Abstract
We determined the sequence of the intergenic spacer (IGS) 1 region, which is located between the 26S and 5S rRNA genes, in 25 species of the genus Trichosporon. IGS 1 sequences varied in length from 195 to 719 bp. Comparative sequence analysis suggested that the divergence of IGS 1 sequences has been greater than that of the internal transcribed spacer regions. We also identified five genotypes of T. asahii, which is a major causative agent of deep-seated trichosporonosis, based on the IGS 1 sequences of 43 strains. Most of the isolates that originated in Japan were of genotype 1, whereas the American isolates were of genotype 3 or 5. Our results suggest that analysis of IGS regions provides a powerful method to distinguish between phylogenetically closely related species and that a geographic substructure may exist among T. asahii clinical isolates.
Fungal rRNA genes are tandemly repeated, with each repeat encoding 18S (small-subunit), 5.8S, and 26S (large-subunit) genes. Two other regions exist in each repeat: the internal transcribed spacer (ITS) region and the intergenic spacer (IGS) region (Fig. 1). Ribosomal DNA (rDNA) has been widely utilized for molecular systematics and the identification of microorganisms. The D1/D2 regions of 26S and ITS sequences have been used mainly to identify pathogenic fungi. At present, the 26S rDNA sequences of almost all yeasts, including nonpathogenic species, have been determined (3, 7, 8). The analysis of ITS sequences has been carried out mainly for pathogenic yeast species (1, 5, 9, 10, 16, 19). Peterson and Kurtzman (13) and Sugita et al. (16) demonstrated that a single species showed less than 1% dissimilarity in either the ITS region or D1/D2 26S rDNA. However, these sequence analyses are sometimes incapable of distinguishing between phylogenetically closely related species, such as the three varieties of Cryptococcus neoformans. Although three varieties within a single species can be distinguished for each varietal level by ITS sequence analysis, the distinction is based on differences of only three or four nucleotides (20). Recently, Diaz et al. (2) and Sugita et al. (17) demonstrated that three varieties of C. neoformans were more clearly distinguished by analysis of IGS 1 and IGS 2 sequences than by ITS sequence analysis. Therefore, IGS sequence analysis appears to be a powerful tool for differentiating between phylogenetically very closely related species.
FIG. 1.
Schematic representation of the rDNA locus in Trichosporon. Boxes indicate coding regions.
The genus Trichosporon currently includes 25 species of basidiomycetous yeasts. Eight of these species are implicated in infectious or allergic diseases. T. asahii, T. asteroides, T. cutaneum, T. inkin, T. mucoides, and T. ovoides are involved in deep-seated or superficial infections (4, 6, 14), and T. asahii, T. domesticum, T. montevideense, and T. mucoides are associated with the allergic disease of summer-type hypersensitivity pneumonitis (12, 15). We have previously presented data on ITS sequences for the molecular identification of all members of the genus Trichosporon. However, this region is so highly homologous across the species that the genus Trichosporon may be considered phylogenetically monophyletic. Consequently, the differentiation of Trichosporon species requires the analysis of genes or regions that have greater divergence than the ITS. This paper describes the application of IGS sequence analysis to the identification of pathogenic species of Trichosporon.
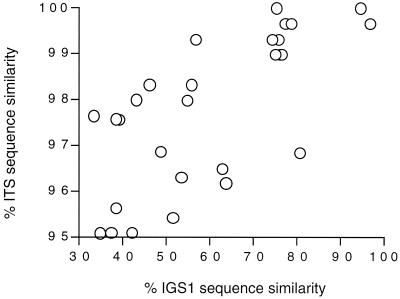
The currently accepted 25 species of the genus Trichosporon were examined as shown in Table 1 Trichosporon DNA was extracted by the method of Makimura et al. (11). The IGS 1 region was amplified by PCR using the following oligonucleotide primers: 26SF, 5′-ATCCTTTGCAGACGACTTGA-3′, and 5SR, 5′-AGCTTGACTTCGCAGATCGG-3′. PCR was performed in a Thermocycler (model 9700; Applied Biosystems, Foster City, Calif.) with an initial 3-min denaturation at 94°C, followed by 30 cycles that consisted of 30 s at 94°C, 30 s at 57°C, and 1 min at 72°C, and a final 10-min extension at 72°C. The PCR products were sequenced with the 26SF and 5SR primers by using the ABI 310 DNA sequencer with an ABI PRISM BigDye Terminator Cycle Sequencing kit (Applied Biosystems) according to the manufacturer's instructions. The lengths of the IGS 1 sequences of 24 Trichosporon species and their respective DDBJ accession numbers are listed in Table 1. The IGS 1 sequences ranged in length from 195 to 719 bp. For some unknown reason, the IGS 1 region of T. loubieri could not be amplified. Figure 2 shows a plot of the sequence similarities in the IGS and ITS regions for pairwise alignments between different species in the genus Trichosporon. The 99% similarity in ITS sequences observed between two species corresponds to approximately 55 to 95% IGS 1 sequence similarity. The 98% ITS sequence similarity in another pairwise comparison corresponds to approximately 45 to 55% IGS 1 sequence similarity. For example, T. asahii (GenBank accession no. AB018013), which is responsible for deep-seated infections, and T. asteroides (AB018017), which is associated with superficial infections, are 98.9% (295 out of 298 bp) similar in their ITS sequences. The similarity in the ITS region between T. asahii and the nonpathogenic species T. coremiiforme (AB018015) is 99.7% (297 out of 298 bp). However, within the IGS 1 region, T. asahii demonstrates 75.1 and 78.8% similarities to T. asteroides and T. coremiiforme, respectively. In addition, since the ITS sequences of T. domesticum and T. montevideense, which are the causative agents of summer-type hypersensitivity pneumonitis, are identical, these species could not be distinguished from one another by ITS sequence analysis (16). However, IGS sequence analysis of these two species reveals 94.6% sequence similarity. It is also noteworthy that the length of the ITS region, including the 5.8S region, ranges from 445 to 470 bp, while that of the IGS 1 region ranges from 195 to 704 bp. Since the members of the genus Trichosporon are phylogenetically very closely related, it appears that IGS sequence analysis is superior to ITS sequence analysis in differentiating Trichosporon species. The IGS sequence is divided into the IGS 1 and 2 regions. The complete IGS sequences of C. neoformans (L27078 and L27079) have been determined, and the IGS 1 and IGS 2 sequences of C. neoformans var. neoformans and C. neoformans var. gattii are 68.1 and 84.2% similar, respectively. The Malassezia IGS 1 sequences are also more divergent than the IGS 2 sequences (unpublished data). We have not yet sequenced the IGS 2 of Trichosporon species, but preliminary results suggest that IGS 1 is more suitable than IGS 2 for the differentiation of phylogenetically closely related species.
TABLE 1.
Strains used and their IGS1 nucleotide sequence accession numbers
| Species | Straina | Source | Length of IGS 1 region (bp) | Accession no. |
|---|---|---|---|---|
| Trichosporon asahii genotype I | CBS 2479T | Skin, Japan | 485 | AB066386 |
| M 9406 | Urine, Japan | 485 | AB066392 | |
| M 9415 | Lung, Japan | 485 | AB066382 | |
| M 9416 | Lung, Japan | 485 | AB066381 | |
| M 9417 | Lung, Japan | 485 | AB066380 | |
| M 9426 | Feces, Japan | 485 | AB066376 | |
| M 9432 | Urine, Japan | 485 | AB066375 | |
| M 9470 | Blood, Japan | 485 | AB066384 | |
| M 9474 | Blood, Japan | 485 | AB066379 | |
| M 9475 | Intestinal fluids, Japan | 485 | AB066393 | |
| M 9483 | Blood, Japan | 485 | AB066389 | |
| M 9485 | Blood, Japan | 485 | AB066388 | |
| M 9486 | Blood, Japan | 485 | AB066390 | |
| M 9496 | Blood, Japan | 485 | AB066387 | |
| M 9927 | Urine, Japan | 485 | AB066377 | |
| M 9928 | Blood, Japan | 485 | AB066383 | |
| M 9929 | Urine, Japan | 485 | AB066391 | |
| M 9930 | Urine, Japan | 485 | AB066378 | |
| M 9937 | Blood, Japan | 485 | AB072602 | |
| M 9938 | Blood, Japan | 485 | AB072601 | |
| M 9939 | Blood, Japan | 485 | AB072599 | |
| M 9940 | Blood, Japan | 485 | AB072603 | |
| M 9947 | Blood, Japan | 485 | AB072604 | |
| M 9948 | Blood, Japan | 485 | AB072605 | |
| M 9950 | Blood, Japan | 485 | AB072600 | |
| M 9951 | Urine, Japan | 485 | AB066385 | |
| Trichosporon asahii genotype 2 | M 9475 | Blood, Japan | 485 | AB072606 |
| Trichosporon asahii genotype 3 | CBS 2530 | Mus musculus, Brazil | 490 | AB066397 |
| CBS 4829 | Feces, Brazil | 490 | AB071385 | |
| M 9402 | Blood, USAb | 490 | AB066396 | |
| M 9403 | Blood, USA | 490 | AB071383 | |
| M 9941 | Blood, USA | 490 | AB072607 | |
| M 9942 | Blood, USA | 490 | AB072608 | |
| M 9943 | Blood, USA | 490 | AB072609 | |
| M 9944 | Blood, USA | 490 | AB072610 | |
| M 9945 | Blood, USA | 490 | AB072611 | |
| Trichosporon asahii genotype 4 | M 9474 | Blood, Japan | 485 | AB066399 |
| M 9949 | Blood, Japan | 485 | AB072612 | |
| Trichosporon asahii genotype 5 | M 9410 | Feces, USA | 490 | AB066401 |
| M 9411 | Sputum, USA | 490 | AB071384 | |
| M 9433 | Urine, Japan | 490 | AB066402 | |
| M 9935 | Blood, USA | 490 | AB071386 | |
| M 9936 | Blood, USA | 490 | AB071387 | |
| Trichosporon aquatile | CBS 5973T | Water | 359 | AB066403 |
| CBS 5988 | Water | 359 | AB066404 | |
| Trichosporon asteroides | CBS 2481T | Skin | 466 | AB066405 |
| Trichosporon coremiiforme | CBS 2482T | Lesion on head | 478 | AB066406 |
| M 9926 | Soil | 478 | AB066409 | |
| M 9932 | Soil | 478 | AB066407 | |
| M 9933 | Soil | 478 | AB066408 | |
| M 9934 | Soil | 478 | AB066410 | |
| Trichosporon debeurumanianum | CBS 1896T | Bronchial secretion | 483 | AB066411 |
| Trichosporon dermatis | CBS 2043T | Skin | 357 | AB066412 |
| M9946 | House | 357 | AB072613 | |
| Trichosporon faecale | CBS 4828T | Feces | 490 | AB066413 |
| Trichosporon brassicae | CBS 6382T | Cabbage | 385 | AB066414 |
| Trichosporon cutaneum | CBS 2466T | Skin lesion | 331 | AB066415 |
| Trichosporon domesticum | M 9401T | House | 705 | AB066416 |
| M 9421 | House | 705 | AB066418 | |
| M 9814 | Cat | 705 | AB066417 | |
| Trichosporon dulcitum | CBS 8257T | Soil | 542 | AB066419 |
| CBS 5785 | Toadstool | 542 | AB066420 | |
| Trichosporon gracile | CBS 8189T | Sour milk | 523 | AB066421 |
| CBS 8193 | Teal | 523 | AB066422 | |
| Trichosporon guehoae | CBS 8521T | Soil | 268 | AB066423 |
| Trichosporon inkin | CBS 5585T | Skin | 489 | AB066424 |
| CBS 7629 | Urine | 489 | AB066425 | |
| Trichosporon japonicum | JCM 8357T | Air | 470 | AB066426 |
| Trichosporon jirovecii | CBS 6864T | Toenail | 328 | AB066427 |
| Trichosporon laibachii | CBS 5790T | Soil | ||
| CBS 2495 | Feces of Rattus rattus, authentic strain of T. multisporum | |||
| Trichosporon loubieri | CBS 7065T | Cow with mastitis | 430 | AB066428 |
| Trichosporon moniliiforme | CBS 2467T | Curdling milk | 436 | AB066429 |
| M 9813 | Bird dropping | 436 | AB066430 | |
| Trichosporon montevideense | CBS 6721T | Water purification tank | 719 | AB066431 |
| CBS 8261 | Feces | 719 | AB066432 | |
| Trichosporon mucoides | CBS 7625T | Meningitis patient | 357 | AB066433 |
| Trichosporon ovoides | CBS 7556T | Scalp | 498 | AB066434 |
| Trichosporon porosum | CBS 2040T | Exudate of yew tree | 228 | AB066435 |
| M 9481 | Soil | 228 | AB066436 | |
| M 9931 | Soil | 228 | AB066437 | |
| Trichosporon sporotrichoides | CBS 8246T | Soil | 302 | AB066438 |
| Trichosporon veenhuisii | CBS 7136T | Buffalo dung | 401 | AB066439 |
| Trichosporon sp. | CBS 8645 | Moist humus, around roots | 195 | AB066440 |
CBS, Centraalbureau voor Schimmelcultures, Delft, The Netherlands; JCM, Japan Collection of Microorganisms, Saitama, Japan; M, Meiji Pharmaceutical University, Tokyo, Japan.\
USA, United States.
FIG. 2.
Sequence similarities between IGS 1 and ITS regions “% ITS sequence similarity” indicates similarity between combined ITS 1 and ITS 2 sequences.
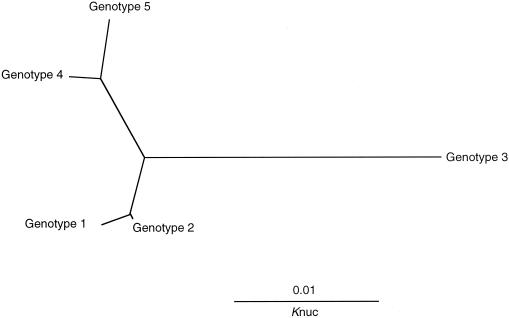
Forty-three isolates of T. asahii, which is the major cause of deep-seated trichosporonosis, were obtained from various sources and geographic locations (Japan and the United States) and analyzed (Table 1). The IGS 1 sequences ranged in length from 485 to 490 bp and were divided into five genotypes (Fig. 3). The genotypes shared between 95.1 and 98.8% similarity. Of the isolates that originated in Japan, 26 of 30 (87%) were genotype 1, while all 13 isolates from the United States were either genotype 3 or genotype 5. Genotypes 2 and 4 were found in only three isolates from Japan. No genotype 1 strains were found among the American isolates. Diaz et al. (2) found a geographic substructure among strains of C. neoformans var. gattii. Of the three genotypes, two corresponded to strains found in the United States, and the third represented Asian strains. We also found a correlation between the serotypes and genotypes of C. neoformans var. gattii strains in an analysis of both the IGS 1 and IGS 2 regions (17). Of the three genotypes, two consisted solely of serotype B strains, while the third consisted of both serotype B and serotype C strains. Although our study dealt with a limited number of strains, the IGS sequence analysis suggests that there is a correlation between the genotype and the geographical substructure of the T. asahii clinical isolates. Unfortunately, we could not obtain T. asahii clinical isolates from European countries. A comparison of the genotypes of strains from Europe should prove interesting.
FIG. 3.
A phylogenetic tree of five IGS 1 genotypes of T. asahii. The sequences were aligned using CLUSTAL W (version 1.8) software (18), and the tree was constructed using TreeView (version 1.6.2) (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
We examined the IGS sequences of all members of the genus Trichosporon and concluded that IGS sequence analysis was superior to ITS sequence analysis in differentiating phylogenetically closely related species. IGS sequence analysis also shows great potential as a new epidemiological tool.
Acknowledgments
We thank the physicians who provided the clinical isolates of T. asahii.
This study was supported in part by a Grant for the Promotion of the Advancement of Education and Research in Graduate Schools by the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barret, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz, M. R., T. Boekhout, B. Theelen, and J. W. Fell. 2000. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast, Cryptococcus neoformans. Syst. Appl. Microbiol. 23:535-545. [DOI] [PubMed] [Google Scholar]
- 3.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. E vol. Microbiol. 50:1351-1571. [DOI] [PubMed] [Google Scholar]
- 4.Guého, E., L. Improvisi, G. S. de Hoog, and B. Dupont. 1994. Trichosporon on humans: a practical account. Mycoses 37:3-10. [DOI] [PubMed] [Google Scholar]
- 5.Gupta, A. K., Y. Kohli, and R. C. Summerbell. 2000. Molecular differentiation of seven Malassezia species. J. Clin. Microbiol. 38:1869-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbrecht, R., H. Koening, K. Waller, L. Liu, and E. Guého. 1993. Trichosporon infections: clinical manifestations and treatment. J. Mycol. Med. 3:129-136. [Google Scholar]
- 7.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 9.Lott, T. J., B. M. Burns, R. Zancope-Oliveira, C. M. Elie, and E. Reiss. 1998. Sequence analysis of the internal transcribed spacer 2 (ITS2) from yeast species within the genus Candida. Curr. Microbiol. 36:63-69. [DOI] [PubMed] [Google Scholar]
- 10.Makimura. K., Y. Tamura, M. Kudo., K. Uchida, H. Saito, and H. Yamaguchi. 2000. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J. Med. Microbiol. 49:29-35. [DOI] [PubMed] [Google Scholar]
- 11.Makimura, K., S. Y. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 12.Nishiura, Y., K. Nakagawa-Yoshida, M. Suga, T. Shinoda, E. Guého, and M. Ando. 1997. Assignment and serotyping of Trichosporon species: the causative agents of summer-type hypersensitivity pneumonitis. J. Med. Vet. Mycol. 35:45-52. [DOI] [PubMed] [Google Scholar]
- 13.Peterson, S. W., and C. P. Kurtzman. 1991. Ribosomal RNA sequence divergence among sibling species of yeasts. Syst. Appl. Microbiol. 14:124-129. [Google Scholar]
- 14.Sugita, T., A. Nishikawa, T. Shinoda, and H. Kume. 1995. Taxonomic position of deep-seated, mucosa-associated, and superficial isolates of Trichosporon cutaneum from trichosporonosis patients. J. Clin. Microbiol. 33:1368-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugita, T., A. Nishikawa, T. Shinoda, K. Yoshida, and M. Ando. 1995. A new species, Trichosporon domesticum, isolated from the house of a summer-type hypersensitivity pneumonitis patient in Japan. J. Gen. Appl. Microbiol. 41:429-436. [Google Scholar]
- 16.Sugita, T., A. Nishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugita, T., R. Ikeda, and T. Shinoda. 2001. Diversity among strains of Cryptococcus neoformans var. gattii as revealed by sequence analysis of multiple genes and chemotype analysis of capsular polysaccharide. Microbiol. Immunol. 45:757-768. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu, J., R. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]