Abstract
We genetically analyzed field isolates of the Newcastle disease (ND) virus isolated in Japan from 1930 to 2001. The coding region of the fusion protein was amplified by reverse transcriptase PCR and directly sequenced. Phylogenetic analysis revealed the presence of viruses belonging to six of the eight known genotypes. It can be concluded from this study that ND outbreaks in Japan have been of multiple etiologies.
Newcastle disease (ND), which is caused by the ND virus (NDV), is one of the most serious diseases in the poultry industry (1). Also called avian paramyxovirus type 1 (APMV1), NDV is an enveloped, negative-stranded RNA virus containing a genome of approximately 15 kb (20). NDV isolates across wide ranges of pathogenicity and virulence, from apathogenic to extremely pathogenic, have been described (1). NDV can infect a great variety of poultry or free-living birds (1), and such infections apparently play a role in the spread of ND. For example, the ND outbreaks that occurred in Great Britain in 1984 are thought to have stemmed from feed contaminated by infected pigeons (1), and the outbreaks in cormorants in the United States, which occurred from 1989 to 1996, were traced to infected exotic psittacines (32, 33).
In all, three major panzootics of ND have been recorded (1). The first began in 1926 in Southeast Asia and spread to most regions of the world. The second panzootic began in the Middle East in the late 1960s and had spread to other countries by 1973. The third panzootic, which was caused by the neurotropic form of NDV, termed the pigeon paramyxovirus type 1 virus, apparently also started in the Middle East, though in the late 1970s. By 1981, it had reached Europe, and thereafter spread rapidly throughout the world.
A comparison of the nucleotide sequences among the different strains of NDV revealed two major groups (31), which could be further divided into three lineages (7, 30, 38) or eight genotypes (3, 13, 14, 44). Three different NDV genotypes, II, III, and IV, were involved in the first panzootic of ND and were restricted to the specific geographic region in which the outbreak began. In the late 1960s, NDV genotypes V and VI emerged and caused the second panzootic. After that, two novel NDV genotypes, VII and VIII, were found in Asia, southern Africa, and a number of European countries (13, 14, 18, 23, 43, 44). Genotype VII was mainly responsible for recent outbreaks in the neighboring countries of Taiwan and China (18, 21, 43, 44). Thus, phylogenetic analysis of NDV is a powerful tool for investigating epidemiological relationships among the NDV isolates present in various parts of the world.
After the first outbreak of ND in Japan was recorded in 1930 (26), large outbreaks continued to occur until the ND live vaccine (Hitchner B1/47 strain) was applied in 1967. Fewer outbreaks have occurred in Japan since then, and those that have occurred have erupted mainly in small flocks that were not vaccinated against the disease or had been vaccinated incorrectly. However, a complete epidemiological analysis of the NDV isolates in Japan has not been conducted, except for a few strains (30, 38), and the relationship between the Japanese NDV isolates and the many strains isolated from other regions has remained unknown.
To define the epidemiology of NDV in Japan, we determined the nucleotide sequences of NDV isolates isolated in Japan between 1930 and 2001 using the reverse transcriptase PCR method coupled with direct sequencing and analyzed the sequences phylogenetically.
A total of 61 NDV isolates, which were collected in Japan between 1930 and 2001, were employed in this study and are listed in Table 1. Most of the NDV isolates were obtained from regional laboratories. Most of the specimens of NDV were isolated by two or three passages with embryonated specific- pathogen-free eggs or tissue cultures. The materials were submitted to our laboratory and were propagated once in specific- pathogen-free eggs. The velogenicity of each isolate was judged on the basis of the time it took for the embryo to be killed and on the basis of plaque and syncytium formation in the chicken embryo fibroblast cultures, as described previously (12, 22).
TABLE 1.
Phylogenetic examination of NDV isolates from Japan
| Isolate | Abbreviation | Genotype | Virulencea | Cleavage site (112RRQKRF117)b | DDBJ accession no. |
|---|---|---|---|---|---|
| APMV1/chicken/Japan/Sato/30 | JP/Sato/30 | III | V | ———R—— | AB070382 |
| APMV1/chicken/Japan/Miyadera/51 | JP/Miyadera/51 | II | V | —————— | AB070383 |
| APMV/chicken/Japan/Sagamihara/53 | JP/Sagamihara/53 | II | V | —————— | AB070384 |
| APMV1/chicken/Japan/Ishii/62 | JP/Ishii/62 | I | L | GK—G—L | AB070385 |
| APMV1/chicken/Japan/Narashino/67 | JP/Narashino/67 | VI | V | —————— | AB070386 |
| APMV1/chicken/Japan/Chiba/69 | JP/Chiba/69 | VI | V | —————— | AB070387 |
| APMV1/chicken/Japan/Chiba/81 | JP/Chiba/81 | VI | V | —————— | AB070388 |
| APMV1/chicken/Japan/Chiba/84 | JP/Chiba/84 | VI | V | —————— | AB070389 |
| APMV1/pigeon/Japan/FK-1/84 | JP/FK-1-pg/84 | VI | M | G————— | AB070390 |
| APMV1/pigeon/Japan/Toyama/84 | JP/Toyama/84 | VI | M | G————— | AB070391 |
| APMV1/pigeon/Japan/Ibaraki/84 | JP/Ibaraki-pg/84 | VI | M | G————— | AB070392 |
| APMV1/pigeon/Japan/Nagano-8/84 | JP/Nagano-8-pg/84 | VI | M | G————— | AB070393 |
| APMV1/chicken/Japan/TY-1/85 | JP/TY-1/85 | VI | M | —————— | AB070394 |
| APMV1/pheasant/Japan/Gunma/85 | JP/Gunma-ph/85 | VII | V | —————— | AB070395 |
| APMV1/chicken/Japan/Yamanashi/85 | JP/Yamanashi/85 | VII | V | —————— | AB070396 |
| APMV1/chicken/Japan/Tochigi/85 | JP/Tochigi/85 | VI | V | —————— | AB070397 |
| APMV1/pheasant/Japan/Tochigi/85 | JP/Tochigi-ph/85 | VI | V | —————— | AB070398 |
| APMV1/chicken/Japan/Ibaraki/85 | JP/Ibaraki/85 | VI | V | —————— | AB070399 |
| APMV1/chicken/Japan/Saitama/85 | JP/Saitama/85 | VII | V | —————— | AB070400 |
| APMV1/chicken/Japan/Shizuoka/85 | JP/Shizuoka/85 | VII | V | —————— | AB070401 |
| APMV1/chicken/Japan/Chiba/85 | JP/Chiba/85 | VII | V | —————— | AB070402 |
| APMV1/chicken/Japan/Niigata/85 | JP/Niigata/85 | VII | V | —————— | AB070403 |
| APMV1/chicken/Japan/Hyogo/85 | JP/Hyogo/85 | VII | V | —————— | AB070404 |
| APMV1/chicken/Japan/Wakayama/85 | JP/Wakayama/85 | VII | V | —————— | AB070405 |
| APMV1/chicken/Japan/Chiba/86 | JP/Chiba/86 | VII | V | —————— | AB070406 |
| APMV1/pigeon/Japan/Tochigi/86 | JP/Tochigi-pg/86 | VI | M | W————— | AB070407 |
| APMV1/chicken/Japan/Chiba/87 | JP/Chiba/87 | VII | V | —————— | AB070408 |
| APMV1/pigeon/Japan/Niigata/88 | JP/Niigata-pg/88 | VI | M | G————— | AB070409 |
| APMV1/chicken/Japan/Niigata/89 | JP/Niigata/89 | VII | V | —————— | AB070410 |
| APMV1/chicken/Japan/Nara/89 | JP/Nara/89 | VII | V | —————— | AB074011 |
| APMV1/pigeon/Japan/Kushiro/91 | JP/Kushiro-pg/91 | VI | M | G————— | AB074012 |
| APMV1/pigeon/Japan/Tokachi/91 | JP/Tokachi-pg/91 | VI | M | G————— | AB074013 |
| APMV1/chicken/Japan/Kagoshima/91 | JP/Kagoshima/91 | VIII | V | —————— | AB074014 |
| APMV1/chicken/Japan/Okinawa/91 | JP/Okinawa/91 | VIII | V | —————— | AB074015 |
| APMV1/pigeon/Japan/Ehime/93 | JP/Ehime-pg/93 | VI | M | G————— | AB074016 |
| APMV1/pigeon/Japan/Kumamoto/95 | JP/Kumamoto-pg/95 | VI | M | G————— | AB074017 |
| APMV1/chicken/Japan/MET/95 | JP/MET/95 | II | L | G——G—L | AB074018 |
| APMV1/pigeon/Japan/Tochigi/95 | JP/Tochigi-pg/95 | VI | M | ——K——— | AB074019 |
| APMV1/pigeon/Japan/Utsunomiya/95 | JP/Utsunomiya-pg/95 | VI | M | ——K——— | AB074020 |
| APMV1/chicken/Japan/Tokyo/96 | JP/Tokyo/96 | VII | V | —————— | AB074021 |
| APMV1/pigeon/Japan/Shiga/96 | JP/Shiga-pg/96 | VI | M | ——K——— | AB074022 |
| APMV1/pigeon/Japan/Fukushima/96 | JP/Fukushima-pg/96 | VI | M | ——K——— | AB074023 |
| APMV1/pheasant/Japan/Ibaraki/97 | JP/Ibaraki-ph/97 | VII | V | —————— | AB074024 |
| APMV1/parakeet/Japan/Chiba/97 | JP/Chiba-pa/97 | VI | V | —————— | AB074025 |
| APMV1/pigeon/Japan/Saitama/97 | JP/Saitama-pg/97 | VI | M | ——K——— | AB074026 |
| APMV1/chicken/Japan/Chiba-222/99 | JP/Chiba-222/99 | VII | V | —————— | AB074027 |
| APMV1/chicken/Japan/Chiba-223/99 | JP/Chiba-223/99 | VII | V | —————— | AB074028 |
| APMV1/chicken/Japan/Chiba-224/99 | JP/Chiba-224/99 | VII | V | —————— | AB074029 |
| APMV1/chicken/Japan/Ibaraki-1/99 | JP/Ibaraki-1/99 | VII | V | —————— | AB074030 |
| APMV1/chicken/Japan/Ibaraki-2/99 | JP/Ibaraki-2/99 | VII | V | —————— | AB074031 |
| APMV1/chicken/Japan/Kanagawa/99 | JP/Kanagawa/99 | VII | V | —————— | AB074032 |
| APMV1/chicken/Japan/Chiba/2000 | JP/Chiba/2000 | VII | V | —————— | AB074033 |
| APMV1/pigeonJapan/Gunma/2000 | JP/Gunma-pg/2000 | VI | M | ——K——— | AB074034 |
| APMV1/chicken/Japan/Ibaraki/2000 | JP/Ibaraki/2000 | VII | V | —————— | AB074035 |
| APMV1/chicken/Japan/Ibaraki/2000 | JP/Ibaraki-254/2001 | VII | V | —————— | AB074036 |
| APMV1/chicken/Japan/Ibaraki/2000 | JP/Ibaraki-258/2001 | VII | V | —————— | AB074037 |
| APMV1/chicken/Japan/Ibaraki/2000 | JP/Ibaraki-266/2001 | VII | V | —————— | AB074038 |
| APMV1/quail/Japan/Ibaraki/2000 | JP/Chiba-qa/2001 | VII | V | —————— | AB074039 |
| APMV1/chicken/Japan/Ibaraki/2000 | JP/Ibaraki-16/2001 | VII | V | —————— | AB074040 |
| APMV1/chicken/Japan/Ibaraki/2000 | JP/Ibaraki-17/2001 | VII | V | —————— | AB074041 |
| APMV1/chicken/Japan/Gunma/2000 | JP/Gunma/2001 | VII | V | —————— | AB074042 |
V, velogenic; M, mesogenic; L, lentogenic.
Genetic identity with consensus is marked with dashes.
Viral RNA was extracted directly from infected allantoic fluid using a commercial kit (ISOGEN; Nippon Gene, Tokyo, Japan). Afterwards, the reverse transcriptase reaction was carried out with Superscript II reverse transcriptase (Life Technologies, Gaithersburg, Md.) with random 9-mers, and the cDNA was amplified by PCR. The first quarter of the coding region of the fusion (F) gene, which includes important structures such as the cleavage site (11, 23, 37), was selected for this analysis. A region comprising the 3′ end of the matrix (M) gene and the 5′ end of the F gene was amplified for sequencing in either one or two steps. We designed two sets of PCR primers based on a comparison of the M and F gene sequences of several known NDV strains (34, 38). In the first step, a 921-bp primary product was obtained (between position 914 of the M gene and position 592 of the F gene) by using external primers M1, 5′ TTC-TCT-AGC-AGT-GGG-ACA-GC 3′ (nucleotides [nt] 914 to 933 of the M gene; sense), and F1, 5′ CAT-CTT-CCC-AAC-TGC-CAC-TG 3′ (nt 592 to 573 of the F gene; antisense). If not enough products were obtained, then a second amplification was performed as the second step. In that case, a 766-bp product was obtained using the internal primers M2, 5′ TGG-AGC-CAA-ACC-CGC-ACC-TGC-GG 3′ (nt 980 to 1003 of the M gene; sense), and F2, 5′ GGA-GGA-TGT-TGG-CAG-CAT-T 3′ (nt 503 to 485 of the F gene; antisense), and was synthesized in the same way as in the first amplification.
The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. The purified PCR products were used as templates for sequencing on an Applied Biosystems 373S automated DNA sequencer using a cycle sequencing dye terminator chemistry kit (Perkins-Elmer/Applied Biosystems, Foster City, Calif.). The purified PCR products were sequenced from both directions.
The determined nucleotide sequences were analyzed with the GENETYX-Mac program (version 10.0; Software Development Corp., Tokyo, Japan). The phylogenetic analysis was conducted with the Clustal X program (36), and the tree based on the nucleotide sequences from a portion (nt 47 to 420) of the F gene were constructed by the neighbor-joining method (29).
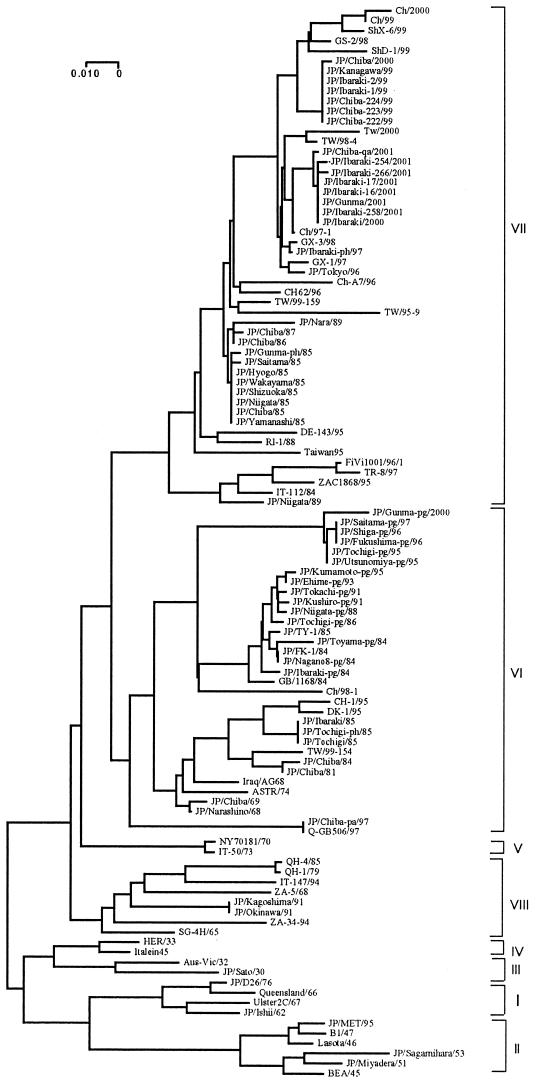
The 61 NDV isolates collected in Japan from 1930 to 2001 were classified into six clusters, and within these, the presence of viruses belonging to six of the eight known genotypes was revealed (Table 1) (Fig. 1). Most of the NDVs isolated in Japan before 1985 corresponded to genotypes from the previous worldwide panzootics (II, III, and VI). One lentogenic chicken isolate (JP/Ishii/62) (17) was classified as NDV genotype I. However, the more recently isolated (in 1995) lentogenic chicken isolate (JP/MET/95) (25) was classified as NDV genotype II. This isolate may be a derivative of the B1/47 strain, to which it shows high genetic similarity (98.4%), like the lentogenic viruses which are spreading in other countries (19, 24, 41).
FIG. 1.
Phylogenetic tree of NDV strains based on nucleotide sequences from a portion (nt 47 to 420) of the F genes. The sequences of the strains from other regions were obtained from GenBank (for the accession numbers, see references 13, 14, 18, 21, 23, 43, and 44). Horizontal distances are proportional to the minimum number of nucleotide differences required to join nodes and sequences. The provisional designations of the genotypes are indicated on the right.
After 1985, NDV genotype VII began to replace genotype VI as the most prevalent genotype in domestic fowl in Japan. The NDVs from the outbreaks in domestic fowl in Japan from the mid-1980s to the present belong mainly to NDV genotype VII, like those found in Taiwan (18, 43, 44). However, NDVs of genotype VIII were found in Japan in 1991. The Japanese genotype VIII NDV isolates showed a 93.9% sequence homology to the SG-4H/65 strain isolated in Singapore. This genotype was mainly enzootic in Southern Africa (13) and has not been reported in neighboring countries such as Taiwan, so we speculate that this genotype VIII NDV originated in Southeast Asia and was then introduced into Japan by an unknown route.
To elucidate the close relationship between many strains isolated from other regions and Japanese strains, it is interesting to examine the recent isolate from parakeets (JP/Chiba-pa/97) that we have been studying. This strain was isolated from parakeets imported from Pakistan in 1997. This strain and the Q-GB506/97 strain were isolated at the same time (2), and both strains fell into the same cluster of NDV genotype VI, together with the IT-148/94 strain isolated in Italy as an exotic isolate (14). This clearly suggested that imported caged birds have contributed to the distribution of NDV, including the outbreaks involving NDV genotype V, which was introduced by imported infected psittacines (39). Many NDVs have been isolated from caged birds worldwide (4, 5, 8, 15, 28, 35). Since the trading of these birds across regional and international boundaries is extensive (over 400,000 nonpoultry birds have been imported into Japan each year for the last 5 years), the risk of worldwide dissemination of potentially virulent NDVs is considerable. Researchers have speculated that the various types of NDV have been introduced via the importation of various kinds of birds, since NDV can persist in birds in an inapparent carrier state (5, 9, 10, 40).
NDV isolate JP/Ibraki-ph/97 was recently isolated from pheasants that were kept together for a while with partridges imported from China. The nucleotide sequence of this strain has a high similarity (>99%) to that of the GX-3/98 strain (21). The isolation of NDV from vaccinated flocks in China and Taiwan has been reported previously (44), and these isolates were found to be closely related genetically to recently isolated NDV from chickens in Japan. In Japan, however, recent outbreaks of ND were thought to be found mainly in flocks that had not been vaccinated or had been improperly vaccinated. Further analyses of the antigenicity of recently isolated NDV and the efficacy of the currently available vaccines are needed.
While the outbreaks in domestic fowl have been limited and sporadic, the outbreaks in pigeons have continued from 1984 to the present. All the Japanese NDVs isolated from pigeons from 1984 to 1995 fell into a subgroup of NDV genotype VI. A novel NDV subgroup of genotype VI has emerged and has been circulating in pigeons since 1995. Many isolates obtained from pigeons before 1995 have 112GRQKRF117 sequences at the cleavage site in the F protein gene, but all isolates obtained from pigeons after 1995 have 112RRKKRF117 sequences, which is typical of virulent NDV strains (6, 11, 37). This motif was a prerequisite for high virulence, but these isolates from pigeons, like recent German and Finnish isolates (16, 27, 42), were not so virulent according to the pathogenicity index test (data not shown). Therefore, some other property might be influencing the full expression of virulence.
It can be concluded from the present study that characteristic multiple genotypes of NDV are present in Japan. Since feral birds or captive birds transmit NDV across regional and international boundaries, multiple genotypes of NDV might have been introduced into Japan by such birds. Thus, the surveillance of NDV in various kinds of birds should be mandatory in order to improve our understanding of the epidemiology of NDV.
Nucleotide sequence accession numbers.
All sequences used in this study were sent to DDBJ and assigned accession numbers AB070382 to AB074042.
Acknowledgments
We thank the veterinary officials of Chiba, Ibaraki, Tochigi, Gunma, Saitama, Kanagawa, Tokyo, Niigata, Fukushima, Shizuoka, Yamanashi, Nagano, Toyama, Shiga, Hyogo, Wakayama, Nara, Ehime, Kumamoto, Kagoshima, Okinawa, and Hokkaido prefectures for their cooperation in the collection of the viral samples. We are also grateful to Y. Murakawa (The Chemo-Sero-Therapeutic Research Institute, Kumamoto, Japan) for providing us with the APMV1/Japan/MET/95 strain.
REFERENCES
- 1.Alexander, D. J. 1991. Newcastle disease and other paramyxovirus infections, p. 496-519. In B. W. Calnek, H. J. Barnes, C. W. Beard, W. M. Reid, and H. W. Yoder, Jr. (ed.), Diseases of poultry. Iowa State University Press, Ames.
- 2.Alexander, D. J., J. Banks, M. S. Collins, R. J. Manvell, K. M. Frost, E. C. Speidel, and E. W. Aldous. 1999. Antigenic and genetic characterization of Newcastle disease viruses isolated from outbreaks in domestic fowl and turkeys in Great Britain during 1997. Vet. Rec. 145:417-421. [DOI] [PubMed] [Google Scholar]
- 3.Ballagi-Pordany, A., E. Wehmann, J. Herczeg, S. Belak, and B. Lomniczi. 1996. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch. Virol. 141:243-261. [DOI] [PubMed] [Google Scholar]
- 4.Brunung-Fann, C., J. Kaneene, and J. Heamon. 1992. Investigation of an outbreak of velogenic viscerotropic Newcastle disease in pet birds in Michigan, Indiana, Illinois, and Texas. J. Am. Vet. Med. Assoc. 201:1709-1714. [PubMed] [Google Scholar]
- 5.Clavijo, A., Y. Robinson, T. Booth, and F. Munroe. 2000. Velogenic Newcastle disease in imported caged birds. Can. Vet. J. 41:404-406. [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, M. S., I. Strong, and D. J. Alexander. 1994. Evaluation of the molecular basis of pathogenicity of the variant Newcastle disease viruses termed “pigeon PMV-1 viruses.” Arch. Virol. 134:403-411. [DOI] [PubMed] [Google Scholar]
- 7.Collins, M. S., I. Strong, and D. J. Alexander. 1996. Pathogenicity and phylogenetic evaluation of the variant Newcastle disease viruses termed “pigeon PMV-1 viruses” based on the nucleotide sequence of the fusion protein gene. Arch. Virol. 141:635-647. [DOI] [PubMed] [Google Scholar]
- 8.Eaves, F. W., and T. M. Grimes. 1978. The isolation and characterization of a Newcastle disease virus from an exotic parrot. Aust. Vet. J. 54:534-537. [DOI] [PubMed] [Google Scholar]
- 9.Erickson, G. A., C. J. Mare, G. A. Gustafson, L. D. Miller, S. J. Proctor, and E. A. Carbrey. 1977. Interactions between viscerotropic velogenic Newcastle diseases virus and pet birds of six species. I. Clinical and serologic responses, and viral excretion. Avian Dis. 21:642-654. [PubMed] [Google Scholar]
- 10.Erickson, G. A., M. Brugh, and C. W. Beard. 1980. Viscerotropic velogenic Newcastle disease in pigeons: clinical disease and immunization. Avian Dis. 24:257-267. [Google Scholar]
- 11.Glickman, R. L., R. J. Syddall, R. M. Iorio, J. P. Sheehan, and M. A. Bratt. 1988. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J. Virol. 62:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson, R. P. 1980. Newcastle disease, p. 63-66. In S. Hitchner, H. G. Purchase, and J. E. Williams (ed.), Isolation and identification of avian pathogens. American Association of Avian Pathologists, Kennett Square, Pa.
- 13.Herczeg, J., E. Wehmann, R. R. Bragg, P. M. Travassos Dias, G. Hadjiev, O. Werner, and B. Lomniczi. 1999. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch. Virol. 144:2087-2099. [DOI] [PubMed] [Google Scholar]
- 14.Herczeg, J., S. Pascucci, P. Massi, M. Luini, L. Selli, I. Capua, and B. Lomniczi. 2001. A longitudinal study of velogenic Newcastle disease virus genotypes isolated in Italy between 1960 and 2000. Avian Pathol. 30:163-168. [DOI] [PubMed] [Google Scholar]
- 15.Hirai, K., T. Yamashita, H. Sawa, M. Takahashi, S. Shimakura, T. Masegi, and M. Inoue. 1980. Isolation of Newcastle disease virus from imported parrots (Katakoe sulpurea). J. Vet. Med. Sci. 42:381-385. [DOI] [PubMed] [Google Scholar]
- 16.Huovilainen, A., C. Ek-Kommone, R. Manvell, and L. Kinnunen. 2001. Phylogenetic analysis of avian paramyxovirus 1 strains isolated in Finland. Arch. Virol. 146:1775-1785. [DOI] [PubMed] [Google Scholar]
- 17.Ishida, M., K. Nerome, M. Matsumoto, T. Mikami, and A. Oya. 1985. Characterization of reference strains of Newcastle disease virus (NDV) and NDV-like isolates by monoclonal antibodies to HN subunits. Arch. Virol. 85:109-121. [DOI] [PubMed] [Google Scholar]
- 18.Ke, G. M., H. J. Liu, M. Y. Lin, J. H. Chen, S. S. Tsai, and P. C. Chang. 2001. Molecular characterization of Newcastle disease viruses isolated from recent outbreaks in Taiwan. J. Virol. Methods 97:1-11. [DOI] [PubMed] [Google Scholar]
- 19.King, D. J., and B. S. Seal. 1998. Biological and molecular characterization of Newcastle disease virus (NDV) field isolates with comparisons to reference NDV strains. Avian Dis. 42:507-516. [PubMed] [Google Scholar]
- 20.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 557-604. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven, Philadelphia, Pa.
- 21.Liang, R., D. J. Cao, J. Q. Li, J. Chen, X. Guo, F. F. Zhuang, and M. X. Duan. 2002. Newcastle disease outbreaks in western China were caused by the genotypes VIIa and VIII. Vet. Microbiol. 87:193-203. [DOI] [PubMed] [Google Scholar]
- 22.Lomniczi, B. 1973. Studies on interferon production and interferon sensitivity of different strains of Newcastle disease virus. J. Gen. Virol. 21:305-313. [DOI] [PubMed] [Google Scholar]
- 23.Lomniczi, B., E. Wehmann, J. Herczeg, A. Ballagi-Pordany, E. F. Kaleta, O. Werner, G. Meulemans, P. H. Jorgensen, A. P. Mante, A. L. Gielkens, I. Capua, and J. Damoser. 1998. Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII). Arch. Virol. 143:49-64. [DOI] [PubMed] [Google Scholar]
- 24.Marin, M. C., P. Villegas, J. D. Bennett, and B. S. Seal. 1996. Virus characterization and sequence of the fusion protein gene cleavage site of recent Newcastle disease virus field isolates from the southeastern United States and Puerto Rico. Avian Dis. 40:382-390. [PubMed] [Google Scholar]
- 25.Murakawa, Y., K. Takase, K. Sakamoto, M. Suesoshi, and H. Nagatomo. 2000. Characterization of a lentogenic Newcastle disease virus isolated from broiler chickens in Japan. Avian Dis. 44:686-690. [PubMed] [Google Scholar]
- 26.Nakamura, J., S. Oyama, K. Fukusyo, and N. Tomonaga. 1933. Vergleichende immunobiologische untersuchengen des Korea-Huhenerseuchen-virus und des Japnischen geflugel-pestivirus zuglech uber die beziehung zum virus der Newcastle disease. J. Jpn. Vet. Sci. 12:135-146. [Google Scholar]
- 27.Oberdorfer, A., and O. Werner. 1998. Newcastle disease virus: detection and characterization by PCR of recent German isolates differing in pathogenicity. Avian Pathol. 27:237-243. [DOI] [PubMed] [Google Scholar]
- 28.Panigrahy, B., D. A. Senne, J. E. Pearson, M. A. Mixson, and D. R. Cassidy. 1993. Occurrence of velogenic viscerotropic Newcastle disease in pet and exotic birds in 1991. Avian Dis. 37:254-258. [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sakaguchi, T., T. Toyoda, B. Gotoh, N. M. Inocencio, K. Kuma, T. Miyata, and Y. Nagai. 1989. Newcastle disease virus evolution. I. Multiple lineages defined by sequence variability of the hemagglutinin-neuraminidase gene. Virology 169:260-272. [DOI] [PubMed] [Google Scholar]
- 31.Seal, B. S., D. J. King, and J. D. Bennett. 1995. Characterization of Newcastle disease virus isolates by reverse transcription-PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J. Clin. Microbiol. 33:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seal, B. S. 1996. Analysis of matrix protein gene nucleotide sequence diversity among Newcastle disease virus isolates demonstrates that recent disease outbreaks are caused by viruses of psittacine origin. Virus Genes 11:217-224. [DOI] [PubMed] [Google Scholar]
- 33.Seal, B. S., D. J. King, D. P. Locke, D. A. Senne, and M. W. Jackwood. 1998. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J. Clin. Microbiol. 36:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seal, B. S., D. J. King, and R. J. Meinersmann. 2000. Molecular evolution of the Newcastle disease virus matrix protein gene and phylogenetic relationships among the paramyxoviridae. Virus Res. 66:1-11. [DOI] [PubMed] [Google Scholar]
- 35.Senne, D. A., J. E. Pearson, L. D. Miller, and G. A. Gustafson. 1983. Virus isolations from pet birds submitted for importation into the United States. Avian Dis. 27:731-744. [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyoda, T., T. Sakaguchi, K. Imai, N. M. Inocencio, B. Gotoh, M. Hamaguchi, and Y. Nagai. 1987. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology 158:242-247. [DOI] [PubMed] [Google Scholar]
- 38.Toyoda, T., T. Sakaguchi, H. Hirota, B. Gotoh, K. Kuma, T. Miyata, and Y. Nagai. 1989. Newcastle disease virus evolution. II. Lack of gene recombination in generating virulent and avirulent strains. Virology 169:273-282. [DOI] [PubMed] [Google Scholar]
- 39.Utterback, W. W., and J. H. Schwartz. 1973. Epizootiology of velogenic viscerotropic Newcastle disease in southern California, 1971-1973. J. Am. Vet. Med. Assoc. 163:1080-1088. [PubMed] [Google Scholar]
- 40.Vickers, M. L., and R. P. Hanson. 1979. Experimental Newcastle disease virus infections in three species of wild birds. Avian Dis. 23:70-79. [PubMed] [Google Scholar]
- 41.Wehmann, E., J. Herczeg, J. Tanyi, E. Nagy, and B. Lomniczi. 1999. Lentogenic field isolates of Newcastle disease virus isolated in Canada and Hungary are identical with the vaccine type used in the region. Avian Pathol. 28:6-12. [DOI] [PubMed] [Google Scholar]
- 42.Werner, O., A. Romer-Oberdorfer, B. Kollner, R. J. Manvell, and D. J. Alexander. 1999. Characterization of avian paramyxovirus type I strains isolated in Germany during 1992 to 1996. Avian Pathol. 28:79-88. [DOI] [PubMed] [Google Scholar]
- 43.Yang, C. Y., H. K. Shieh, Y. L. Lin, and P. C. Chang. 1999. Newcastle disease virus isolated from recent outbreaks in Taiwan phylogenetically related to viruses (genotype VII) from recent outbreaks in western Europe. Avian Dis. 43:125-130. [PubMed] [Google Scholar]
- 44.Yu, L., Z. Wang, Y. Jiang, L. Chang, and J. Kwang. 2001. Characterization of newly emerging Newcastle disease virus isolates from the People's Republic of China and Taiwan. J. Clin. Microbiol. 39:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]