Abstract
Segmental duplicons (>1 kb) of high sequence similarity (>90%) covering >5% of the human genome are characterized by complex sequence variation. Apart from a few well-characterized regions (MHC, β-globin), the diversity and linkage disequilibrium (LD) patterns of duplicons and the role of gene conversion in shaping them have been poorly studied. To shed light on these issues, we have re-sequenced the human Luteinizing Hormone/Chorionic Gonadotropin β (LHB/CGB) cluster (19q13.32) of three population samples (Estonians, Mandenka, and Han). The LHB/CGB cluster consists of seven duplicated genes critical in human reproduction. In the LHB/CGB region, high sequence diversity, concentration of gene-conversion acceptor sites, and strong LD colocalize with peripheral genes, whereas central loci are characterized by lower variation, gene-conversion donor activity, and breakdown of LD between close markers. The data highlight an important role of gene conversion in spreading polymorphisms among duplicon copies and generating LD around them. The directionality of gene-conversion events seems to be determined by the localization of a predicted recombination “hotspot” and “warm spot” in the vicinity of the most active acceptor genes at the periphery of the cluster. The data suggest that enriched crossover activity in direct and inverted segmental repeats is in accordance with the formation of palindromic secondary structures promoting double-strand breaks rather than fixed DNA sequence motifs. Also, this first detailed coverage of sequence diversity and structure of the LHB/CGB gene cluster will pave the way for studying the identified polymorphisms as well as potential genomic rearrangements in association with an individual's reproductive success.
Segmental duplications represent direct and/or inverted low-copy repeats consisting of arrayed genes/pseudogenes and other repeated chromosomal segments. For human, the genome-wide frequency of segmental duplications (>1 kb, >90% identity) has been estimated, by computational analysis, at 5%–10% (Bailey et al. 2002). It has been suggested that these regions may underlie a greater amount of human phenotypic variation and disease than was previously recognized (Stankiewicz and Lupski 2002; Iafrate et al. 2004; Sebat et al. 2004). However, apart from a couple of well-characterized loci—MHC (Horton et al. 2004), β-globin (Papadakis and Patrinos 1999)—there is little knowledge about the nature and driving forces of sequence variability and linkage disequilibrium (LD) structure in duplicons. Frequently, it appears that the SNPs in databases mapped in the segmental duplications are, in fact, variants in paralogous sequences (Estivill et al. 2002). Fredman et al. (2004) defined a new type of polymorphism—multisite variation (MSV)—representing the sum of the signals from many individual duplicon copies that vary in sequence content because of duplication, deletion, or gene conversion, and attributed 28% of the SNPs in duplicons to MSVs. Meiotic gene conversion, favored by high sequence homology between tandem-arrayed or inverted DNA segments, leads to concerted evolution of duplicons (Hurles 2001; Bettencourt and Feder 2002) and the spread of mutations (Tusié-Luna and White 1995; Papadakis and Patrinos 1999; Boocock et al. 2003). Highly similar DNA segments also favor nonallelic homologous crossing-over coupled with rearrangements, leading to genomic disorders (Stankiewicz and Lupski 2002; Shaw and Lupski 2004).
Interestingly, there is a nonrandom distribution of the functions of human segmentally duplicated genes within the proteome (Bailey et al. 2002). Several genes associated with female or male reproduction have been shown to be duplicated during primate evolution as well as evolving under positive Darwinian selection (Wyckoff et al. 2000; Bailey et al. 2002; Nahon 2003). One of the gene families that has evolved in the primate lineage is the gonadotropin hormone β-subunit (GtHB) family, represented in human by seven duplicated Luteinizing Hormone β (LHB)/Chorionic Gonadotropin β (CGB) genes located at 19q13.32 and two single-copy genes, FSHB at 11p13–p14 and TSHB at 1p13.2. Consistent with an essential role in reproduction, all of the few described nonsynonymous mutations lead to either infertility or reduced gonadal function (Themmen and Huhtaniemi 2000). The ancestral LHB gene has duplicated several times during primate evolution, giving rise to a new gene, CGB, differing from LHB both in the time (pregnancy vs. adult lifetime) and tissue (placenta vs. pituitary) of expression as well as mRNA stability (Policastro et al. 1986; Maston and Ruvolo 2002).
In order to study fine-scale sequence variation and LD structure in duplicated regions, we have applied the LHB/CGB gene cluster as a model and re-sequenced population samples from three continents. We explore the following questions: (1) What is the role of gene conversion in shaping the diversity and LD patterns in duplicons? (2) What potentially determines the distribution of crossovers and gene conversion in duplicated regions?
In addition, as an important contribution to human reproductive genetics, this is the first survey of sequence variation in human LHB/CGB genes, essential for successful fertilization and pregnancy. The detailed knowledge of the structure and diversity of the LHB/CGB gene cluster paves the way for studying an individual's general or reduced (e.g., susceptibility to spontaneous abortions) reproductive success in association with identified sequence variants of LHB/CGB genes as well as potential genomic rearrangement patterns (insertions, deletions, duplications, etc.) between homologous regions within the cluster.
Results
Fine-scale structure and features of the human LHB/CGB cluster
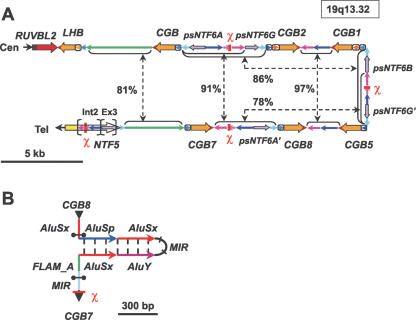
Although the exact order of duplication events is still to be determined, the fine-scale structure of the human LHB/CGB cluster in Figure 1A provides hints for putative past evolutionary events. Previous reports have indicated the duplication of the ancestral LHB gene in the common ancestor of anthropoid primates (Policastro et al 1986; Maston and Ruvolo 2002). Our detailed analysis of the structure of the human LHB/CGB cluster revealed that in addition to LHB, the initial duplication apparently involved a part of the neutrophin 5 (NTF5) gene (3′-UTR, exon 3, and intron 2) and the LHB–NTF5 intergenic region. The next rounds of duplication included only LHB/CGB genes and the duplicated segment of NTF5. LHB gave rise to six human CGB genes (CGB, CGB2, CGB1, CGB5, CGB7, CGB8), and exon 3 of NTF5 to five NTF6 pseudogenes (psNTF6A, 6G, 6B, 6G′, 6A′). An Alu-rich fragment, containing also recombination and a gene-conversion-associated χ-site (GCTGGTGG) (Smith 1988), spread from intron 2 of NTF5 to intergenic regions of LHB/CGB genes (current Alu-sequence content from 10% up to 56%). Both Alu-sequences and the presence of a χ-site could have been stimulators of direct and inverted duplications within the cluster (Bailey et al. 2003). An insertion upstream of consensus exon 1 distinguishes CGB1 and CGB2, providing an alternative exon 1 as well as a new potential promoter segment (Figs. 1A, 2A; Supplemental Fig. S1). BLAST analysis indicated the possible origin of this inserted fragment in noncoding regions of Chromosomes 2, 3, or 19.
Figure 1.
(A) Structure of the human LHB/CGB gene cluster. Identical color codes refer to highly homologous DNA sequences within the cluster. Genes (RUVBL2, LHB/CGBs, NTF5) are depicted as wide arrows in the direction of transcription with promoters as boxed arrows 5′ of the genes. Intergenic areas are marked as narrow lines, indicating also the direction of a segment, and broken arrows unite intergenic regions that share >75% of sequence identity. χ denotes the localization of consensus Escherichia coli χ-sequence (GCTGGTGG) (Smith 1988) associated with crossing-over and gene-conversion activity. Int2 and Ex3 refer to intron 2 and exon3 in the NTF5 gene; the former is the source of intergenic regions between CGB genes, and the latter has given rise to NTF6 pseudogenes. (B) Prediction of single-stranded DNA secondary structure for the estimated recombination hotspot (bordered by black brackets) between CGB8 and CGB7. The hotspot is flanked by double inverted Alu-sequences forming the stem (625 bp) of the palindrome, and its center falls within the loop (222 bp). (MIR) Mammalian-wide interspersed repeat; (FLAM_A) Alu-element-like repeat.
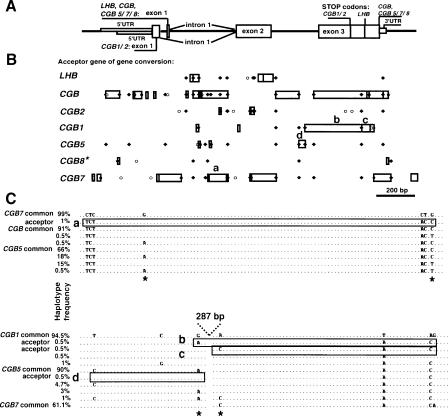
Figure 2.
(A) Consensus exon–intron structure of LHB/CGB genes presented in parallel with (B) the summary distribution of gene-conversion acceptor sites and multisite variants (MSVs) within LHB/CGB genes. A minimum gene-conversion site (□) was defined as a segment within an acceptor gene including ≥2 associated SNPs, for which a potential donor gene could be identified (detailed information in Table 3). Typically, for each gene-conversion site, several converted tracts could be determined (overlapping □). MSV1 (○) is defined as an SNP in one of the duplicate genes, which is also represented as a paralogous sequence variant among homologous genes (Fredman et al. 2004). MSV2 (♦) is an SNP that is present as a polymorphism at the identical sequence position in several duplicated genes. Several gene-conversion sites overlap with MSVs, indicating their origin from a gene-conversion event, rather than parallel mutation. Lettering (a, b, c, d) refers to gene-conversion sites, which are described in detail in C. CGB8* data are from the analysis of 11 Estonian individuals; the analysis of the rest of the genes is based on a combined sample set of 95 Estonian (n = 47), Han (n = 25), and Mandenka (n = 23) individuals. (C) Examples of gene-conversion segments (boxed sequences a–d) and their potential donor genes. Haplotype frequencies of each segment variant are derived from the combined sample (n = 95) of Estonians, Mandenka, and Han Chinese. Segment a within CGB7 (frequency 1%) originates most probably from a common variant of CGB (91%) or CGB5 (66%). Segments b (0.5%) and c (0.5%) within CGB1 were derived by gene conversion from common variants of CGB5 (90%) and CGB7 (61.1%), respectively. Reciprocally, a common variant of CGB1 (94.5%) or CGB7 (61.1%) is the most apparent donor of segment d to CGB5 (0.5%). “Shared SNPs” or MSV2 between gene pairs (*) are colocalized with gene-conversion sites.
Among the genes coding the β subunit of hCG hormone (CGB, CGB5, CGB7, and CGB8), there is 97%–99% DNA sequence identity, whereas their identity with the functionally divergent LHB gene is 92%–93% and with the CGB1 and CGB2 genes, 85% (Supplemental Fig. S1). Despite primary DNA sequence homology with the rest of the genes, CGB1 and CGB2 have possibly diverged in function (although a protein is still uncharacterized) through the use of an alternative exon 1 as well as a shifted open reading frame (Fig. 2A; Supplemental Figs. S1, S2). Whereas among the hCG hormone β-subunit-coding genes the protein identity is 98%–100%, and to LHβ 85%, there is no significant amino acid sequence similarity between hCGβ coded by CGB, CGB5, CGB7, CGB8, and LHβ on the one side, and the predicted protein for CGB1 and CGB2 on the other side (Supplemental Fig. S2). In addition to the high DNA sequence homology among LHB/CGB genes, also the intergenic regions within the cluster reveal pairwise sequence similarity from 81% up to 97% (Fig. 1A). The whole genomic region is extremely high in G+C content (≥55%), with individual genes exceeding 60% (Table 1).
Table 1.
Summary statistics of sequence variation data for individual LHB/CGB genes and predicted hotspot region
| Haplotypesh
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Analyzed region
|
Segregating sites (S)
|
MSVd
|
MAF ≥10%
|
|||||||||||||
| Length (bp) | GC % | Pa | All | ≥10% MAFc | Singletons | Type 1 | Type 2 | % | SNPs in dbSNP | πe | θf | Dg | All No. | No. | Carriers (%) | |
| Individual genes | ||||||||||||||||
| LHB | 1541 | 63 | E | 20 | 15 | 2 | 0 | 7 | 35.0 | 12 | 0.00371 | 0.00254 | 1.35 | 14 | 2 | 65.9 |
| H | 17 | 10 | 1 | 0 | 6 | 35.3 | 12 | 0.00348 | 0.00246 | 1.28 | 7 | 2 | 84 | |||
| M | 20 | 10 | 5 | 1 | 7 | 40.0 | 11 | 0.00313 | 0.00295 | 0.20 | 16 | 2 | 50 | |||
| All | 27 | 8 | 1 | 10 | 55.0 | 12 | 0.00375 | 0.00301 | ||||||||
| CGB | 1543 | 64 | E | 18 | 14 | 2 | 7 | 72.2 | 2 | 0.00406 | 0.00228 | 2.24* | 15 | 2 | 74.5 | |
| H | 20 | 16 | 2 | 8 | 8 | 80.0 | 2 | 0.00530 | 0.00304 | 2.39* | 13 | 2 | 72 | |||
| M | 28 | 23 | 3 | 10 | 13 | 82.1 | 5 | 0.00666 | 0.00413 | 2.06* | 14 | 3 | 60.8 | |||
| All | 35 | 6 | 11 | 16 | 77.1 | 5 | 0.00545 | 0.00401 | ||||||||
| CGB2 | 1521 | 63 | E | 8 | 4 | 2 | 2 | 3 | 62.5 | 2 | 0.00115 | 0.00103 | 0.29 | 10 | 4 | 92.6 |
| H | 12 | 3 | 6 | 2 | 6 | 66.7 | 2 | 0.00101 | 0.00176 | -1.26 | 8 | 3 | 80 | |||
| M | 24 | 4 | 10 | 2 | 9 | 45.8 | 2 | 0.00237 | 0.00359 | -1.12 | 12 | 2 | 56.5 | |||
| All | 34 | 15 | 5 | 13 | 52.9 | 2 | 0.00145 | 0.00384 | ||||||||
| CGB1 | 1510 | 63 | E | 5 | 4 | 0 | 0 | 1 | 20.0 | 1 | 0.00095 | 0.00065 | 0.99 | 6 | 3 | 82.9 |
| H | 12 | 4 | 5 | 2 | 1 | 25.0 | 1 | 0.00119 | 0.00177 | -0.98 | 8 | 3 | 78 | |||
| M | 14 | 5 | 3 | 2 | 2 | 28.6 | 1 | 0.00157 | 0.00226 | -0.95 | 9 | 3 | 75.9 | |||
| All | 20 | 5 | 2 | 5 | 35.0 | 1 | 0.00124 | 0.00239 | ||||||||
| CGB5 | 1661 | 64 | E | 13 | 7 | 2 | 0 | 8 | 61.5 | 2 | 0.00155 | 0.00153 | 0.03 | 15 | 2 | 73.4 |
| H | 13 | 6 | 5 | 1 | 5 | 46.2 | 2 | 0.00117 | 0.00175 | -0.99 | 9 | 2 | 78 | |||
| M | 13 | 10 | 1 | 2 | 8 | 76.9 | 2 | 0.00213 | 0.00178 | 0.59 | 11 | 4 | 78.3 | |||
| All | 25 | 7 | 2 | 13 | 60.0 | 2 | 0.00165 | 0.00259 | ||||||||
| CGB7 | 2233 | 63 | E | 30 | 27 | 1 | 6 | 6 | 40.0 | 4 | 0.00552 | 0.00271 | 3.18** | 31 | 2 | 47.8 |
| H | 29 | 27 | 0 | 4 | 10 | 48.3 | 4 | 0.00484 | 0.00300 | 2.04* | 16 | 2 | 52 | |||
| M | 41 | 25 | 1 | 8 | 13 | 51.2 | 5 | 0.00429 | 0.00418 | 0.09 | 21 | 2 | 45.6 | |||
| All | 50 | 1 | 9 | 14 | 46.0 | 5 | 0.00550 | 0.00385 | ||||||||
| Predicted recombination hotspot regionb | ||||||||||||||||
| All region | 8338 | 56 | E | 64 | 42 | 14 | 0.00212 | 0.00196 | 0.32 | |||||||
| Intergenic CGB5/8 | 1879 | 52 | E | 17 | 12 | 2 | 0.00257 | 0.00219 | 0.61 | |||||||
| CGB8 | 2156 | 62 | E | 15 | 6 | 7 | 1 | 5 | 42.9 | 6 | 0.00152 | 0.00178 | -0.54 | 10 | 3 | 50 |
| Intergenic CGB8/7 | 4303 | 54 | E | 32 | 24 | 5 | 0.00227 | 0.00208 | 0.35 | 19 | ||||||
(E) Estonians (n=47); (H) Han (n=25); (M) Mandenka (n=23).
Data for CGB8 are from 11 Estonian individuals from the resequencing of potential recombination hotspot (see text for explanation).
(MAF) Minor allele frequency.
Multisite variation (Fredman et al. 2004): (MSV1) SNPs that are also represented as paralogous sequence variants (PSVs) among the duplicons. (MSV2) SNPs present in >1 duplicated gene.
Estimate of nucleotide diversity per site from average pairwise difference among individuals.
Estimate of nucleotide diversity per site from number of segregating sites (S).
Significance level of Tajima's D statistics: (**) p < 0.01; (*) p < 0.05.
Haplotype distribution is the estimate by PHASE algorithm (Stephens et al. 2001).
Sequence diversity of LHB, CGB, CGB2, CGB1, CGB5, and CGB7 genes
We re-sequenced six of the seven LHB/CGB genes (in total 10,009 kb) in population samples from three continents: Europe (Estonians, n = 47), Africa (Mandenka, n = 23), and Asia (Chinese Han, n = 25) (Supplemental Table S1). In total we identified 191 SNPs: 62 (32%) were spread in three continents, 21 (11%) were shared by two populations, and 108 (57%) were population-specific SNPs (Table 1; Supplemental Table S2). Consistent with other genomic regions (for review, see Tishkoff and Verrelli 2003; Crawford et al. 2004), African chromosomes were characterized by the highest diversity as well as by a high fraction of private SNPs (n = 62) (Table 1). Only a small fraction of these SNPs are currently represented in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/index.html). Compared to a data set of 74 genes (Crawford et al. 2004), characterized by a mean nucleotide diversity parameter π = 0.0010 for African Americans and π = 0.0008 for European Americans, LHB/CGB genes exhibit high diversity. Interestingly, there is a clear decrease in diversity levels toward the center of the cluster: the peripheral loci CGB7 (mean π = 0.0055), LHB (0.0038), and CGB (0.0040) being highly diverse compared to the central loci CGB2 (0.0015), CGB1 (0.0012), and CGB5 (0.0017) (Table 1). Positive Tajima D-values (Table 1) point out the excess of high-frequency SNPs for CGB7, CGB, and LHB. In contrast, for CGB1, CGB2, and CGB5, Tajima D-values are mostly negative, and the frequency distributions tend to be skewed toward rare variants. However, as theoretical simulations have shown that statistical tests of neutrality based on the standard coalescent theory for a single-copy gene may not be appropriate for duplicated genes (Innan 2003), the Tajima test results should be interpreted with caution.
A new type of polymorphism—multisite variation (MSV) (Fredman et al. 2004)—has been described for SNPs in duplicons. Of the SNPs determined in individual LHB/CGB genes, 35%–77% were actually MSVs (Table 1; Supplemental Table S2)—SNPs that are also represented as paralogous sequence variants (PSVs) among duplicons (MSV1) or “shared” SNPs located at the same position in several duplicated genes (MSV2). The spread of MSV2 ranged from only a pair of genes “sharing SNPs” up to polymorphisms mapping at the same position for six genes (Fig. 2B; Supplemental Table S2). MSVs and high sequence diversity are explainable by gene conversion spreading SNPs among paralogous genes. An alternative scenario, parallel de novo mutations in several gene copies, is less likely but cannot be ruled out.
For LHB and four hCG β-subunit coding genes, only a few nonsynonymous mutations were identified: two signal peptide (LHB) and eight mature peptide variants (two for LHB, one for CGB5, four for CGB7, and one shared by CGB, CGB5, and CGB7) (Supplemental Table S3). Two of the variants overlap with the previously characterized mutations: (1) Ala-3Thr in the signal peptide of LHβ (Jiang et al. 2002) identified with a low frequency (2.2%) in Mandenka; and (2) a worldwide-spread Trp8Arg variant (Estonians 12%, Mandenka 7%, and Han 4%) in LHβ mature protein (Pettersson et al. 1994; Nilsson et al. 1997).
Traces of gene conversion among LHB/CGB genes
When polymorphism data are obtained from duplicate loci, gene conversion between copies is visually detectable if polymorphisms are shared between the loci. We aligned complete nucleotide sequence variants for all possible gene pairs to identify clustered polymorphism motifs potentially generated by gene conversion between duplicate genes (Fig. 2B,C; Table 2). Altogether 27 gene-conversion sites were identified (each might be a target for 1 + n gene-conversion events) with a minimum observed tract of 2–387 bp (mean 57 bp, median 23 bp) and maximum extension up to 796 bp (mean and median for maximum tract length across sites 229 and 138 bp, respectively). The highest number of acceptor sites was identified for CGB (eight sites) and CGB7 (seven sites); fewer converted segments were determined within LHB (two sites) and CGB2 (two sites), CGB1 (three sites) and CGB5 (three sites) (Fig. 2B). For some acceptor sites, there were multiple potential donor genes; for other sites, the donor gene could be unequivocally determined (Table 2; Fig. 2C). Gene conversion events involving the 5′-UTR up to +60 were identified only between CGB, CGB5, and CGB7, all coding the hCG β-subunit (Table 2). In the case of the functionally divergent LHB (specificity defined by exon 3) and CGB1/2 (different ORF), detectable acceptor sites are clustered mostly in the middle of the gene sequence. Of 29 “shared” polymorphic sites or MSV2, several were identified as part of minimum gene-conversion tracts, supporting the hypothesis that they were derived from gene-conversion events rather than being just highly mutable positions (Fig. 2B,C).
Table 2.
Minimal gene conversion (GC) tracts identified within LHB/CGB genes and potential donor genes of a converted segment
|
Acceptor GC site
|
Potential donor gene
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | No. | LHB | CGB | CGB2 | CGB1 | CGB5 | CGB7 | CGB8a |
| LHB | 1 | +122; +125 | +122; +125 | +122; +125 | +122; +125 | +122; +125 | +122; +125 | |
| +122; +154 | ||||||||
| 2 | +466; +546 | +466; +546 | +466; +546 | +466; +546 | +466; +546 | +466; +546 | ||
| +484; +546 | +484; +546 | +484; +546 | +484; +546 | +484; +546 | +484; +546 | |||
| CGB | 1 | -328; -268 | ||||||
| 2 | -185; -180 | -185; -180 | -185; -180 | |||||
| -185; -144 | ||||||||
| 3 | -111; -109 | -111; -109 | ||||||
| 4 | -77; -76 | -77; -76 | ||||||
| 5 | +88; +101 | +88; +101 | +88; +101 | +88; +101 | +88; +101 | |||
| 6 | +126; +168 | +155; +168 | +155; +168 | |||||
| +163; +168 | +163; +168 | +163; +168 | +163; +168 | |||||
| 7 | +554; +674 | |||||||
| 8 | +751; +1137 | +751; +1026 | ||||||
| +1026; +1038 | +1026; +1137 | +1026; +1038 | ||||||
| +1038; +1109 | ||||||||
| CGB2 | 1 | +353; +354 | +353; +354 | +353; +354 | +353; +354 | +353; +354 | +353; +354 | |
| 2 | +507; +531 | +507; +531 | ||||||
| CGB1 | 1 | +219; +241 | ||||||
| 2 | +452; +458 | +452; +458 | +452; +458 | +452; +458 | +452; +458 | +452; +458 | ||
| 3 | +794; +1150 | +1087; +1150 | ||||||
| +1136; +1150 | +1136; +1150 | +1136; +1150 | +1136; +1150 | |||||
| CGB5 | 1 | No GC tracts identified | +151; +160 | |||||
| 2 | +179; +186 | +179; +186 | ||||||
| 3 | +673; +704 | +673; +704 | +673; +704 | +673; +704 | ||||
| CGB7 | 1 | -394; -396 | -394; -396 | |||||
| 2 | -379; -357 | |||||||
| 3 | -58; -46 | -58; -46 | -58; -46 | |||||
| -58; +60 | ||||||||
| 4 | +198; +200 | +198; +301 | +198; +200 | +198; +200 | +198; +301 | |||
| +298; +301 | +298; +301 | +298; +299 | ||||||
| 5 | +422; +423 | +422; +423 | +417; +423 | +417; +423 | +417; +423 | +422; +423 | ||
| +417; +553 | +417; +553 | |||||||
| 6 | +1061; +1151 | |||||||
| 7 | +1241; +1278 | |||||||
| CGB8a | 1 | No GC tracts identified | No GC tracts identified | No GC tracts identified | -268; -265 | |||
| 2 | +1146; +1147 | +1146; +1147 | ||||||
Exact position of GC site (beginning; end) is defined by its localization in acceptor gene relative to ATG. Distribution of GC acceptor sites relative to exon—intron structure of the genes is presented in Figure 2.
CGB8 data are from the analysis of 11 Estonian individuals; the analysis of the rest of the genes is based on the combined sample set of 95 Estonian, Han, and Mandenka individuals.
Alternatively, Sawyer's gene-conversion detection algorithm (Sawyer 1989), implemented using the GENECONV computer program, was used. This algorithm does not rely on the sample's polymorphism data, but detects whether individual pairs of sequences share unusually long stretches of similarity given their overall similarity. The analysis predicts the fragments likely to have been converted between gene pairs, but the results do not give information about the direction of the gene-conversion events. Altogether, the GENECONV algorithm estimated 398 conversion events between all pairs of genes in the LHB/CGB cluster (Supplemental Table S4). No preference was detected in the involvement of a gene pair respective to their mutual orientation (either direct or inverted duplicates). The length of estimated gene-conversion tracts ranged from 35 to 1055 bp (mean 313 bp, median 291 bp), somewhat longer than the estimates based on shared polymorphisms. The difference could arise, as the GENECONV algorithm is also able to detect gene-conversion events that did not lead to “shared” SNPs. The analysis highlighted CGB2 as the most active participant (either donor or acceptor) in gene-conversion events, in contrast to the least involved functionally divergent LHB (Supplemental Table S4). The maximum number of estimated between-loci events reached 49 and 44 for CGB2–CGB and CGB2–CGB7 gene pairs, respectively. An association was detected between the number of conversion events estimated by the GENECONV algorithm and the number of shared SNPs (MSV2) between gene pairs (Pearson's correlation coefficient 0.44, p = 0.044) (Supplemental Fig. S3).
Estimation of population crossing-over and linkage disequilibrium parameters
Two approaches were used to characterize the decay of linkage disequilibrium between SNPs across the LHB/CGB region. First, we quantified the levels of LD by estimating the population crossing-over parameter ρ/bp=4Necbp, where Ne is the effective population size and cbp is the crossing-over rate per generation between adjacent nucleotide positions. The parameter ρ is a key determinant of LD patterns, with the strength of LD decreasing when ρ increases. We used two alternative algorithms: (1) the Li and Stephens (2003) “product of approximate conditionals” (PAC) likelihood method based on simultaneous analysis of all loci; and (2) Hudson's (2001) “composite likelihood” (CL) method based on multiplying together the likelihoods for every pair of sites. The first method has the advantage of allowing variation of recombination rate across the region of interest (Li and Stephens 2003). The extension of the second approach has the advantage of allowing simultaneous estimation of ρCL and f, where f is the ratio of gene-conversion to crossing-over events (Frisse et al. 2001). The average recombination rate calculated across the studied region for SNPs with MAF > 10% (Estonians, ρPAC = 4.43 × 10–4, ρCL = 6.13–7.40 × 10–4; Han, ρPAC = 2.34 × 10–4, ρCL = 6.42–7.06 × 10–4; Mandenka, ρPAC = 8.93 × 10–4, ρCL = 1.011–1.492 × 10–3) falls in the range published for a large set of 74 genes (Table 3A; Crawford et al. 2004). Higher ρ values for Africans are consistent with the idea that African populations maintained a larger long-term effective population size Ne than did non-African ones (Frisse et al. 2001). When two ρ estimates are compared, ρCL provides generally higher estimates than ρPAC. Also, ρCL based on two-locus sampling distribution seems to be somewhat less influenced by demography (closer estimates for different population samples) and less biased by SNP frequencies (less variation in estimates using all or only common, >10% MAF, SNPs) than multiloci-based ρPAC (Table 3A). When gene conversion was incorporated into the model, ρCL estimates for the LHB/CGB region decreased, but were independent of the length of assumed conversion tract (L) for any given sample. In contrast, the estimated ratio of gene-conversion to crossing-over rate depended inversely on the conversion tract length: As the length of the tract decreases, the estimated rate of gene conversion increases. Including only common SNPs, the maximum likelihood estimate for L = 30 bp ranged from 6 (Han) to 16 (Mandenka); whereas for L = 500 bp, f is 0.5 (Han) to 1.5 (Mandenka). This difference in estimates of f could result from the fact that effects of high gene-conversion rates with small tracts are similar to the effects of lower conversion rates with longer tracts. Recently, Ptak et al. (2004a) reported similar estimates of f for L = 500 (African-Americans, f ∼ 1; CEPH, f ∼ 0.25) obtained from joint analysis of 84 genomic regions. Reports from single sperm analysis seem to support much shorter conversion tract lengths, 54–132 bp for HLA-DPB1 (Zangenberg et al. 1995) and 55–290 bp for DNA3 loci (Jeffreys and May 2004). As the tract lengths identified for LHB/CGB genes are consistent with the single sperm data, we suggest f to range from 2 to 16.
Table 3.
Estimation of population crossing-over and gene conversion rates
|
A. Whole LHB/CGB region
|
B. Recombination hotspot
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Estonians
|
Han
|
Mandenka
|
Estonians
|
||||||
| All SNPs | ≥10% MAFd | All SNPs | ≥10% MAF | All SNPs | ≥10% MAF | All SNPs | >10% MAF | ||
| Maximum PAC likelihood estimates (Li and Stephens 2003) | |||||||||
| ρa (× 104) | 4.16 | 4.43 | 2.91 | 2.34 | 17.00 | 8.93 | 43.21 | 35.91 | |
| Composite likelihood estimates (Frisse et al. 2001; Hudson 2001) | |||||||||
| L = 0b | ρ (× 104) | 8.02 | 7.40 | 7.21 | 7.06 | 17.63 | 14.92 | 93.42 | 68.05 |
| L = 30 | ρ (× 104) | 6.38 | 6.25 | 6.60 | 6.53 | 12.29 | 10.38 | 68.05 | 56.17 |
| fc | 14 | 12.5 | 6 | 6 | 13 | 16 | 3 | 2 | |
| L = 50 | ρ (× 104) | 6.36 | 6.21 | 6.55 | 6.53 | 12.29 | 10.32 | 67.36 | 54.63 |
| f | 9 | 8 | 4 | 3.5 | 8 | 10 | 2 | 1.5 | |
| L = 100 | ρ (× 104) | 6.41 | 6.23 | 6.55 | 6.49 | 12.12 | 10.17 | 57.96 | 52.83 |
| f | 4.5 | 4 | 2 | 2 | 4.5 | 5.5 | 2 | 1 | |
| L = 250 | ρ (× 104) | 6.41 | 6.13 | 6.82 | 6.68 | 12.18 | 10.11 | 35.96 | 34.58 |
| f | 2 | 0.5 | 0.5 | 2.5 | 2 | 2.5 | 3 | 2 | |
| L = 500 | ρ (× 104) | 6.49 | 6.20 | 6.49 | 6.42 | 11.80 | 10.25 | 16.11 | 17.74 |
| f | 1 | 1 | 0.5 | 0.5 | 1 | 1.5 | 6.5 | 4 | |
The population crossing-over rate per base pair, ρ = 4Necbp.
L is the length of the gene conversion tract.
f = g/c is the ratio of gene conversion to the crossing-over rate.
SNPs with minor allele frequency ≥ 10%.
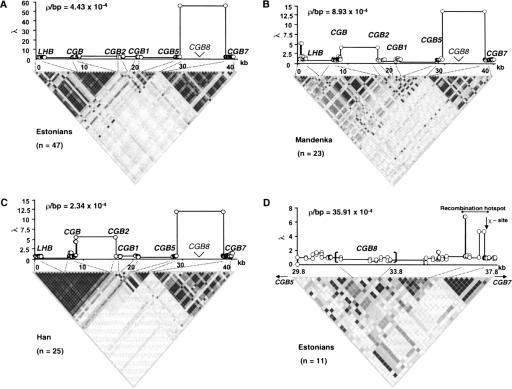
In addition to ρ estimation, we used a descriptive approach relying on summarizing LD by a pairwise summary, r2, which measures the correlation between alleles. In order to overcome the sensitivity to allele frequencies, we included only SNPs with MAF > 10%. Consistent with previous studies (for review, see Tishkoff and Verrelli 2003), the LD in African Mandenka does not extend as far as in non-African Han Chinese (represents an Asian population) and Estonians (represents a typical European population) (Fig. 3A,B,C; Dawson et al. 2002). In general, strong allelic associations at the periphery (LHB, CGB, CGB7) and breakdown of the LD toward the center (CGB1, CGB2, CGB5) characterize the LD structure of the LHB/CGB cluster in all studied population samples. Three arbitrary “LD blocks” could be defined: strongly associated LHB and CGB; CGB5 weakly associated with CGB1; and the strong block of CGB7. In all three populations, LD breaks sharply down between CGB5 and CGB7. Using the Li and Stephens (2003) algorithm, a recombination “hotspot” was estimated between CGB5 and CGB7 genes in all studied populations (λ = 57.1 for Estonians, 11.6 for Han, and 13.6 for Mandenka) (Fig. 3A,B,C). Another, “warm spot” was identified between CGB and CGB2 (λ = 2.36 for Estonians, 5.47 for Han, and 4.17 for Mandenka). Consistently, the predicted 8.3-kb hotspot is colocalized with the strongest LD-breakdown region within the cluster (Fig. 3A,B,C). No recombination hotspots were predicted within the LHB/CGB genes, shown above as active in gene conversion.
Figure 3.
LD structure by r2-blot and estimation of crossing-over activity (measured by ρ =4Necbp) and potential recombination hotspots (measured by [λ = ρ between a locus pair]/[average ρ for the analyzed region]) by the PHASE algorithm (Stephens et al. 2001) across the LHB/CGB genome cluster for SNPs (○) with MAF > 10%: (A) Estonians; (B) Mandenka; and (C) Han Chinese samples. Regions of predicted high crossover activity colocalize with the breakdown (white) LD areas and low recombination rate with strong allelic associations (black/dark gray). (D) Refinement of recombination hotspot by resequencing genomic region between CGB5 and CGB7 genes in an Estonian sample as a model. The recombination hotspot colocalizes with LD breakdown and is located between CGB8 and CGB7 within a <1-kb region next to a crossover-associated Escherichia coli χ-sequence.
We also explored the relationship of gene-conversion acceptor-site distribution and linkage disequilibrium (LD) patterns at the LHB/CGB gene cluster. In contrast to the predictions (Andolfatto and Nordborg 1998; Ardlie et al. 2001; Frisse et al. 2001), most gene-conversion acceptor sites are located in high-LD regions within peripheral genes (Figs. 2B and 3A,B,C; Supplemental Fig. S4). Thus, direction of gene-conversion activity from the center (mainly donor genes) toward the periphery of the cluster (acceptor genes) has led not only to high sequence diversity but also generated strong LD for the peripheral genes. New alleles copied from the donor to the acceptor gene have been acting like novel mutations increasing variation and creating LD around the recipient site.
Structural analysis of the potential recombination hotspot
In order to narrow down the potential hotspot region, we resequenced the 8.3-kb region from CGB5 up to CGB7, using an Estonian sample (n = 11) as a model. The sequence diversity parameters as well as the gene-conversion activity of the included CGB8 gene were similar to neighboring CGB5 (Table 1; Fig. 2B). Apparently owing to the small sample size and fewer SNPs, both estimates of recombination rate parameters ρPAC and ρCL exhibit more variation compared to the analysis of the whole LHB/CGB region, depending on SNP allele frequencies as well as the assumed length of the gene-conversion tract (for ρCL) (Table 3B). However, despite both approaches' struggle to provide accurate estimates of ρ, they are consistent in that the average recombination rate of the potential hotspot region is ∼10 times higher compared to the rest of the LHB/CGB cluster (Table 3B). The estimates of f (ranging from 1 to 6.5 compared to the ρCL estimate across the 8.3-kb region) suggest that this region has also gene-conversion activity as high as or even higher than the LHB/CGB genes (Table 3B). When pairwise LD patterns were studied, strong associations were detected only between a few scattered loci, except in the region adjacent to CGB7 (Fig. 3D). The potential recombination hotspot was narrowed within a <1-kb region colocalizing with LD breakdown between CGB8 and CGB7, embedded within an Alu-rich (∼75% Alu-sequences) segment and 90–100 bp from a recombination-associated χ-sequence (Fig. 3D). The hotspot exhibited 6.9 times higher ρPAC compared to an average of the 8.3-kb region. As the recombination rate between CGB5–CGB7 was estimated to be ∼10 times higher compared to the whole LHB/CGB region, the crossing-over rate of the hotspot exceeds ∼70 times the background rate in the gene cluster.
Further sequence analysis revealed inverted Alu repeats (625 bp) that could give rise to stem–loop secondary structure formation (Fig. 1B). The single-stranded loop segment (222 bp), located exactly at the center of the predicted hotspot, could be sensitive to chromatin-altering factors and thus promote double-strand breaks (DSBs) and recombination/gene conversion (Akgün et al. 1997). The stem–loop structure might also disrupt DNA synthesis during replication, generating an unbound 3′-tail of the nascent strand that could invade homologous regions in the center of the cluster (Lobachev et al. 1998). The hypotheses of chromosome-altering factors and stalled replication are both sufficient to explain the direction of gene conversion from the center toward the periphery of the cluster, the invading strand always acting as the recipient during a gene-conversion event (Akgün et al. 1997). A region between CGB and CGB2 predicted as a warm spot in recombination analysis involves also inverted repeats (2094 bp), but with a much longer spacer between them (2788 bp), which might hinder the formation of a stem–loop secondary structure as stable as predicted for the hotspot (Lobachev et al. 1998). A segment homologous to hot and warm spots, located between CGB1 and CGB5, has undergone an inversion resulting in the partial loss of a palindromic DNA fragment preceding an NTF6G′ pseudogene (Fig. 1A), thus prohibiting the formation of a proper stem–loop structure.
Discussion
Gene conversion generates high diversity and strong LD for acceptor sites
Re-sequencing LHB/CGB genes in a sample set from populations of three continents revealed a high number of SNPs, altogether 206 found for all genes in a pooled sample. Few regions in the human genome re-sequenced so far have exceeded nucleotide diversity estimates π > 0.002: Examples are ABO and KNG (Crawford et al. 2004) and the X-linked long-wave “red” opsin gene OPN1LW, arisen from the tandem duplication ∼30–40 million years ago (Mya) among Old World primates and shaped by gene conversion between OPN1LW and OPN1MW (Verrelli and Tishkoff 2004). Only one study (Bosch et al. 2004) has reported a diversity level as high as we determined for CGB7 (π = 0.00550): for the distal Y-chromosomal direct HERV repeats (π = 0.00544), shaped by directional gene conversion from proximal (π = 0.0016) to distal repeat. In LHB/CGB genes, part of the high diversity is due to multisite variations (MSVs): SNPs located at the same position in several genes or represented also as paralogous sequence variants. Although parallel de novo mutations cannot be ruled out as the source for MSVs, gene conversion between highly homologous LHB/CGB genes is a more likely scenario. As a support to this scenario, we identified MSVs within multiple gene-conversion tracts between gene pairs, detected by alignment of gene variants as well as by computational analysis using the GENECONV algorithm. Directional gene conversion has shaped the diversity patterns of the LHB/CGB region. Central genes of the cluster were characterized with mainly gene donor activity and lower variation, in contrast to highly diverse peripheral genes rich in acceptor sites.
It has been suggested that over short distances, gene conversion, rather than crossing over, is likely to be the dominant force that breaks up associations among sites (Andolfatto and Nordborg 1998; Ardlie et al. 2001; Frisse et al. 2001). Results from the LHB/CGB cluster did not support this hypothesis. A majority of gene-conversion recipient sites colocalized with high-LD regions in the peripheral loci of the region, whereas the middle of the cluster was characterized by LD breakdown and lower gene-conversion acceptor activity.
These observations are consistent with theoretical simulations showing that relative to a single-gene model, polymorphism may be elevated and positive LD created at duplicated genes due to gene conversion (Innan 2002, 2003).
Functional consequences of gene conversion in LHB/CGB genes
For duplicated genes, gene conversion has been shown to be an essential source for spreading disease mutations. For example, Boocock et al. (2003) showed that in the case of Shwachman-Diamond syndrome (SDS), 85% of patients carried a mutation in the SBDS gene originating from a neighboring pseudogene copy, SBDSP. To what extent has gene conversion shaped variation of coding sequences for LHB/CGB genes? The worldwide-spread Trp8Arg variant (Nilsson et al 1997) as well as the neighboring His10Arg change (Supplemental Table S3) in exon 2 of LHB most probably originate from one of the hCG β-subunit-coding genes having in these positions conserved arginines. Another example is a widely spread variant of CGB7 (41.5% in Estonians, 19.6% in Mandenka, and 28% in Han), consisting of three polymorphisms in exon 2 (+417, +422, and +423) and coding two associated amino acid changes: Arg2Lys and Met4Pro (Supplemental Table S3). Arg2–Met4 combination is unique to the CGB7 gene, whereas CGB, CGB5, and CGB8 carry the Lys2–Pro4 variant. As this segment in the CGB7 gene has also been found to be within a gene-conversion acceptor site (Fig. 2B; Table 2), we interpret that the Lys2–Pro4 variant in CGB7 originates from another CGB gene. Possible sources for novel variants in LHB and CGB, CGB5, CGB7, and CGB8 are CGB1/CGB2, which have a 1-bp-shifted ORF and divergence in 5′- and 3′-UTRs. Thus, a neutral polymorphism of CGB1/CGB2 could cause an amino acid change in a duplicate gene if spread by gene conversion. As an example, we have identified a rare Asp117Ala change in exon 3 of CGB and CGB5, potentially originating from a common variant in the 3′-UTR region of CGB2 (SNP at position +1087) (Supplemental Table S2).
Variation patterns in LHB/CGB region and population demography
Current patterns of human genetic variation reflect not only crossover history, but also past demographic processes as well as possible selective pressures on studied genes. In the LHB/CGB region, higher diversity and number of SNPs, shorter range of LD and less pronounced LD structure, as well as higher estimated recombination rates (measured by ρ =4Necbp) were detected for the Mandenka compared to the Estonian and the Han samples. These differences are likely to be explained by the distinct demographic histories of African and non-African populations: The former are older and have maintained larger Ne, and the latter have experienced a bottleneck event during the expansion of modern humans out of Africa within the past 100,000 years (for review, see Tishkoff and Verelli 2003). As a result of the bottleneck, non-African populations represent only a subset of African diversity and exhibit longer LD created during the founding event and maintained in rapidly expanding populations. It is noteworthy that despite high diversity as well as differences in variation and LD levels across the genes and populations, each LHB/CGB gene has only two to four major haplotypes in a population (Table 1). The joint bottleneck in the history of non-African populations is supported by mostly shared common gene variants between Estonians and Han, whereas Mandenka have several population-specific high-frequency haplotypes (data not shown). This observation has an important implication for LD-based mapping, suggesting that core variants in highly diverse regions may also be “tagged” by a few SNPs when an appropriate marker density is chosen.
Although putative hotspots and warm spots of recombination were predicted within the same regions in all studied samples, the estimated crossover intensity at these spots differs severalfold among populations (Fig. 3). If the background recombination rate (ρPAC) is taken into account (Table 3A), the hotspot crossover intensity for the Estonians exceeds approximately twice the estimation for the Mandenka and 10-fold for the Han. In contrast, fourfold higher activity is predicted for the warm spot in the Mandenka compared to the other populations. This is consistent with a suggestion that local recombination rates can vary among human populations because of differences in their allele frequencies or in historical factors affecting Ne in local regions of the genome (Jeffreys and Neumann 2002; Crawford et al. 2004; Ptak et al. 2004b; Evans and Cardon 2005).
Recombination and gene-conversion activity are potentially associated with palindrome sequences
In yeast, recombination activity has been associated with high G+C content, nuclease-sensitive chromatin, and transcription factor binding sites. Although no sequence motifs are known to predict recombination hotspots in humans, putative crossover-initiating motifs have been identified in other species (Petes 2001; De Massy 2003). The LHB/CGB gene cluster has all the properties described for a recombination-active region: extremely C+G-rich, Alu-richness, and the presence of several χ-sequence motifs, associated with crossover activity in several species. Despite high gene-conversion activity among the LHB/CGB genes, we estimated only one recombination hotspot within an intergenic region. Apparently, gene conversion between duplicons in the human genome also occurs without crossovers, consistent with the synthesis-dependent strand-annealing (SDSA) pathway described for yeast (Allers and Lichten 2001). What could be the determinants of the estimated potential recombination hotspot within the LHB/CGB cluster? It is probably not defined by primary DNA sequence, as there are two other highly homologous segments (Fig. 1A) located within the cluster. It has been suggested that double-stranded breaks, which are prerequisites for crossover initiation, are stimulated by the formation of palindromic secondary structures (Krawinkel et al. 1986; Akgün et al. 1997; Lobachev et al. 1998). Indeed, a stable stem–loop is formed around the center of the predicted hotspot (Fig. 1B), which is not the case for the two homologous regions because of minor rearrangements of these DNA segments. The hypothesis of palindrome sequences stimulating DSBs and crossover activity is supported by direct sperm analysis—a structurally similar recombination hotspot, bordered by inverted Alu-motifs and characterized by crossover asymmetry, has been identified for the MHC hotspot DNA2 (Jeffreys and Neumann 2002). We suggest that a high recombination rate and low LD, but also high gene-conversion activity in segmental duplications, could be favored by secondary structures formed by palindrome sequences. The abundance of direct and inverted repeats common in segmental duplications provides the basis for DNA secondary structure formations, initiating DSBs.
Methods
In silico analysis of human LHB/CGB genome cluster
The structure of the LHB/CGB genome cluster (#MIM 152780, 118860, 608823–608827; http://www.ncbi.nlm.nih.gov/Omim/) has been reconstructed by Web-based global alignment (http://www.ebi.ac.uk/clustalw/; CLUSTALW) and BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) tools. For the analysis, we used the sequence obtained from the NCBI GenBank database (http://www.ncbi.nlm.nih.gov; locus no NG_000019; June 26, 2002 release). Prediction of palindromic features was performed using the EMBOSS einverted program (http://emboss.sourceforge.net/; Rice et al 2000).
Population samples
The study has been approved by the Ethics Committee of Human Research of the University Clinic of Tartu, Estonia (permission no. 117/9, 16.06.03). For re-sequencing of six genes (LHB, CGB, CGB1, CGB2, CGB5, and CGB7), in total 95 DNA samples from three continents were used: 47 Estonian (Europe), 23 Mandenka (Africa), and 25 Chinese Han (Asia) individuals. The Estonian sample represents a typical European population (Dawson et al. 2002). Mandenka and Han samples were obtained from the HGDP-CEPH Human Genome Diversity Cell Line Panel (http://www.cephb.fr/HGDP-CEPH-Panel/; Cann et al 2002). The detailed analysis of the predicted recombination hotspot was conducted using a sample of 11 Estonian individuals.
Gene-specific and long-range PCR
A total of 12 PCR primers for the LHB, CGB, CGB1, CGB2, CGB5, and CGB7 genes were designed based on the human chorionic gonadotropin β region sequence (NCBI Refseq NG_000019) using the Web-based version of the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). While designing PCR primers aiming to result in gene-specific amplification products, we relied on the detailed structure of the LHB/CGB region (Fig. 1). The uniqueness of all PCR primers was checked using BLAST, and only primer pairs with at least one primer being unique in the human genome were regarded suitable for amplification. The six genes were amplified to cover the entire coding sequence and part of flanking regions; amplified fragments were 1599–2364 bp long. Specificity of the PCR products was controlled in three steps: (1) design of unique primer pairs capable to amplify only one of the duplicated genes; (2) verification of monomorphic status for gene-specific positions used as markers for each individual gene (Supplemental Fig. S1); (3) test for Hardy-Weinberg Equilibrium for each identified SNP.
Amplification of genomic DNA (100 ng) was performed using the Long PCR Enzyme Mix (MBI Fermentas) by the standard protocol recommended by the manufacturer. Amplifications were performed in a PTC-200 thermal cycler (MJ Research). The reactions were initiated with a denaturation at 95°C for 5 min, followed by 10 cycles of denaturation at 95°C for 20 sec, annealing at 68°C for 30 sec (decrease of temperature 1°C per cycle), elongation at 68°C for 2 min; 10 cycles of 95°C (20 sec), 56°C (30 sec), 68°C (2 min); 10 cycles of 95°C (20 sec), 54°C (30 sec), 68°C (2 min); 10 cycles of 95°C (20 sec), 51°C (30 sec), and 68°C (2 min). A final extension step was performed at 68°C for 10 min.
The potential hotspot region between CGB5 and CGB7 was amplified in two stages. First, a long-range PCR was conducted that yielded a product of 8.3 kb. Second, six inner fragments (1193–1675 bp) were reamplified by nested PCR. Amplifications of 100 ng of genomic DNA (Long PCR Enzyme Mix; MBI Fermentas) were performed in a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems). The reactions were initiated with a denaturation at 94°C for 5 min, followed by four cycles of denaturation at 94°C for 20 sec, annealing at 68°C for 30 sec (decrease of temperature 1°C per cycle), elongation at 68°C for 8 min, 11 cycles at 94°C (20 sec), 64°C (30 sec), 68°C (8 min); 25 cycles at 94°C (20 sec), 64°C (30 sec), and 68°C (8 min + 5 sec per cycle). A final extension step was performed at 68°C for 10 min. All primer sequences are available upon request.
Re-sequencing
To remove unincorporated PCR primers and mononucleotides, PCR products were treated with exonuclease I (1 U; MBI Fermentas) and shrimp alkaline phosphatase (1.5 U; USB) and incubated in a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems) at 37°C for 20 min followed by enzyme inactivation at 80°C for 15 min. Purified PCR product (1.5–3 μL) served as a template in sequencing reactions (10 μL) with sequencing primer (2 pmol) and DYEnamic ET Terminator Cycle Sequencing Kit reagent premix (Amersham Biosciences Inc.) as recommended by the supplier. LHB, CGB, CGB1, CGB2, CGB5, and CGB7 genes were sequenced from both strands and using six different sequencing primers. Altogether 20 sequencing primers for LHB, CGB, CGB1, CGB2, CGB5, and CGB7 genes (a set of six primers for every gene) and 36 primers for 8.3-kb hotspot region (six for each nested PCR product) were designed as described above for resequencing of both strands. Sequencing reactions (1.5 μL) were run on an ABI 377 Prism automated DNA sequencer (Applied Biosystems) using ReproGel 377 gels (Amersham Biosciences Inc.).
For each gene and each population, the sequence data were assembled into a contig using phred and phrap software (Ewing et al 1998), and the contig was edited in a consed package (Gordon et al. 1998) to ensure that the assembly was accurate (http://www.phrap.org/phredphrapconsed.html). Polymorphisms were identified using the polyphred program (Version 4.2) (Nickerson et al. 1997) and confirmed by manual checking. A genetic variant was verified only if it was observed in both the forward and the reverse orientations. Allele frequencies were estimated and conformance with HWE was computed by an exact test (α = 0.05) using HaploView (http://www.broad.mit.edu/mpg/haploview/index.php; Barrett et al. 2005) program. In total, six rare SNPs for Mandenka or Han were found to be deviating from HWE, apparently because of small sample size.
Statistical analysis
Sequence diversity parameters were calculated by DnaSP software (Version 4.0) (http://www.ub.es/dnasp/; Rozas and Rozas 1999). The direct estimate of per-site heterozygosity (π) was derived from the average pairwise sequence difference, and Watterson's θ (Watterson 1975) represents an estimate of the expected per-site heterozygosity based on the number of segregating sites (S). Tajima's D (DT) statistic (Tajima 1989) was performed to determine if the observed patterns of diversity in the three studied population samples are consistent with the standard neutral model. Significant positive DT values may indicate an excess of intermediate-frequency SNPs consistent with balancing selection as well as population bottlenecks or subdivision, whereas significant negative DT values indicate an excess of low-frequency SNPs consistent with recent directional selection or population expansion. Haplotypes were inferred from unphased genotype data using the Bayesian statistical method in the program PHASE 2.1 (http://www.stat.washington.edu/stephens/; Stephens et al. 2001). For haplotype reconstruction, the model allowing recombination was used. Running parameters for PHASE are described below.
Detection of gene-conversion events
Gene sequence variants derived from estimated haplotypes were used for gene-conversion analysis. For manual detection of gene-conversion sites between a pair of LHB/CGB genes, the derived complete sequence variants were aligned using Web-based ClustalW. A minimum gene-conversion site was defined as a region within an acceptor gene with ≥2 associated, motif-forming polymorphisms for which a potential donor gene could be defined. The maximum possible gene-conversion tract covers the identical sequence between two compared genes on both sides of the minimum gene-conversion tract. Alternatively, the aligned sequences of all possible gene pairs were analyzed for evidence of gene conversion using Stanley Sawyer's gene-conversion detection method as implemented in his GENECONV program (Version 1.81) (http://www.math.wustl.edu/∼sawyer/geneconv/; Sawyer 1989). Sawyer's gene-conversion detection algorithm detects whether pairs of sequences share unusually long stretches of similarity given their overall similarity. The GENECONV program computes global and pairwise p-values and allows mismatches within converted regions. Global and pairwise p-values are calculated using two methods. The first method is based on (10,000) permutations of the original data, and the second is based on a method similar to that used by the BLAST database-searching algorithm. Here, we only used p-values from permutations (simulations) because they are more conservative and accurate. We also only considered p-values (p < 0.05) from global fragments because their p-values are corrected for multiple comparisons whereas the p-values of pairwise fragments are not. Alignments were analyzed using the most stringent “g0” parameter, meaning that mismatches within fragments are not allowed.
Measures of linkage disequilibrium
The descriptive statistic of linkage disequilibrium (LD), r2 (Hill and Robertson 1968), was calculated for pairs of markers and summarized using Haploview software (Barrett et al. 2005). Reliable LD patterns were achieved by inclusion of only common SNPs with minor allele frequency (MAF) >10%. To locate gene-conversion acceptor sites at the LD landscape of LHB/CGB cluster, we calculated pairwise LD for all identified SNPs, as several converted SNPs represent low allele frequencies.
Another way to quantify levels of LD is to estimate the population crossing-over parameter ρ =4Necbp, where Ne equals effective population size and cbp the crossing-over rate per base pair per generation. We estimated ρ using two alternative algorithms. The Li and Stephens (2003) method is based on a “Product of Approximate Conditionals” (PAC) model considering all loci simultaneously, allowing variation of recombination rate across the region of interest and thus estimation of putative recombination hotspots. Average background recombination rate (ρPAC) and the factor (λ) by which the recombination rate between loci exceeds the average background rate were estimated from unphased genotype data using the PHASE 2.1 software (http://www.stat.washington.edu/stephens/; Stephens et al. 2001; Li and Stephens 2003). Within this model, a λ value of 1 corresponds to an absence of recombination rate variation, while values of λ >1 indicate increase crossover activity. The value of 1< λ < 10 is considered a recombination “warm spot,” and the value of λ > 10 is considered a recombination “hotspot” (Crawford et al. 2004). For hotspot estimation, only common SNPs (MAF > 10%) were included in the analysis. The running parameters were number of iterations = 1000, thinning interval = 1, burn-in = 100; for increasing the number of iterations of the final run of the algorithm the -X10 parameter, making the final run 10 × longer than other runs, was used. To relax the assumption of stepwise mechanism inappropriate for triallelic SNPs, the -d1 option was used. For each sample set, we ran the algorithm 10 times, resulting in identical outputs of the parallel analysis; thus we used the median of the values obtained from one of the runs.
Alternatively, we used the “composite likelihood” (CL) method by Hudson (2001) to estimate simultaneously the population recombination parameter ρCL and f, where f is the ratio of gene conversion to crossing-over events (Frisse et al. 2001). Hudson's method is based on multiplying together likelihoods for every pair of sites genotyped, in which these pairwise likelihoods are computed via simulation, assuming an “infinite-sites” model. The method assumes that gene conversion and crossing-over are alternative solutions of a Holliday junction and that the conversion-tract length is geometrically distributed with mean length L. We obtained maximum likelihood estimates for ρCL and f from unphased data using MAXDIP (http://genapps.uchicago.edu/maxdip/index.html) with the following running parameters: starting value of ρ = 0.0002; f ranging from 0 to 30, with the intervals of 0.5. The analysis was run for gene-conversion-tract lengths L = 30, 50, 100, 250, and 500. The choice of the L values was based on reports from human single-sperm analysis (Zangenberg et al. 1995; Jeffreys and May 2004) and lengths of gene-conversion tracts identified for LHB/CGB genes.
Acknowledgments
We thank Howard Cann for providing Mandenka and Chinese Han DNA samples from the CEPH Human Genome Diversity Cell Line Panel; Roger Horton, Robert K. Campbell, Elin Lõhmussaar, Kristiina Rull, Tarmo Annilo, and Laura Sedman for comments on the manuscript; and Viljo Soo for technical help. The study has been supported by the Wellcome Trust International Senior Research Fellowship (grant no. 070191/Z/03/Z) in Biomedical Science in Central Europe (M.L.), Estonian Science Foundation grant no. 5796 (M.L.), and personal stipends from the Artur Lind Gene Technology Fund (P.H.) and Estonian-Revelia Harald Raudsepp Academic Fund (P.H. and L.N.).
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to dbSNP under accession nos. LHB ss48399882–ss48399908, CGB ss48399909–ss48399943, CGB1 ss48399944–ss48399963, CGB2 ss48399964–ss48399997, CGB5 ss48399998–ss48400022, CGB7 ss48400023–ss48400071, CGB8 ss48399818–ss48399832, CGB5-CGB8 intergenic region ss48399833–ss48399849, CGB8-CGB5 intergenic region ss48399850–ss48399881. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: H. Cann.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4270505. Freely available online through the Genome Research Immediate Open Access option.
References
- Akgün, E., Zahn, J., Baumes, S., Brown, G., Liang, F., Romanienko, P.J., Lewis, S., and Jasin, M. 1997. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17: 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers, T. and Lichten, M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P. and Nordborg, M. 1998. The effect of gene conversion on intralocus association. Genetics 148: 1397–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie, K., Liu-Cordero, S.N., Eberle, M.A., Daly, M., Barrett, J., Winchester, E., Lander, E.S., and Kruglyak, L. 2001. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am. J. Hum. Genet. 69: 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Liu, G., and Eichler, E.E. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 73: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, J.C., Fry, B., Maller, J., and Daly, M.J. 2005. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Bettencourt, B.R. and Feder, M.E. 2002. Rapid concerted evolution via gene conversion at the Drosophila hsp70 genes. J. Mol. Evol. 54: 569–586. [DOI] [PubMed] [Google Scholar]
- Boocock, G.R., Morrison, J.A., Popovic, M., Richards, N., Ellis, L., Durie, P.R., and Rommens, J.M. 2003. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 33: 97–101. [DOI] [PubMed] [Google Scholar]
- Bosch, E., Hurles, M.E., Navarro, A., and Jobling, M.A. 2004. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 14: 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann, H.M., de Toma, C., Cazes, L., Legrand, M.F., Morel, V., Piouffre, L., Bodmer, J., Bodmenr, W.F., Bonne-Tamir, B., Cambon-Thomsen, A., et al. 2002. Human genome diversity cell line panel. Science 296: 261–262. [DOI] [PubMed] [Google Scholar]
- Crawford, D.C., Bhangale, T., Li, N., Hellenthal, G., Rieder, M.J., Nickerson, D.A., and Stephens, M. 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36: 700–706. [DOI] [PubMed] [Google Scholar]
- Dawson, E., Abecasis, G.R., Bumpstead, S., Chen, Y., Hunt, S., Beare, D.M., Pabial, J., Dibling, T., Tinsley, E., Kirby, S., et al. 2002. A first-generation linkage disequilibrium map of human chromosome 22. Nature 418: 544–548. [DOI] [PubMed] [Google Scholar]
- De Massy, B. 2003. Distribution of meiotic recombination sites. Trends Genet. 19: 514–522. [DOI] [PubMed] [Google Scholar]
- Estivill, X., Cheung, J., Pujana, M.A., Nakabayashi, K., Scherer, S.W., and Tsui, L.-C. 2002. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms SNPs correlate with segmental duplications in the human genome. Hum. Mol. Genet. 11: 1987–1995. [DOI] [PubMed] [Google Scholar]
- Evans, D.M. and Cardon, L.R. 2005. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am. J. Hum. Genet. 76: 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M., and Green, P. 1998. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Fredman, D., White, S.J., Potter, S., Eichler, E.E., Den Dunnen, J.T., and Brookes, A.J. 2004. Complex SNP-related sequence variation in segmental genome duplications. Nat. Genet. 36: 861–866. [DOI] [PubMed] [Google Scholar]
- Frisse, L., Hudson, R.R., Bartoszewicz, A., Wall, J.D., Donfack, J., and Di Rienzo, A. 2001. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69: 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Hill, W.G. and Robertson, A. 1968. The effects of inbreeding at loci with heterozygote advantage. Genetics 60: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R., Wilming, L., Rand, V., Lovering, R.C., Bruford, E.A., Khodiyar, V.K., Lush, M.J., Povey, S., Talbot Jr., C.C., Wright, M.W., et al. 2004. Gene map of the extended human MHC. Nat. Rev. Genet. 5: 889–899. [DOI] [PubMed] [Google Scholar]
- Hudson, R.R. 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles, M.E. 2001. Gene conversion homogenizes the CMT1A paralogous repeats. BMC Genomics 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate, A.J., Feuk, L., Rivera, M.N., Listewnik, M.L., Donahoe, P.K., Qi, Y., Scherer, S.W., and Lee, C. 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36: 949–951. [DOI] [PubMed] [Google Scholar]
- Innan, H. 2002. A method for estimating the mutation, gene conversion and recombination parameters in small multigene families. Genetics 161: 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H. 2003. The coalescent and infinite-site model of a small multigene family. Genetics 163: 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A.J. and May, C.A. 2004. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36: 151–156. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A.J. and Neumann, R. 2002. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat. Genet. 31: 267–271. [DOI] [PubMed] [Google Scholar]
- Jiang, M., Lamminen, T., Pakarinen, P., Hellman, J., Manna, P., Herrerra, R.J., and Huhtaniemi, I. 2002. A novel Ala–3Thr mutation in the signal peptide of human luteinizing hormone β-subunit: Potentiation of the inositol phosphate signaling pathway and attenuation of the adenylate cyclase pathway by recombinant variant hormone. Mol. Hum. Reprod. 8: 201–212. [DOI] [PubMed] [Google Scholar]
- Krawinkel, U., Zoebelein, G., and Bothwell, A.L.M. 1986. Palindromic sequences are associated with sites of DNA breakage during gene conversion. Nucleic Acids Res. 14: 3871–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. and Stephens, M. 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165: 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev, K.S., Shor, B.M., Tran, H.T., Taylor, W., Keen, J.D., Resnick, M.A., and Gordenin, D.A. 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148: 1507–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston, G.A. and Ruvolo, M. 2002. Chorionic gonadotropin has a recent origin within primates and an evolutionary history of selection. Mol. Biol. Evol. 19: 320–335. [DOI] [PubMed] [Google Scholar]
- Nahon, J.-L. 2003. Birth of `human-specific' genes during primate evolution. Genetica 118: 193–208. [DOI] [PubMed] [Google Scholar]
- Nickerson, D.A., Tobe, V.O., and Taylor, S.L. 1997. Polyphred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, C., Pettersson, K., Millar, R.P., Coerver, K.A., Matzuk, M.M., and Huhtaniemi, I.T. 1997. Worldwide frequency of a common genetic variant of luteinizing hormone: An international collaborative research. International Collaborative Research Group. Fertil. Steril. 67: 998–1004. [DOI] [PubMed] [Google Scholar]
- Papadakis, M.N. and Patrinos, G.P. 1999. Contribution of gene conversion in the evolution of the human β-like globin gene family. Hum. Genet. 104: 117–125. [DOI] [PubMed] [Google Scholar]
- Petes, T.D. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360–369. [DOI] [PubMed] [Google Scholar]
- Pettersson, K., Mäkelä, M.M., Dahlen, P., Lamminen, T., Huoponen, K., and Huhtaniemi, I. 1994. Genetic polymorphism found in the LH β gene of an immunologically anomalous variant of human luteinizing hormone. Eur. J. Endocrinol. 130 [Suppl 2]: 65.8124480 [Google Scholar]
- Policastro, P.F., Daniels-Mcqueen, S., Carle, G., and Boime, I. 1986. A map of the hCG-LHB gene cluster. J. Biol. Chem. 261: 5907–5916. [PubMed] [Google Scholar]
- Ptak, S.E., Voelpel, K., and Przeworski, M. 2004a. Insights into recombination from patterns of linkage disequilibrium in humans. Genetics 167: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak, S.E., Roeder, A.D., Stephens, M., Gilad, Y., Pääbo, S., and Przeworski, M. 2004b. Absence of the TAP2 human recombination hotspot in chimpanzees. PLOS Biol. 2: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, P., Longden, I., and Bleasby, A. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Rozas, J. and Rozas, R. 1999. DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526–538. [DOI] [PubMed] [Google Scholar]
- Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528. [DOI] [PubMed] [Google Scholar]
- Shaw, C.J. and Lupski, J.R. 2004. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 13: R57–R64. [DOI] [PubMed] [Google Scholar]
- Smith, G.R. 1988. Homologous recombination in prokaryotes. Microbiol. Rev. 52: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz, P. and Lupski, J.R. 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18: 74–82. [DOI] [PubMed] [Google Scholar]
- Stephens, M., Smith, N.J., and Donnelly, P. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themmen, A.P.N. and Huhtaniemi, I.T. 2000. Mutations of gonadotropins and gonadotropin receptors: Elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocrine Rev. 21: 551–583. [DOI] [PubMed] [Google Scholar]
- Tishkoff, S. and Verrelli, B.C. 2003. Patterns of human genetic diversity: Implications for human evolutionary history and disease. Annu. Rev. Genomics Hum. Genet. 4: 293–340. [DOI] [PubMed] [Google Scholar]
- Tusié-Luna, M.-T. and White, P.C. 1995. Gene conversions and unequal crossovers between CYP21 steroid 21-hydroxylase gene and CYP21P involve different mechanisms. Proc. Natl. Acad. Sci. 92: 10796–10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli, B.C. and Tishkoff, S. 2004. Signatures of selection and gene conversion associated with human color vision variation. Am. J. Hum. Genet. 75: 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G.A. 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Wyckoff, G.J., Wang, W., and Wu, C.-I. 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403: 304–309. [DOI] [PubMed] [Google Scholar]
- Zangenberg, G., Huang, M.M., Arnheim, N., and Erlich, H. 1995. New HLA-DPB1 alleles generated by interallelic gene conversion detected by analysis of sperm. Nat Genet. 10: 407–414. [DOI] [PubMed] [Google Scholar]
Web site references
- http://emboss.sourceforge.net/; EMBOSS einverted Software.
- http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; Primer3 software.
- http://genapps.uchicago.edu/maxdip/index.html; MAXDIP application.
- http://www.broad.mit.edu/mpg/haploview/index.php; Haploview software.
- http://www.cephb.fr/HGDP-CEPH-Panel/; HGDP-CEPH Human Genome Diversity Cell Line Panel.
- http://www.ebi.ac.uk/clustalw/; ClustalW software.
- http://www.math.wustl.edu/∼sawyer/geneconv/; GENECONV computer program.
- http://www.ncbi.nlm.nih.gov/BLAST/; Basic Local Alignment Search Tool.
- http://www.ncbi.nlm.nih.gov/Omim/; Online Mendelian Inheritance in Man (OMIM).
- http://www.ncbi.nlm.nih.gov/SNP/index.html; dbSNP.
- http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information.
- http://www.phrap.org/phredphrapconsed.html; Phred, Phrap, Consed software.
- http://www.stat.washington.edu/stephens/; PHASE 2.1 software.
- http://www.ub.es/dnasp/; DNAsp software.