Abstract
To discern expression patterns of individual storage-protein genes in hexaploid wheat (Triticum aestivum cv Chinese Spring), we analyzed comprehensive expressed sequence tags (ESTs) of common wheat using a bioinformatics technique. The gene families for α/β-gliadins and low molecular-weight glutenin subunit were selected from the EST database. The alignment of these genes enabled us to trace the single nucleotide polymorphism sites among both genes. The combinations of single nucleotide polymorphisms allowed us to assign haplotypes into their homoeologous chromosomes by allele-specific PCR. Phylogenetic analysis of these genes showed that both storage-protein gene families rapidly diverged after differentiation of the three genomes (A, B, and D). Expression patterns of these genes were estimated based on the frequencies of ESTs. The storage-protein genes were expressed only during seed development stages. The α/β-gliadin genes exhibited two distinct expression patterns during the course of seed maturation: early expression and late expression. Although the early expression genes among the α/β-gliadin and low molecular-weight glutenin subunit genes showed similar expression patterns, and both genes from the D genome were preferentially expressed rather than those from the A or B genome, substantial expression of two early expression genes from the A genome was observed. The phylogenetic relationships of the genes and their expression patterns were not correlated. These lines of evidence suggest that expression of the two storage-protein genes is independently regulated, and that the α/β-gliadin genes possess novel regulation systems in addition to the prolamin box.
Comprehensive analyses of expressed sequence tags (ESTs) have been carried out among various plant species to provide powerful tools for functional genomics, such as DNA microarray, gene chips, databases for comparative genomics, and single nucleotide polymorphism (SNP) analysis (Ewing et al.,1999; Asamizu et al., 2000; Ogihara et al., 2003; Vettore et al., 2003; Asamizu et al., 2004; Sterky et al., 2004; Zhang et al., 2004; Pavy et al., 2005), even in plants whose complete genome sequences are not available. Wheat is one of the most important staple food crops in the world and is an appropriate model for functional genomics, as the largest EST sequences have been collected from this plant (dbEST, National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). Using this substantial EST database, we have developed an efficient method to distinguish transcripts from the individual homoeologous loci of hexaploid wheat (Triticum aestivum cv Chinese Spring [CS]; Mochida et al., 2003; Ogihara et al., 2003). This method includes (1) grouping large numbers of ESTs into contigs that correspond to homoeoloci, (2) efficient detection of SNP sites among the corresponding homoeoloci, and (3) digital display of expression patterns for individual homoeoloci in the wheat life cycle.
The gliadins and the glutenins are major components of the storage proteins in wheat endosperm. Wheat gluten is composed of a protein complex of monomeric gliadins and polymeric glutenins, and this complex also plays a substantial role in determining processed food quality (for review, see Shewry et al., 2003). Gliadin proteins, which are extracted into the alcohol-soluble fraction of gluten, were further separated into three groups based on electrophoretic mobility: α/β-gliadin, γ-gliadin, and ω-gliadin (Jackson et al., 1983). Glutenin proteins, which are extracted into alcohol-insoluble fractions of gluten, were classified as high molecular-weight glutenin subunits (HMW-GS) and low molecular-weight glutenin subunits (LMW-GS; Jackson et al., 1983). The chromosome loci of each protein fraction were determined. The α/β-gliadins are located on the Gli-2 loci of the short arm of the homoeologous group 6 chromosomes (Payne, 1987), the γ- and ω-gliadins are tightly linked and are located on the Gli-1/Gli-3 loci of the short arm of the homoeologous group 1 chromosomes (Payne et al., 1984), HMW-GS is located on the Glu-1 loci of the long arm of the homoeologus group 1 chromosomes (Payne et al., 1982), and LMW-GS is located on the Glu-3 loci of the short arm of the homoeologous group 1 chromosomes (Gupta and Shepherd, 1990). These genes comprise a multigene family in the wheat genome.
Estimated copy numbers of storage-protein genes in the hexaploid wheat genome differ among the cultivars from >100 (Okita et al., 1985) to 150 (Anderson et al., 1997) for the α/β-gliadins, 17 to 39 for the γ-gliadins (Sabelli and Shewry, 1991), 15 to 18 for the ω-gliadins (Sabelli and Shewry, 1991), 22 to 39 (Sabelli and Shewry, 1991) and 30 to 40 (Cassidy et al., 1998) for the LMW-GS genes, and >6 for the HMW-GS genes (Thompson et al., 1983). The high copy numbers of storage-protein genes complicate the elucidation of the expression and function of individual storage-protein genes. Although allelic variations in storage-protein genes have been shown to be major factor(s) in determining the properties of wheat flour (Gupta and Shepherd, 1990; Masci et al., 1998; D'Ovidio et al., 1999), functional analyses for expression of these multigenes are difficult using traditional methods.
Here, we carefully traced the expression patterns of individual genes encoding the α/β-gliadins and LMW-GS from hexaploid wheat, as obtained from a comprehensive EST database. The genes for the α/β-gliadins and LMW-GS were selected as multigene models in hexaploid wheat. Both the α/β-gliadins and LMW-GS are important for gluten quality, but the characterization of each multigene is not simple, because a number of multigenes are transcribed, translated, and modified posttranslationally in the seed maturation process (Shewry et al., 2003). Therefore, discriminating the expression patterns of individual seed storage-protein genes must be the first step in understanding the rapid evolution and complex expression system(s) of the multigene family. This information could contribute to breeding programs for improving gluten quality. We extracted ESTs homologous to genes for the α/β-gliadin and LMW-GS, and classified these into contigs corresponding to each homoeologous gene. Chromosome locations of these contigs were determined using an allele-specific PCR method (Moczulski and Salmanowicz, 2003; Zhang et al., 2003). We first confirmed that the α/β-gliadin genes showed two distinct expression patterns during the course of seed maturation, and that early expression genes among the α/β-gliadin and LMW-GS genes were preferentially expressed from the D genome during the seed maturation process. On the other hand, two late expression α/β-gliadin genes from the A genome were strongly expressed, although a certain number of other late expression α/β-gliadin genes were weakly expressed from the three genomes. The evolutionary relationships of the multigenes and their expression patterns are also discussed. The present functional genomics data should provide valuable information regarding the improvement of wheat flour quality.
RESULTS
Assembling Multigenes for α/β-Gliadin and LMW-GS
Through computer analysis, 361,180 ESTs derived from 32 cDNA libraries that confer more than 10,000 ESTs (Supplemental Table I) were collected and grouped into 53,976 contigs by the phrap method (University of Washington Genome Center; http://www.genome.washington.edu/UWGC). The phrap parameters were sufficiently strict to classify ESTs into each multigene from three homoeologous genomes (Mochida et al., 2003; Ogihara et al., 2003). The contigs homologous to genes encoding α/β-gliadin and LMW-GS were selected by BLASTN search against the 53,976 contigs. Consequently, 36 and 15 contigs homologous to α/β-gliadins and LMW-GS, respectively, were obtained as multigenes. Copy number of α/β-gliadin genes in CS wheat was estimated to be 60 by genomic Southern hybridization (Anderson et al., 1997), and thus approximately 60% of the α/β-gliadins in the CS genome were expressed. This supports a previous estimation that about half of the α/β-gliadin genes were pseudogenes (Anderson and Greene, 1997).
On the other hand, the number of genes for LMW-GS in hexaploid wheat was previously estimated to be 22 to 39 (Sabelli and Shewry, 1991) and 30 to 40 (Cassidy et al., 1998) by genomic Southern-hybridization analyses. ESTs homologous to LMW-GS were grouped into 15 contigs, and thus about half of the LMW-GS genes were expressed from three homoeologous genomes.
The number of EST members comprising each contig varied from two to 114 for α/β-gliadin and from one to 104 for LMW-GS. In total, the 36 α/β-gliadin contigs included 1,008 ESTs, and the 15 LMW-GS contigs included 468 ESTs. It was thus concluded that the α/β-gliadin gene expression is 2-fold higher than LMW-GS gene expression in terms of number of genes and expression levels.
Phylogenetic Analysis of α/β-Gliaidin and LMW-GS Genes and Chromosome Locations
To assess the phylogenetic relationships among the individual genes for both storage proteins, we aligned the coding regions of respective contigs with previously reported counterpart genes (Table I). Dendrograms showing the phylogenetic relationships of both genes were constructed using Clustal W (Thompson et al., 1994).
Table I.
α/β-Gliadin and LMW-GS genes registered in public DNA database
Asterisks indicate direct submission (A.E. Blechl and O.D. Anderson, direct submission to GenBank U08287, 1994).
α/β-Gliadin Genes
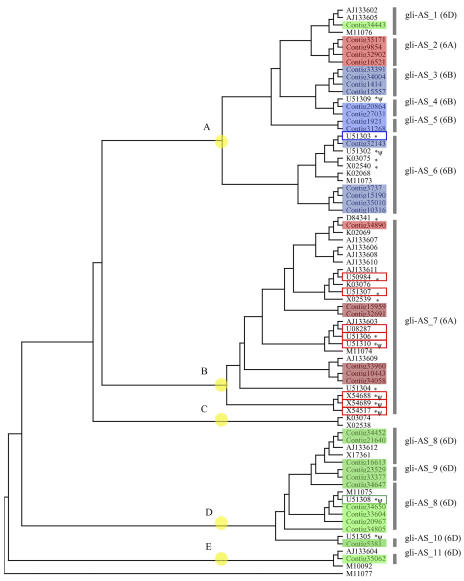
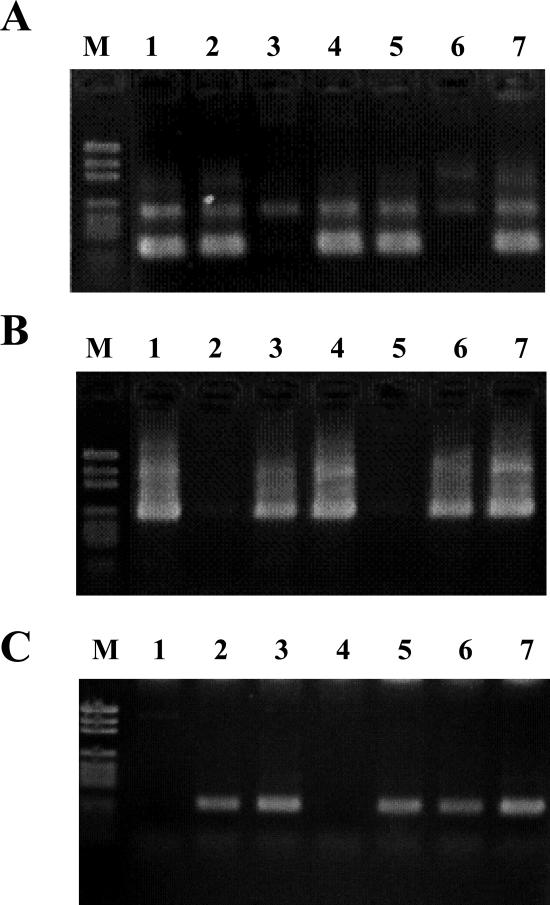
Because the α/β-gliadin genes comprise a multigene family and are highly variable (Gu et al., 2004), the approximately 380 bp at the 3′ site of the conserved coding region of the α/β-gliadin genes were used for dendrogram construction. The resultant dendrogram is shown in Figure 1. The expressed contigs were classified into five major groups. Each major group included contigs identified in this study and their counterparts registered in DNA databases, except for group C, which only contained singleton ESTs from the DNA databases. Novel classes of genes consisting of independent branches within the major classes were obtained in this study (Fig. 1), indicating the effectiveness of comprehensive EST analysis for accumulation of expressed genes. The chromosome locations of all contigs were determined by allele-specific PCR (Table II) in combination with aneuploids of CS wheat. Typical agarose gel electrophoresis patterns are depicted in Figure 2. All contigs were located on the short arm of chromosome group 6, indicating that the contigs are expressed from the Gli-2 loci (Payne, 1987). While the major group (group A) included genes expressed from three genomes, namely A, B, and D, the other groups (groups B, D, and E) harbored genes derived from one of three genomes (genome A for group B and genome D for groups D and E), as shown in Figure 1.
Figure 1.
Phylogenetic tree of α/β-gliadin genes. Accession numbers indicate the genes registered in the DDBJ (Table I). Numbers in open boxes indicate genes previously assigned to the chromosome. Contig numbers in colored boxes indicate genes identified by EST analysis in this study. Boxes in red, blue, and green indicate genes from 6AS, 6BS, and 6DS chromosomes, respectively. Yellow circle indicates cluster groups, namely, A, B, C, D, and E. Specific primers are indicated by gli-AS_1 to 10 (Table II) for the genes in the gray bar. Assigned chromosome locations are given in parentheses. Accession numbers marked with asterisks indicate genes obtained from genomic DNA, and ψ indicates genes determined to be pseudogenes. γ-Gliadin (M11077) is used as the out-of-group gene.
Table II.
Primer sets for assigning contigs to respective chromosomes
| Gene | Primer Set | Chromosome | Primer 1 (5′→3′) | Primer 2 (5′→3′) |
|---|---|---|---|---|
| α/β-Gliadin | ||||
| gli-AS_1 | 6D | CCGCTACAACGATCAAACTGTCGAT | CAATATCCATCAGGCCAGGGCTC | |
| gli-AS_2 | 6A | CAACGACCAAACCATGGACTAAGAGC | GCCCAGGGCTTTGTCCAACC | |
| gli-AS_3 | 6B | TCACCGCTACAACGACCAAACCATGTTT | GCAACCATTTCTGCCACAACTACCAT | |
| gli-AS_4 | 6B | CCTAGGCCTATGGGTTCTGCTGAGA | GCAACCACAGTATCCGCAACCAC | |
| gli-AS_5 | 6B | AATGACCAAACCATGGACTAAGAAAAGA | CCCCAATTTGAGGAAATAAGGAACC | |
| gli-AS_6 | 6B | GCGCAATGGTGGTCGAGCAACG | GCCAGGTCTCCTTCCAACAGCC | |
| gli-AS_7 | 6A | AGTTAGTACCGAAGATGCCAAATGGCGGG | GCAACTACCATATTCGCAGCCACAA | |
| gli-AS_8 | 6D | AGTACCGAAGATGCCAACTGGA | TCAGCAAAACCCACAGGCCC | |
| gli-AS_9 | 6D | TGCACATTGCAGGTAGCGTCTC | CAAGTTCCATTGGTACAACAACAGC | |
| gli-AS_10 | 6D | ATGGCTGGAAGGAGCCTTGG | CCAGCTGGTGTAACAATTGTGTTG | |
| gli-AS_11 | 6D | GCTGAGATGGTTGGAAGAAGCCCT | AACAACCATATCCACAGCCGCAAC | |
| LMW-GS | ||||
| Glu-AS_1 | 1B | AATCCCCGAACAATCACGCTACG | CATCGTTGGCAGGGTACGAAGTG | |
| Glu-AS_2 | 1B | GGCACAGGGTACCTTTTTGCATC | ATACAAGGGCACATTGACACGGC | |
| Glu-AS_3 | 1B | AGGTTCGGGGTTCCATCCAAACT | TAGGCACCAACTCCAGTGCCAAC | |
| Glu-AS_4 | 1A | GGTCACAAATGTTGCAGCAGAGGAT | TCTGGTGTGGCTGCAAAAAGGT |
Figure 2.
Example of haplotype-specific PCR using aneuploid lines of CS wheat. A, B, and C indicate haplotype-specific PCR using primer sets gli-AS_1, gli-AS_2, and gli-AS_3, respectively (Table II). 1, N6AT6B; 2, N6BT6A; 3, N6DT6B; 4, DT6AL; 5, DT6BL; 6, DT6DL; 7, CS; and M, pGEM DNA markers (Promega).
LMW-GS Genes
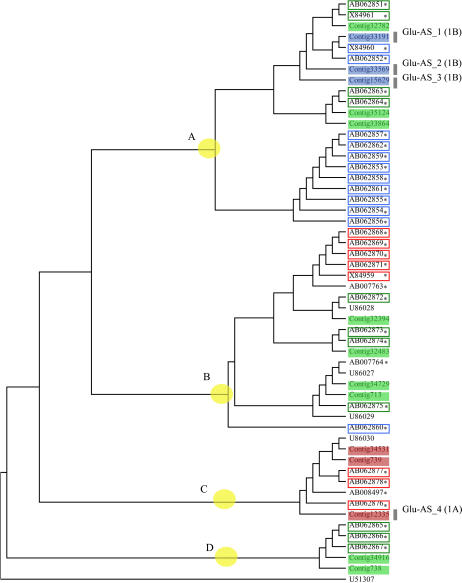
LMW-GS genes also comprise a multigene family and are highly variable (Ikeda et al., 2002). Accordingly, the 228 bp of the C-terminal-conserved domain was used for nucleotide alignments. The constructed dendrogram is depicted in Figure 3. The expressed contigs were classified into four major groups. All expressed genes identified in this study had their counterparts registered in the DNA databank. Some genome sequences, mainly derived from cultivar Norin 61 (Ikeda et al., 2002), comprised subbranches without any transcripts (Fig. 3). Because the contigs in this study had closely related counterparts whose chromosome locations had been determined, the chromosome locations of the contigs were predicted by their counterparts. Furthermore, certain contigs whose chromosome locations remained uncertain were subjected to allele-specific PCR (Table II; Fig. 3). Thus, all contigs in this study were confirmed to be located on the short arm of homoeologous chromosome group 1, indicating that the contigs were expressed from the Glu-3 locus (Gupta and Shepherd, 1990). The major group (group A) included genes expressed from two genomes (genomes B and D). Although group B harbored genes predicted from the genome sequences of genomes A, B, and D, only genes from genome D were expressed. The constituents of groups C and D consisted of members from only a single genome (A and D, respectively).
Figure 3.
Phylogenetic tree of LMW-GS genes. Accession numbers indicate genes registered in the DDBJ (Table I). Numbers in open boxes indicate genes previously assigned to chromosomes. Contig numbers in colored boxes indicate genes identified by EST analysis in this study. Boxes in red, blue, and green represent genes from 1AS, 1BS, and 1DS chromosomes, respectively. Yellow circles indicate cluster group, namely A, B, C, and D. Specific primers are indicated by Glu-AS_1 to 4 (Table II) for the genes in the gray bar. Assigned chromosome locations are given in parentheses. Accession numbers marked with asterisks indicate genes obtained from genomic DNA. α/β-Gliadins (U51307) is used as the out-of-group gene.
The Two Storage-Protein Genes Are Specifically Expressed during Seed Development
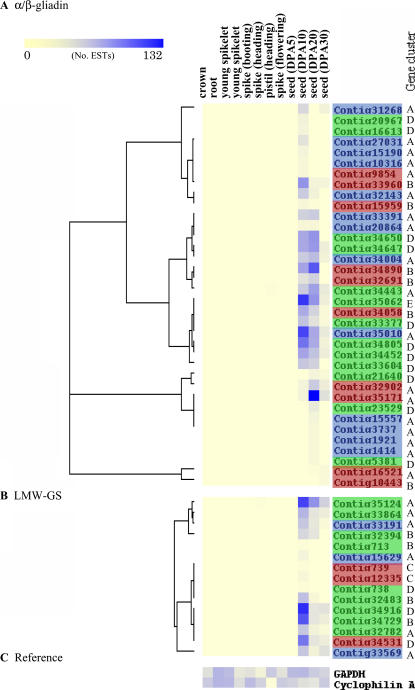
By counting the constituents of each contig in various tissues, relative expression patterns of each contig (gene) can be monitored during the wheat life cycle (Ogihara et al., 2003; Mochida et al., 2005). This method was applied to the two storage proteins, as shown in Figure 4. The expression patterns of each contig from the two storage-protein genes were clustered with correlation coefficients according to the expression frequencies among 12 tissues (Eisen et al., 1998). Figure 4 clearly shows that the two storage-protein genes are specifically expressed during the late stages of seed development. Because contigs from distinct groups (Figs. 1 and 3) were classified into groups showing similar expression patterns during seed maturation (Fig. 4), the α/β-gliadin and LMW-GS genes showing phylogenetic similarities were not expressed with similar gene expression controls during seed development.
Figure 4.
Hierarchical clustering of gene expression patterns at 12 stages of wheat life cycle. A, Thirty-six α/β-gliadin genes. B, Fifteen LMW-GS genes. C, Reference genes (glyceraldehyde-3-P dehydrogenase and cyclophilin A) as constantly expressed throughout the life cycle. Contig names colored in red, blue, and green indicate the assigned to A, B, and D genomes, respectively. Letters on the right of the contig names indicate clusters grouped by evolutional relationship in Figures 1 (α/β-gliadins) and 3 (LMW-GS).
Expression Profiles of the Two Storage-Protein Genes during Seed Maturation
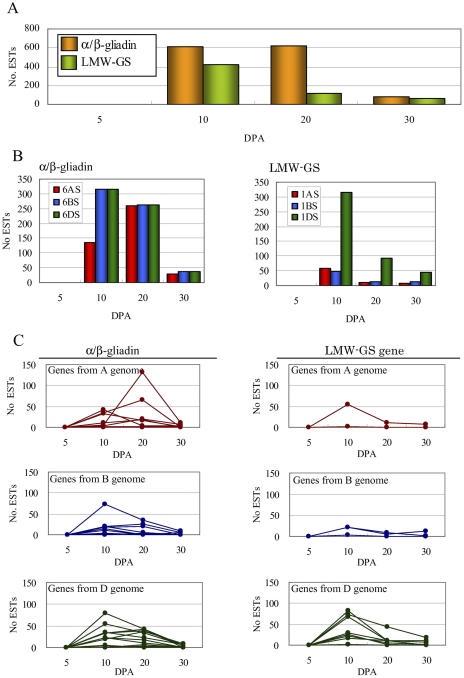
The two storage-protein genes were abundantly expressed during specific seed maturation stages. As presented in Figure 5A, expression of the two storage-protein genes was substantially induced at 10 DPA, and gradually decreased as seeds matured. In fact, 4.2%, 4.2%, and 0.6% of all ESTs at 10 DPA, 20 DPA, and 30 DPA corresponded to α/β-gliadin genes, while 2.9%, 0.8%, and 0.4% of all ESTs at 10 DPA, 20 DPA, and 30 DPA corresponded to LMW-GS gene expression. A remarkable feature of the expression patterns of the two storage-protein genes was the high expression of the α/β-gliadin genes at 20 DPA, whereas LMW-GS gene expression was largely absent at this time point.
Figure 5.
Gene expression patterns of multigenes encoding two storage proteins during seed development. A, Total number of ESTs for α/β-gliadins and LMW-GS expressed during the course of seed maturation. B, Number of ESTs corresponding to each locus for α/β-gliadin and LMW-GS genes. C, Number of ESTs corresponding to each multigene for α/β-gliadin and LMW-GS gene loci from three genomes. Number of ESTs presented in the figure was normalized to equal the original size of cDNA libraries.
α/β-Gliadin Genes Show Two Distinct Expression Patterns during Seed Maturation
Each contig is distinguishable from its locus among the three homoeologous chromosomes and the expression patterns of individual genes from the three homoeoloci are shown in Figure 5B. Genes from the 6DS were preferentially expressed in comparison to other genes. Expression of genes from 6BS appeared to be suppressed (Table III); the number of contigs was similar among the three homoeoloci (11 loci for 6AS, 13 for 6BS, and 12 for 6DS). However, the number of ESTs varied among the three homoeoloci (327 ESTs for 6AS, 216 for 6BS, and 465 for 6DS). Although the expression of genes from 6BS and 6DS were mostly observed at 10 DPA and gradually decreased during maturation, gene expression from 6AS increased at 20 DPA and decreased dramatically at 30 DPA. Subsequently, the expression patterns of each contig were carefully traced (Figs. 4 and 5C). It is remarkable that two expression patterns for the α/β-gliadin genes can be distinguished. Expression of some genes peaked at 10 DPA, after which expression levels decreased with maturation, while other genes peaked at 20 DPA. The former were designated as early expression genes, and the latter as late expression genes. The expression patterns of the early expression and late expression genes were simultaneously detected in three homoeologous genes (Fig. 5C). However, the expression levels of the two late expression genes from 6AS were extremely high at 20 DPA: contig35171 (group A) and contig34890 (group B). These expression levels are attributed to the high expression of α/β-gliadin genes from genome A at 20 DPA. While the late expression α/β-gliadin genes from genome A were preferentially expressed, and those from genome D were expressed to some extent (Table III), those from genome D were apparently suppressed (33% relative to genome A).
Table III.
Number of storage-protein genes expressed in developing seeds
En dashes indicate that all LMW-GS genes were early expression genes.
| Gene
|
Chromosome Location
|
Total No. of Genes
|
Expressed Genes Showing Peak at DPA10
|
Expressed Genes Showing Peak at DPA20
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Contig No. | EST No. | Average EST No. per Contig | Contig No. | EST No. | Average EST No. per Contig | Contig No. | EST No. | Average EST No. per Contig | ||
| α/β-Gliadin | 6AS | 10 | 325 | 32.5 | 4 | 83 | 20.8 | 6 | 242 | 40.3 |
| 6BS | 13 | 216 | 16.6 | 6 | 135 | 22.5 | 7 | 81 | 11.6 | |
| 6DS | 13 | 467 | 35.8 | 7 | 285 | 40.7 | 6 | 182 | 30.3 | |
| Total | 36 | 1,008 | 28.0 | 17 | 503 | 29.6 | 19 | 505 | 26.6 | |
| LMW-GS | 1AS | 3 | 58 | 19.3 | 3 | 58 | 19.3 | – | – | – |
| 1BS | 3 | 60 | 20.0 | 3 | 60 | 20.0 | – | – | – | |
| 1DS | 9 | 350 | 38.9 | 9 | 350 | 38.9 | – | – | – | |
| Total | 15 | 468 | 31.2 | 15 | 468 | 31.2 | – | – | – | |
LMW-GS Genes Reveal Uniform Expression Pattern in Seed Maturation
The LMW-GS genes from 1DS were preferentially expressed, as depicted in Figure 5B. Nine contigs located on 1DS were detected (Table III), while only three contigs from 1AS and 1BS were identified. About 60 ESTs were involved in each contig of 1AS and 1BS, while 350 ESTs were included in the contigs of 1DS. Genes were markedly induced at 10 DPA, after which expression levels decreased rapidly. All contigs for LMW-GS revealed similar expression patterns during seed maturation, irrespective of expression levels (Fig. 5C).
Early Expression α/β-Gliadin Genes Show Similar Expression Patterns as LMW-GS Genes
The expression patterns of early expression α/β-gliadin genes showed similar expression patterns as the LMW-GS genes. The number of contigs belonging to the early expression α/β-gliadin genes is presented in Table III. The number of ESTs per contig is also shown in Table III. The number of contigs (genes) corresponding to genomes A and B were similar (about 20), and were equivalent to that of the LMW-GS genes (Table III). On the other hand, the number of contigs (genes) in genome D was almost double (approximately 40) that of genomes A and B. It can be concluded that the expression patterns of the early expression α/β-gliadin and LMW-GS genes are similar and that genes from the D genome are preferentially expressed in CS wheat.
DISCUSSION
Phylogenetic Relationships between Expressed α/β-Gliadin and LMW-GS Genes in CS Wheat
ESTs of CS wheat homologous to α/β-gliadin and LMW-GS genes were selected from the comprehensive EST database (Mochida et al., 2005). A total of 36 expressed genes for α/β-gliadin and 15 expressed genes for LMW-GS were obtained (Table III). The α/β-gliadin genes were confirmed to be expressed from the Gli-2 locus located on the short arm of homoeologous chromosome 6 (Payne, 1987), and the LMW-GS genes were expressed from the Glu-3 locus located on the short arm of homoeologous chromosome 1 (Gupta and Shepherd, 1990). The copy numbers of the α/β-gliadin and LMW-GS genes were estimated to be approximately 100 (Okita et al., 1985; Anderson et al., 1997) and 30 (Cassidy et al., 1998), and thus about one-half to one-third of the genes were expressed in CS wheat. As grouped in Figures 1 and 3, new classes of expressed genes were identified in this study. It is well known that storage-protein genes, such as α/β-gliadin and LMW-GS genes, comprise a multigene family and are hypervariable in terms of insertions/deletions and base substitutions (Wicker et al., 2003; Gu et al., 2004). Figures 1 and 3 show that the α/β-gliadin and LMW-GS genes harbored two types of genes, i.e. relatively conserved genes and rapid divergent genes; α/β-gliadin group A in the dendrogram contained genes from three genomes (A, B, and D), while groups B, D, and E had genes from a single genome (A or D), indicating the rapid evolution of respective genes after differentiation of wheat genomes (Fig. 1). With regard to the LMW-GS genes (Fig. 3), groups A and B contained genes from two genomes (genomes B and D for group A; genomes A and D for group B), while groups C and D harbored genes from a single genome (A and D, respectively), indicating the rapid evolution of these genes.
Expression Patterns of α/β-Gliadin and LMW-GS Genes in Wheat Seed Maturation
We have presented evidence that the α/β-gliadin genes showed two distinct expression patterns during the course of wheat seed maturation: early expression genes, which showed higher expression level at 10 DPA, and late expression genes, which showed higher expression level at 20 DPA (Fig. 5C). The classification of comprehensive ESTs into specific contigs enables us to distinguish expression patterns of each multigene from the three homoeologues (Mochida et al., 2003). Although the temporal resolution of the developmental expression of the genes analyzed here is limited for the number of constructed libraries during seed maturation, the expression pattern of early expression genes was similar to that of LMW-GS genes (both genes from genome D were preferentially expressed over those from genomes A and B) in terms of gene number and expression activity (Table III; Fig. 5). This strongly suggests that early expression α/β-gliadin and LMW-GS genes are controlled by common regulatory elements, such as the prolamin box (Hammond-Kosack et al., 1993; Mueller and Knudsen, 1993). Furthermore, distinct members showing higher expression level at 20 DPA (designated as late expression genes) were recognized among the α/β-gliadin genes (Figs. 4 and 5). Although the number of genes from the three genomes equally contributed to the late expression genes, two α/β-gliadin genes from genome A were markedly expressed and those from genome B were more or less suppressed (Table III). These lines of evidence strongly suggest novel controlling system(s) for expression of α/β-gliadin genes in addition to the prolamin box (Shewry et al., 2003). Genomic clones of CS wheat corresponding to individual contigs are required to carry out promoter analyses (Shewry et al., 2003).
Phylogenetic Relationships of Multigenes Are Not Correlated to Expression Patterns during Seed Maturation
It is well known that plant storage-protein genes exist as multigene families and evolved rapidly, even among related plant species (Shewry et al., 2003). The structural features of the wheat α/β-gliadin and LMW-GS genes obtained to date are disrupted by repetitive sequences, such as retrotransposons (Wicker et al., 2003; Gu et al., 2004; Johal et al., 2004), such that adjacent members are separated by tens of kilobase pairs. This genomic situation contrasts to the case of zein (maize [Zea mays]) and secalin (rye [Secale cereale]), in which gene members are arranged almost head to tail (Clarke et al., 1996; Song et al., 2001). Therefore, the Gli-2 (Payne, 1987) and Glu-3 (Gupta and Shepherd, 1990) loci must share relatively large regions on the 6S and 1S chromosomes, respectively. Meanwhile, the groupings of genes classified by their expression patterns are not correlated to their phylogenetic relationships (Fig. 4). In fact, genes classified into the same phylogenetic groups were assigned into different groups showing distinct expression patterns, for example, early expression and late expression α/β-gliadin genes (Fig. 4). This strongly suggests that the expression of the α/β-gliadin and LMW-GS multigenes are independently regulated, irrespective of their phylogenetic relationships, in response to wheat seed maturation, as is the case in other plant multigenes (Duan and Schuler, 2005; Kirch et al., 2005).
Because a large number of wheat ESTs has been pooled in the public domain (National Center for Biotechnology Information), bioinformatics studies enable us to monitor wheat expression patterns during the wheat life cycle and in response to environmental stresses. Profiling of expression patterns of storage-protein genes using a comprehensive EST database provides a powerful tool to analyze the structure and expression of individual multigenes and to manipulate their functions. Wheat EST databases are now indispensable to the study of functional wheat genomics.
MATERIALS AND METHODS
Collection of EST Sequences Encoding Two Wheat Storage Proteins
A total of 361,180 EST sequences were collected from the 32 cDNA libraries constructed from various tissues obtained throughout the life cycle and from abiotically stressed tissues of common wheat (Triticum aestivum cv CS, T. aestivum cv Kitakei 1354, and T. aestivum cv Valuevskaya; Ogihara et al., 2003; Mochida et al., 2005). Each library contained more than 10,000 EST sequences. The ESTs were grouped into contigs using the phrap method (University of Washington Genome Center; http://www.genome.washington.edu/UWGC). The Sequence Retrieval System (SRS) of the DNA Data Bank of Japan (DDBJ; http://srs.ddbj.nig.ac.jp/index-j.html) was adopted for selecting the annotated α/β-gliadin and LMW-GS genes (Table I). Sequences were used as queries for the BLASTN search (Altschul et al., 1990) against our wheat EST database.
Construction of Dendrogram Showing Phylogenetic Relationships of the Two Storage-Protein Genes
The nucleotide sequences of EST contigs for α/β-gliadin and LMW-GS were independently aligned along with the sequences registered in the DDBJ. For the construction of the dendrogram, 380 bp at the 3′-site of the coding region of α/β-gliadins and 228 bp at the C-terminal-conserved domain of LMW-GS were used. Each dendrogram was constructed using ClustalW software (Thompson et al., 1994). As out of group genes, the sequences for γ-gliadin (M11077) and α/β-gliadin (U51307) were clustered along with α/β-gliadin and LMW-GS, respectively.
Haplotype-Specific PCR for Assigning ESTs to the Three Homoeologous Chromosomes
Total DNA was isolated from seedlings of common wheat (T. aestivum cv CS) and from nullisomic-tetrasomic as well as ditelosomic lines of CS (Sears, 1965), as described previously (Ogihara et al., 1994). By tracing the SNP sites in the contigs, primer sets were designed using Primer3 software (Rozen and Skaletsky, 2000) so as to specifically amplify each multigene (Table II). PCR was performed using 20 ng of total DNA and KOD-plus (Toyobo) in 20 μL of reaction mixture following the manufacturer's instructions. The thermal cycling program was as follows: 94°C for 2 min followed by 35 cycles of 94°C for 15 s, 65°C for 30 s, and 68°C for 40 s, or 94°C for 2 min followed by 35 cycles of 94°C for 15 s and 68°C for 40 s. Amplification of PCR products was confirmed by 2% agarose gel electrophoresis.
Profiling of Gene Expression Patterns by Hierarchical Clustering
Because the sequenced cDNAs were more or less changed in their libraries, the number of ESTs was normalized among the 32 libraries. The expression pattern of each gene in certain tissues and/or after certain treatments was monitored by counting the constituents involved in each contig. The contigs were clustered using Pearson's correlation coefficient (Eisen et al., 1998). The expression patterns of each contig were displayed for 12 stages of wheat grown under natural conditions (crown, root, young spikelet at early flowering, young spikelet at late flowering, spike at booting stage, spike at heading date, pistil at heading date, spike at flowering date, and seeds at DPA [DPA5, DPA10, DPA20, and DPA30]). The expression patterns of contigs homologous to the genes for glyceraldehyde-3-P dehydrogenase and cyclophilin A were displayed as references.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BJ207047 to BJ323305.
Supplementary Material
Acknowledgments
We thank Dr. Yukiko Yamazaki of the National Institute of Genetics, Japan, for her maintenance of the EST database of wheat, “KOMUGI” (http://shigen.lab.nig.ac.jp/wheat/komugi/ests/tissueBrowse.jsp).
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (“Molecular mechanisms of species differentiation”; no. 14087204) and National Bioresource Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yasunari Ogihara (yogihara@kpu.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070722.
References
- Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Anderson O (1991) Characterization of members of a pseudogene subfamily of the wheat alpha-gliadin storage protein genes. Plant Mol Biol 16: 335–337 [DOI] [PubMed] [Google Scholar]
- Anderson O, Greene F (1997) The α-gliadin gene family. II. DNA and protein sequence vatiation, subfamily structure, and origins of pseudogenes. Theor Appl Genet 95: 59–65 [Google Scholar]
- Anderson O, Litts J, Greene F (1997) The α-gliadin gene family. I. Characterization of ten new wheat α-gliadin genomic clones, evidence for limited sequence conservation of flanking DNA, and Southern analysis of the gene family. Theor Appl Genet 95: 50–58 [Google Scholar]
- Arentz-Hansen EH, McAdam SN, Molberg O, Kristiansen C, Sollid LM (2000) Production of a panel of recombinant gliadins for the characterisation of T cell reactivity in coeliac disease. Gut 46: 46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res 7: 175–180 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2004) Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol Biol 54: 405–414 [DOI] [PubMed] [Google Scholar]
- Cassidy BG, Dvorak J, Anderson OD (1998) The wheat low-molecular-weight glutenin genes: characterization of six new genes and progress in understanding gene family structure. Theor Appl Genet 96: 743–750 [Google Scholar]
- Clarke BC, Mukai Y, Appels R (1996) The Sec-1 locus on the short arm of chromosome 1R of rye (Secale cereale). Chromosoma 105: 269–275 [PubMed] [Google Scholar]
- D'Ovidio R, Marchitelli C, Ercoli Cardelli L, Porceddu E (1999) Sequence similarity between allelic Glu-B3 genes related to quality properties of durum wheat. Theor Appl Genet 98: 455–461 [Google Scholar]
- Duan H, Schuler M (2005) Differential expression and evolution of the Arabidopsis CYP86A subfamily. Plant Physiol 137: 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing R, Ben Kahla A, Poirot O, Lopez F, Audic S, Claverie J (1999) Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res 9: 950–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Maroto F, Marana C, Garcia-Olmedo F, Carbonero P (1990) Nucleotide sequence of a cDNA encoding an alpha/beta-type gliadin from hexaploid wheat (Triticum aestivum). Plant Mol Biol 14: 867–868 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Crossman C, Kong X, Luo M, You FM, Coleman-Derr D, Dubcovsky J, Anderson OD (2004) Genomic organization of the complex alpha-gliadin gene loci in wheat. Theor Appl Genet 109: 648–657 [DOI] [PubMed] [Google Scholar]
- Gupta RB, Shepherd KW (1990) Two-step one-dimensional SDS-PAGE analysis of LMW subunits of glutelin. 1. Variation and genetic control of the subunits in hexaploid wheats. Theor Appl Genet 80: 65–74 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack MCR, Holdsworth MJ, Bevan MW (1993) In vivo footprinting of a low molecular weight glutenin gene (LMWG-1D1) in wheat endosperm. EMBO J 12: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Nagamine T, Fukuoka H, Yano H (2002) Identification of new low-molecular-weight glutenin subunit genes in wheat. Theor Appl Genet 104: 680–687 [DOI] [PubMed] [Google Scholar]
- Jackson EA, Holt LM, Payne IP (1983) Characterisation of high molecular weight gliadin and low-molecular-weight glutenin subunits of wheat endosperm by two-dimensional electrophoresis and the chromosomal localisation of their controlling genes. Theor Appl Genet 66: 29–37 [DOI] [PubMed] [Google Scholar]
- Johal J, Gianibeli MC, Rahman S, Morell MK, Gale KR (2004) Characterization of low-molecular-weight glutenin genes in Aegilops tauschii. Theor Appl Genet 109: 1028–1040 [DOI] [PubMed] [Google Scholar]
- Kasarda D, Okita T, Bernardin J, Baecker P, Nimmo C, Lew E, Dietler M, Greene F (1984) Nucleic acid (cDNA) and amino acid sequences of alpha-type gliadins from wheat (Triticum aestivum). Proc Natl Acad Sci USA 81: 4712–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch H, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D (2005) Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol 57: 315–332 [DOI] [PubMed] [Google Scholar]
- Maruyama N, Ichise K, Katsube T, Kishimoto T, Kawase S, Matsumura Y, Takeuchi Y, Sawada T, Utsumi S (1998) Identification of major wheat allergens by means of the Escherichia coli expression system. Eur J Biochem 255: 739–745 [DOI] [PubMed] [Google Scholar]
- Masci S, D'Ovidio R, Lafiandra D, Kasarda D (1998) Characterization of a low-molecular-weight glutenin subunit gene from bread wheat and the corresponding protein that represents a major subunit of the glutenin polymer. Plant Physiol 118: 1147–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Kawaura K, Shimosak E, Shin-I T, Kohara Y, Yamazaki Y, Ogihara Y (2005) Tissue expression map of comprehensive expressed sequence tags and its application to in silico screening of stress response genes in common wheat. Mol Genet Genomics (in press) [DOI] [PubMed]
- Mochida K, Yamazaki Y, Ogihara Y (2003) Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol Genet Genomics 270: 371–377 [DOI] [PubMed] [Google Scholar]
- Moczulski M, Salmanowicz BP (2003) Multiplex PCR identification of wheat HMW glutenin subunit genes by allele-specific markers. J Appl Genet 44: 459–471 [PubMed] [Google Scholar]
- Mueller M, Knudsen S (1993) The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J 4: 343–355 [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Mochida K, Nemoto Y, Murai K, Yamazaki Y, Shin-I T, Kohara Y (2003) Correlated clustering and virtual display of gene expression patterns in the wheat life cycle by large-scale statistical analyses of expressed sequence tags. Plant J 33: 1001–1011 [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Shimizu H, Hasegawa K, Tsujimoto H, Sasakuma T (1994) Chromosome assignment of four photosynthesis-related genes and their variability in wheat species. Theor Appl Genet 88: 383–394 [DOI] [PubMed] [Google Scholar]
- Okita T, Cheesbrough V, Reeves C (1985) Evolution and heterogeneity of the alpha-/beta-type and gamma-type gliadin DNA sequences. J Biol Chem 260: 8203–8213 [PubMed] [Google Scholar]
- Pavy N, Laroche J, Bousquet J, Mackay J (2005) Large-scale statistical analysis of secondary xylem ESTs in pine. Plant Mol Biol 57: 203–224 [DOI] [PubMed] [Google Scholar]
- Payne IP (1987) Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu Rev Plant Physiol 38: 141–153 [Google Scholar]
- Payne IP, Holt LM, Worland AJ, Law CN (1982) Structural and genetical studies on the high-molecular-weight subunits of wheat glutenin. Part 3. Telocentric mapping of subunit genes on the long arms of the homoeologous group 1 chromosomes. Theor Appl Genet 63: 129–138 [DOI] [PubMed] [Google Scholar]
- Payne IP, Jackson EA, Holt LM, Law CN (1984) Genetic linkage between endosperm storage protein genes on each of the short arms of chromosomes 1A and 1B in wheat. Theor Appl Genet 67: 235–243 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Sabelli P, Shewry PR (1991) Characterization and organisation of gene families at the Gli-1 loci of bread and durum wheats by restriction fragment analysis. Theor Appl Genet 83: 209–216 [DOI] [PubMed] [Google Scholar]
- Sears E (1965) Nullisomic-tetrasomic combinations in hexaploid wheat. In R Riley, K Lewis, eds, Chromosome Manipulation and Plant Genetics. Oliver and Boyd, Edinburgh, pp 29–58
- Shewry PR, Halford NG, Lafiandra D (2003) Genetics of wheat gluten proteins. Adv Genet 49: 111–184 [DOI] [PubMed] [Google Scholar]
- Song R, Llaca V, Linton E, Messing J (2001) Sequence, regulation and evolution of the maize 22-kD alpha zein gene family. Genome Res 11: 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Bhalerao R, Unneberg P, Segerman B, Nilsson P, Brunner A, Charbonnel-Campaa L, Lindvall J, Tandre K, Strauss S, et al (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101: 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner-Smith M, Rafalski J, Sugiyama T, Stoll M, Soll D (1985) Conservation and variability of wheat alpha/beta-gliadin genes. Nucleic Acids Res 13: 3905–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RD, Bartels D, Harberd NP, Flavell RB (1983) Characterization of the multigene family coding for HMW glutenin subunits in wheat using cDNA clones. Theor Appl Genet 67: 87–96 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Campenhout S, Vander Stappen J, Sagi L, Volckaert G (1995) Locus-specific primers for LMW glutenin genes on each of the group 1 chromosomes of hexaploid wheat. Theor Appl Genet 91: 313–319 [DOI] [PubMed] [Google Scholar]
- Vettore A, da Silva F, Kemper E, Souza G, Da Siva A, Ferro M, Henrique-Silva F, Giglioti LM, Coutinho EA, Nobrega LL, et al (2003) Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res 13: 2725–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Yahiaui N, Guyot R, Schlagenhauf E, Liu ZD, Dubkovsky J, Keller B (2003) Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15: 1186–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sreenivasulu N, Weschke W, Stein N, Rudd S, Radchuk V, Potokina E, Scholz U, Schweizer P, Zierold U, et al (2004) Large-scale analysis of the barley transcriptome based on expressed sequence tags. Plant J 40: 276–290 [DOI] [PubMed] [Google Scholar]
- Zhang W, Gianibelli MC, Ma W, Rampling L, Gale KR (2003) Identification of SNPs and development of allele-specific PCR markers for γ-gliadin alleles in Triticum aestivum. Theor Appl Genet 107: 130–138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.