 32.Saavedra, H. I., Maiti, B., Timmers, C., Altura, R., Tokuyama, Y., Fukasawa, K. & Leone, G. (2003) Cancer Cell 3, 333-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.