Abstract
The 2 Mb domain on chromosome 15q11–q13 that carries the imprinted genes involved in Prader–Willi (PWS) and Angelman (AS) syndromes is under the control of an imprinting center comprising two regulatory regions, the PWS-SRO located around the SNRPN promoter and the AS-SRO located 35 kb upstream. Here we describe the results of an analysis of the epigenetic features of these two sequences and their interaction. The AS-SRO is sensitive to DNase I, and packaged with acetylated histone H4 and methylated histone H3(K4) only on the maternal allele, and this imprinted epigenetic structure is maintained in dividing cells despite the absence of clearcut differential DNA methylation. Genetic analysis shows that the maternal AS-SRO is essential for setting up the DNA methylation state and closed chromatin structure of the neighboring PWS-SRO. In contrast, the PWS-SRO has no influence on the epigenetic features of the AS-SRO. These results suggest a stepwise, unidirectional program in which structural imprinting at the AS-SRO brings about allele-specific repression of the maternal PWS-SRO, thereby preventing regional activation of genes on this allele.
Keywords: DNA methylation/DNase sensitivity/histone modification/Prader–Willi/Angelman regional control center
Introduction
Genomic imprinting involves the marking of genes during gametogenesis or early embryo development to achieve monoallelic, parent-of-origin-specific expression. The molecular mechanisms that underlie this complex process are, as yet, not fully understood. Allele-specific DNA methylation is an important feature of imprinted genes and is believed to play an important role in the establishment and maintenance of the imprinted state (Li et al., 1993). Several lines of evidence suggest that chromatin structure (Feil and Khosla, 1999) and asynchronous replication timing (Simon et al., 1999) also take part in this process.
Many imprinted genes are organized in conserved clusters. A typical example is the Prader–Willi/Angel man syndrome (PWS/AS) domain on human chromosome 15q11–q13 and its ortholog on mouse chromosome 7C-D1. The 2 Mb PWS/AS domain contains a group of genes that are paternally expressed, and at least two genes, UBE3A and ATP10C, which are expressed exclusively from the maternal allele (Nicholls et al., 1998; Meguro et al., 2001). Genetic aberrations in this domain result in two clinically distinct neurobehavioral disorders, PWS and AS. PWS is a result of molecular defects that bring about silencing of the paternally expressed genes, while AS comes about because of molecular defects that cause a loss of expression of genes on the maternal copy of this domain.
Studies of spontaneous minideletions in the 15q11–q13 domain in man and induced deletions of the orthologous region on chromosome 7 of the mouse have led to the proposal that the imprinting process is coordinated by an imprinting center (IC) located upstream of the SNRPN gene (Buiting et al., 1995; Bielinska et al., 2000). One region of this IC is required for establishing and maintaining the paternal imprint, and is defined by a series of PWS families in which minideletions are observed on the paternal allele. The shortest region of deletion overlap (PWS-SRO) for this region maps to a 4.3 kb sequence that encompasses the SNRPN promoter and exon 1 (Ohta et al., 1999a). In these PWS families, the paternally expressed genes are all methylated and silenced. On the other hand, families with AS carry minideletions on the maternal chromosome that all overlap a 880 bp sequence (AS-SRO) located 35 kb upstream of the SNRPN gene (Buiting et al., 1999). Defects in the AS-SRO affect the maternal imprint exclusively.
Deletion of both the PWS-SRO and AS-SRO on the same chromosome affects the paternal imprint but not the maternal imprint (Ohta et al., 1999b), suggesting that the AS-SRO must operate by repressing the PWS-SRO on the maternal chromosome (Brannan and Bartolomei, 1999). In order to gain some insight into the details of this mechanism, we used cells derived from AS and PWS families carrying minideletions to analyze the epigenetic features at the AS-SRO and PWS-SRO and determine how these sequences affect each other’s structure.
Results
The epigenetic features of AS-SRO
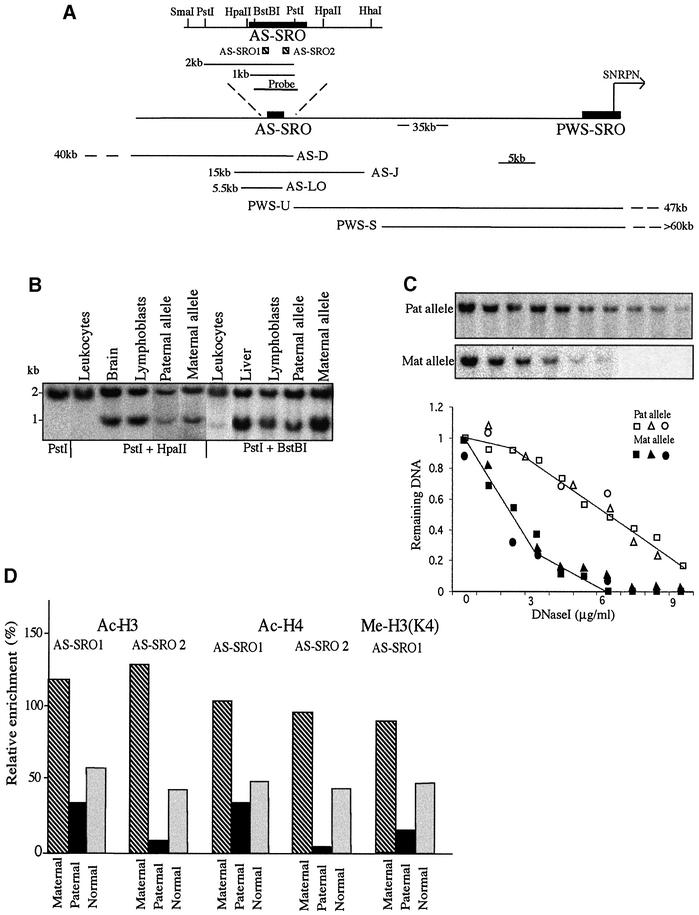
It was shown previously that the PWS-SRO is differentially methylated on the maternal allele in somatic cells (Glenn et al., 1996), while the paternal allele is in a more open chromatin conformation as determined by DNase I sensitivity and histone acetylation (Schweizer et al., 1999; Saitoh and Wada, 2000). Very little is known, however, about the imprinting state of the AS-SRO. As a first step, we assessed the DNA methylation pattern of this region using methyl-sensitive restriction enzymes. Although CpG sites in this region appear to be completely methylated on both alleles in leukocytes (Schumacher et al., 1998; Figure 1B), DNA from normal tissues and lymphoblast cell lines is only partially methylated (60–80%) (Figure 1A and B).
Fig. 1. Epigenetic features of the AS-SRO. (A) Physical map of the AS-SRO and PWS-SRO regions showing restriction sites, the sizes of expected fragments, the positions and extent (kb) of the deletions in the families studied, the AS-SRO probe, the SNRPN transcription start site (horizontal arrow) and the approximate distance between the SNRPN transcription start site and the AS-SRO (Buiting et al., 1999). (B) Southern blot analyses of the AS-SRO methylation pattern in DNA from leukocytes, lymphoblasts, sperm and brain of normal individuals. To demonstrate the methylation status of the paternal allele, DNA from lymphoblasts of an AS patient was used, and the methylation of the maternal allele was tested on lymphoblast DNA of a patient’s mother. DNA samples were digested with PstI alone, PstI + HpaII or PstI + BstBI, electrophoresed, blotted and hybridized with the 1 kb AS-SRO probe. This measures the methylation status of the 5′ HpaII and BstBI sites. The 2 kb band represents the methylated allele while the ∼1 kb band represents the unmethylated allele. The upstream SmaI and downstream HpaII and HhaI sites were also found to be partially methylated biallelically (data not shown). (C) DNase I sensitivity. Nuclei prepared from lymphoblasts of AS-D, AS-J or AS-LO family members including both the AS patient himself (intact paternal allele) and his mother (intact maternal allele) were treated with increasing concentrations of DNase I (0–0.9 µg/ml) and the resulting DNA digested with PstI, Southern blotted and hybridized with the 1 kb AS-SRO probe. A sample blot shows results from the AS-D family. Blots from all of the families (AS-D, squares; AS-J, triangles; AS-LO, circles) were scanned and the data normalized and plotted to show the kinetics of DNase I digestion. (D) Histone H3 and H4 acetylation or H3 Lys4 methylation in the AS-SRO region was determined by ChIP. PCR amplification was carried out on bound and input DNA from lymphoblast mononucleosomes of the AS-D family containing either the maternal (hatched) or paternal (black) allele and normal cells (gray) using primer pairs that amplify two different regions, representing the 5′ part (AS-SRO1) and the 3′ part (AS-SRO2) of the AS-SRO region [see diagram in (A)]. Relative enrichment was calculated with respect to that obtained with GAPDH (defined as 100%).
In order to determine whether this pattern may be imprinted, we carried out blot hybridization on lymphoblast DNA from families carrying single allele deletions of this region. Surprisingly, both maternally and paternally derived alleles appear to be methylated to almost the same extent (Figure 1B). Further analysis showed that other CpG sites covering the surrounding 4 kb region are also partially methylated in a biallelic manner (data not shown). Thus, unlike other imprinting control regions, the AS-SRO does not appear to carry a clearcut differential methylation pattern. Indeed, since the number of CpG residues distributed over the AS-SRO is extremely low (<0.5%), it could be suggested that, in any event, methylation does not play a significant role in this region.
We next investigated whether the AS-SRO may have a differential chromatin structure. To this end, nuclei from lymphoblast cells carrying deletions of this region on either the paternal or maternal copy were isolated and analyzed for DNase I sensitivity. Strikingly, treatment with increasing concentrations of DNase I demonstrated that the maternal AS-SRO allele is vastly more accessible than the paternal allele (Figure 1C). As a further test of structure, we employed antibodies specific to acetylated histones H3 and H4 to carry out chromatin immunoprecipitation (ChIP) analysis on mononucleosomes isolated from these same mutant cells. In keeping with the DNase I results, we found that the maternal allele is differentially enriched for histone acetylation over the full length of the AS-SRO, including both 5′ (AS-SRO1) and 3′ (AS-SRO2) sequences (Figure 1A and D), and slightly enriched for Me-H3(K4) (Figure 1D) which, unlike methylated H3(K9), is characteristic of active chromatin. Taken together with other structural studies (Schweizer et al., 1999), these data demonstrate that the genetically defined AS-SRO domain has a clearcut imprinted conformation in somatic cells. Thus, this structure is similar to that observed for the PWS-SRO (Schweizer et al., 1999; Saitoh and Wada, 2000; Fulmer-Smentek and Francke, 2001), but is set up reciprocally, with the maternal allele being more open.
The effect of AS-SRO on PWS-SRO epigenetic structure
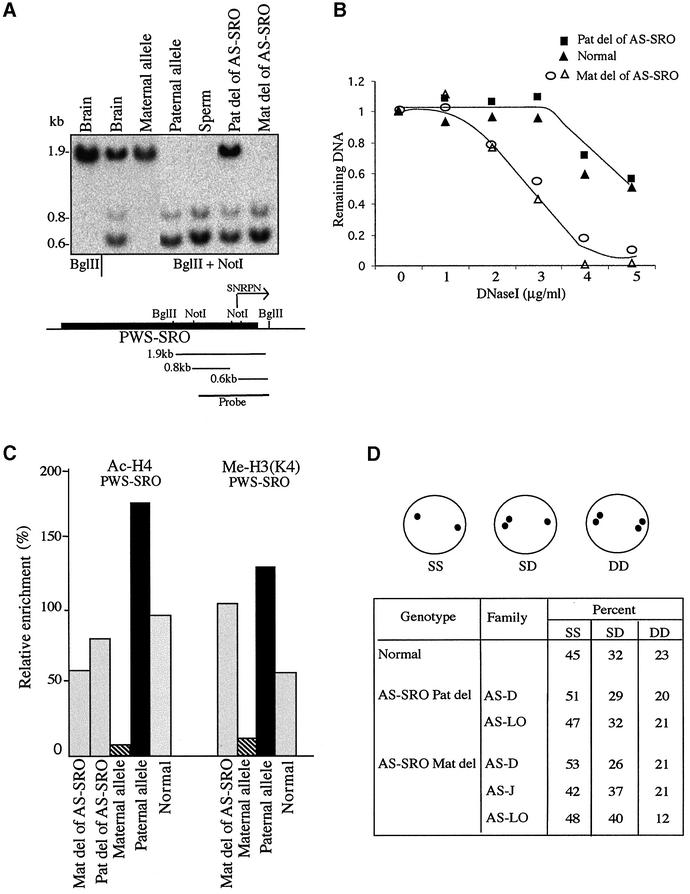
In order to understand how these two control elements may interact with each other to produce regional imprinting, we examined the effect of AS-SRO deletion on the structure of the PWS-SRO. The absence of an AS-SRO from the paternal allele had no obvious influence on the methylation pattern of the PWS-SRO, suggesting that this locus remains unmethylated on the paternal allele (Figure 2A). In contrast, when the AS-SRO is missing from the maternal allele, the neighboring PWS-SRO becomes biallelically unmethylated, clearly implicating this region in the process of de novo methylation which normally occurs on this allele (Figure 2A). In keeping with this, the maternal PWS-SRO has also adopted an open DNase I chromatin conformation (Figure 2B) containing nucleosomes methylated at the K4 residue of histone H3 (Figure 2C). Despite these epigenetic changes, however, deletion of the AS-SRO from either the maternal or paternal allele has no significant effect on the differential pattern of histone acetylation at the PWS-SRO (Figure 2C). These results indicate that while the AS-SRO plays a major role in setting up methylation at the PWS-SRO, it is clearly not the sole determinant of chromatin structure at this site.
Fig. 2. Effect of the AS-SRO on the epigenetic features of the PWS-SRO. (A) The methylation status of the PWS-SRO was determined using DNA from normal tissues, lymphoblast cell lines from a PWS family containing only the maternal or paternal PWS-SRO and lymphoblast cell lines from AS families containing a deletion of the AS-SRO on one of the alleles. DNA was digested with Bglll or Bglll + NotI, Southern blotted and probed with the PWS-SRO probe (see diagram). The 1.9 kb band represents the methylated allele while the 0.8 and 0.6 kb bands represent the unmethylated paternal allele. Note that maternal deletion of the AS-SRO leads to undermethylation of the adjacent PWS-SRO. Similar results were obtained using DNA from three different AS families (AS-D, AS-J and AS-LO) (data not shown). (B) DNase I sensitivity of the PWS-SRO locus was determined on nuclei from lymphoblasts of a normal individual (with two AS-SRO alleles), or lymphoblasts from two different AS families (AS-D and AS-J) containing either maternal or paternal deletions of the AS-SRO, but two copies of the PWS-SRO. Note that the maternal AS-SRO deletion brings about an increase in DNase I sensitivity over the PWS-SRO. (C) The pattern of histone H4 acetylation and H3(K4) methylation at the PWS-SRO locus was determined by ChIP using mononucleosomes from lymphoblasts derived from PWS-S family members containing only the maternal (hatched) or paternal (black) PWS-SRO allele, or from AS-D family members containing a maternal or paternal deletion of the AS-SRO. These AS-SRO deletions had a profound effect on K4 methylation, but did not significantly affect the level of histone H4 acetylation on the PWS-SRO as compared with normal individuals. (D) Replication timing analysis was carried out by FISH using a 11 kb probe that covers the entire PWS-SRO using normal lymphoblasts as well as cells from three different AS families. Single/single (SS), single/double (SD) and double/double (D/D). SD over 20% is considered asynchronous. Note that replication of the first (paternal) allele always occurs at approximately the same time in S phase.
Asynchronous replication timing represents another epigenetic mark that is intimately associated with imprinting (Kitsberg et al., 1993). In the PWS/AS domain, an entire region of 2 Mb normally replicates differentially, with the paternal allele always being the early one in each cell (Kitsberg et al., 1993; Knoll et al., 1994). In order to determine whether the AS-SRO plays a role in setting up this structure, we used fluroescence in situ hybridization (FISH) to analyze replication timing in normal and mutant lymphoblasts. As expected, we observed a large percentage of nuclei with single/double hybridization signals in normal cells, as is indicative of asynchronous replication timing. The same appears to be true even for cells lacking the AS-SRO on either the maternal or paternal allele (Figure 2D). Thus, despite the centrality of the AS-SRO in the control of structure at the PWS locus, this sequence is not involved in the regulation of region-wide allele-specific replication timing.
The effect of PWS-SRO on the epigenetic structure of AS-SRO
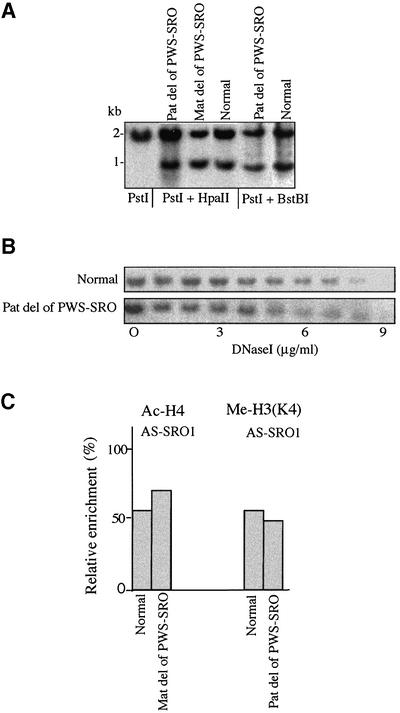
To examine the possibility of a reciprocal relationship between the two control elements, we also tested the effect of PWS-SRO deletion on epigenetic features of the AS-SRO. As seen in Figure 3A, whether this deletion is on the maternal or paternal allele, there is no influence on the methylation pattern of the AS-SRO. Furthermore, absence of the PWS-SRO had no effect on the differential DNase I sensitivity (Figure 3B) or histone modification patterns normally associated with this region (Figure 3C). It thus appears that while the AS-SRO is responsible for setting up allele-specific features of PWS-SRO structure, the reverse is not true.
Fig. 3. Effect of the PWS-SRO on epigenetic features of the AS-SRO. (A) The methylation status of the AS-SRO was determined using DNA from normal tissues and lymphoblasts from the PWS-S family containing either a maternal or paternal deletion of the PWS-SRO. DNA was digested with PstI alone, PstI + HpaII or PstI + BstBI, Southern blotted and probed with the AS-SRO probe (see Figure 1A). Similar results were also obtained for a second family (PWS-U; data not shown). (B) DNase I sensitivity at the AS-SRO region was assayed using nuclei from normal or PWS-S (paternal deletion) lymphoblasts. (C) Histone H4 acetylation and H3(K4) methylation at the AS-SRO region was assayed by ChIP using mononucleosomes from normal lymphoblasts or cells carrying a maternal deletion of the PWS-SRO (PWS-S). No effects of the PW-SRO deletion were observed. See controls in Figure 1D.
Discussion
The AS-SRO has been mapped by genetic studies as an imprinting control region needed for directing the normal pattern of expression on the maternal allele, and its deletion brings about AS. Despite this important function, our experiments and other previous studies in lymphocytes (Schumacher et al., 1998) failed to detect differential cytosine methylation, a normally standard feature of imprinting regulation. In this study, we have demonstrated for the first time that this region does carry a genuine epigenetic imprint in the form of chromatin structure, with the maternal allele in a DNase I-sensitive conformation, and the paternal allele being closed and inaccessible. This differential structure is also reflected in the histone modification pattern, which is characterized by histone H3 and H4 acetylation and H3(K4) methylation exclusively on the maternal allele.
Although it is unusual for an imprinting control region to lack differential methylation cytosine (Razin and Cedar, 1994), there is already a considerable amount of evidence suggesting that there must be additional epigenetic mechanisms that contribute to the regulation of imprinting. Some methyl groups at key imprinting sites, for example, are transiently erased from the genome during pre-implantation development (Shemer et al., 1996), and imprints can be formed on DNA fragments injected into gamete-specific pronuclei even though the actual cytosine methylation is delayed for several cell divisions (Birger et al., 1999). Furthermore, differential methylation is not always correlated with monoallelic gene expression (Szabo and Mann, 1996; Davis et al., 1999), and some genes still retain their imprinted status in Dnmt1-deficient embryos (Caspary et al., 1998; Tanaka et al., 1999). When taken together, these studies suggest that it may be the chromatin structure itself which constitutes the functional mark distinguishing the two alleles at imprinted domains. Developmentally, this may be generated through a number of different molecular pathways, including differential DNA methylation or asynchronous replication timing (Simon et al., 1999).
Even if one allows for the involvement of other epigenetic effectors in the establishment of imprinting, it is usually assumed that DNA methylation is absolutely required for the long-term maintenance of imprinting in dividing cells. Thus, it is interesting that the AS-SRO manages to preserve its differential structure for many cell generations both in vivo and in cell culture. It has been suggested that histone modification patterns can be genocopyed in a semi-conservative manner during replication (Ekwall et al., 1997; Nielsen et al., 2001), and this could serve as an elegant mechanism for the maintenance of allele-specific chromatin structure.
Genetic studies on AS patients with deletions of the AS-SRO have allowed us to define better the function of this region in the imprinting process. These experiments clearly show that the AS-SRO is necessary for maintaining several epigenetic features characteristic of the maternal PWS-SRO. These include DNA methylation, the closed DNase I-insensitive chromatin conformation and undermethylation of histone H3(K4). It is not completely clear how all of these epigenetic features are actually generated in a coordinated manner. One possibility is that the primary role of the AS-SRO is to cause de novo methylation of the PWS-SRO during early development, which could then bring about closure of chromatin and prevent methylation of H3(K4) at the PWS-SRO. Differential methylation of H3(K9) on the maternal allele may also contribute to this structural imprint (Xin et al., 2001). Our results suggest that the AS-SRO is not the sole determinant of PWS-SRO chromatin structure, since the PWS-SRO still maintains its deacetylated histone state even when the AS-SRO is deleted. This does not appear to be a critical functional feature, however, since this underacetylation does not prevent activation of the PWS-SRO.
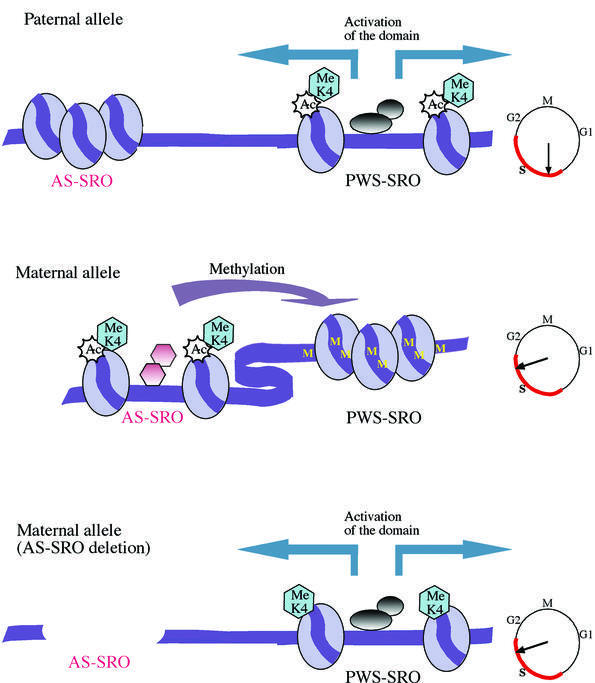
Based on these observations, we propose the following model for how the AS-SRO and PWS-SRO operate in a stepwise and unidirectional manner to generate an imprinted structure at the PWS/AS locus. Our data suggest that the AS-SRO acquires its differential epigenetic makeup prior to the PWS-SRO, probably during gametogenesis. The PWS-SRO, on the other hand, emerges from both gametes unmethylated in its CpG island sequences (El-Maarri et al., 2001). We suggest that it is the conformationally active AS-SRO on the maternal allele which acts in cis as a repressor to bring about de novo methylation of the adjacent PWS-SRO and packaging into a closed chromatin structure (see Figure 4). In contrast, the PWS-SRO on the paternal allele remains unmethylated, presumably because its corresponding AS-SRO is in the off conformation [this had been postulated before by Brannan and Bartolomei (1999), and is substantiated here] (Figure 1). Finally, the open PWS-SRO on the paternal allele operates in cis to bring about structural and transcriptional activation over the entire PWS/AS domain. This model explains why minideletions of the PWS-SRO cause repression of genes on the paternal allele in PWS patients (Buiting et al., 1995), while the absence of the primary imprinting mark at the AS-SRO in AS patients releases the repression normally imposed on the maternal allele (Reis et al., 1994).
Fig. 4. Model of the mechanism of imprinting at the PWS/AS regional control center. The maternal AS-SRO initially acquires a DNase I-sensitive conformation and becomes differentially packaged with acetylated histones. The function of this active AS-SRO is to methylate the adjacent PWS-SRO and put it in an inactive chromatin structure (DNase I insensitive). This epigenetic state fails to form on the PWS-SRO if the AS-SRO is deleted or positioned too far from the PWS-SRO (Buiting et al., 2001). The function of the paternal PWS-SRO is to activate genes in the PWS/AS domain. This does not occur on the maternal allele, since its PWS-SRO has an inactive conformation. Judging from the mouse (Simon et al., 1999), the setting up of asynchronous replication timing at the PWS/AS locus with the paternal allele early and the maternal allele late (see clock) occurs during gametogenesis, and thus probably takes place developmentally upstream of epigenetic fixation at the AS-SRO. Late replication of the maternal allele may in turn be responsible for bringing about differential deacetylation (Rountree et al., 2000) of histones on the PWS-SRO, independently of the AS-SRO (see Figure 2C).
Materials and methods
Biological material
Human lymphoblast cell lines were grown in RPMI supplemented with 10% fetal calf serum, 2 mM glutamine and 1000 U/ml penicillin plus 100 µg/ml streptomycin. Data on the PWS-S patient, the PWS-U family and AS-D, AS-J and AS-LO family members (see Figure 1A) were reported previously (Reis et al., 1994; Buiting et al., 1995, 1999). Lymphoblast cell lines from these families were kindly provided by K.Buiting and R.Nicholls. DNA samples of sperm and other tissues were the same as those described previously (Shemer et al., 1991).
DNase I hypersensitivity analysis
Lymphoblasts (∼2 × 108) were resuspended in nuclei buffer (20 mM Tris pH 7, 3 mM CaCl2, 2 mM MgCl2, 0.3% NP-40) and incubated on ice for 10 min. Nuclei were then resuspended in RSB (10 mM Tris pH 7, 10 mM NaCl, 3 mM MgCl2) to a concentration of 108 nuclei/ml, DNase I was added to final concentrations of 0.1–1.0 µg/ml in 100 µl and incubated at 37°C for 15 min. Extracted genomic DNA (15 µg) was then digested with the appropriate restriction enzymes and Southern blotted.
FISH analysis to determine replication timing
Cells used for FISH analysis were incubated for 1 h in culture with 3 × 10–5 M bromodeoxyuridine (BrdU) to label cells in S phase, treated with hypotonic KCl solution, fixed in methanol:acetic acid (3:1), dropped on slides and hybridized with a human SNRPN probe (–10 to +1 kb) (Buiting et al., 1995) as described previously (Selig et al., 1992). Replication timing profiles are presented as a percentage of chromosomes in the single/single (SS), single/double (SD) or double/double (DD) state as determined by counting a minimum of 100 BrdU-positive nuclei.
ChIP assay for histone modification
Nuclear pellets were treated with 0.1–0.3 U/ml micrococcal nuclease for 10 min at 30°C, followed by nucleosome fractionation on sucrose gradients (Hebbes et al., 1994). Mononucleosomes were divided into two samples, one serving as an immunoprecipitation control (input) and the other sample being exposed to anti-acetylated (K9 and K14) H3 or H4 (K5, K8, K12 and K16) antibody (Upstate Biotechnology; cat. nos 06-599 and 06-598) or anti-dimethyl H3(K4) (Upstate Biotechnology; cat. no. 07-030) at a concentration of 10 µg/60 µg chromatin DNA. DNA was extracted from the input control and the immunoprecipitated samples, and subjected to PCR using appropriate primers as listed below. Two primer pairs were used for the AS-SRO: AS-SRO1 for the 5′ end of the AS-SRO (position 241–300) and AS-SRO2 for the 3′ end (position 820–900). A 1 µl aliquot of [α-32P]dCTP (3 Ci/mmol) was added to the PCR mixture. PCR products were run on 7% acrylamide gels, and autoradiographs were quantitated using the Alpha Imager 2200 Documentation and Analysis System (Alpha Innotech). Primers used were as follows: AS-SRO1, 5′-AGAGCTGAAGCCCAGTTTCA and 5′-CTTGAGGGGGTTTGAGTG TA; AS-SRO 2, 5′-GCTTTGTGAAGGCTTCAGATG and 5′-TCAAGC AAACTCTGCTCACC; PWS-SRO, 5′-CGGTCAGTGACGCGATGG AGCGG and 5′-GCTCCCCAGGCTGTCTCTTGAGAG; GAPDH, 5′-TTCATCCAAGCGTGTAAGGG and 5′-TGGTTCCCAGGACTGGA CTGT; and β-globin, 5′-GACACAACTGTGTTCACTAGC and 5′-AC TTCTCCTCAGGAGTCAGA.
The PCRs were performed using three quantities (1, 3 and 9 µl) of DNA, and the amount of product was averaged. Relative enrichment for each region was calculated as the ratio of bound/input divided by the enrichment obtained with the β-globin primer control. These values were then normalized to GAPDH (set at 100% enrichment).
Acknowledgments
Acknowledgements
We are grateful to T.Hashimshony and J.Zhang for help in the ChIP assay, to M.Goldmit for help in the FISH assay, and to K.Buiting and B.Horsthemke for helpful discussions. This work was supported by the Israel Science Foundation Centers of Excellence Programme, NIH grants to A.R. and H.C., and a grant from the Israel Cancer Research Foundation to H.C. J.P. was partially supported by a Foulkes Foundation Fellowship.
References
- Bielinska B., Blaydes,S.M., Buiting,K., Yang,T., Krajewska-Walasek,M., Horsthemke,B. and Brannan,C.I. (2000) De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet., 25, 74–78. [DOI] [PubMed] [Google Scholar]
- Birger Y., Shemer,R., Perk,J. and Razin,A. (1999) The imprinting box of the mouse Igf2r gene. Nature, 397, 84–88. [DOI] [PubMed] [Google Scholar]
- Brannan C.I. and Bartolomei,M.S. (1999) Mechanisms of genomic imprinting. Curr. Opin. Genet. Dev., 9, 164–170. [DOI] [PubMed] [Google Scholar]
- Buiting K., Saitoh,S., Gross,S., Dittrich,B., Schwartz,S., Nicholls,D.R. and Horsthemke,B. (1995) Inherited microdeletions in the Angelman and Prader–Willi syndromes define an imprinting centre on human chromosome 15. Nat. Genet., 9, 395–400. [DOI] [PubMed] [Google Scholar]
- Buiting K., Lich,C., Cottrell,S., Barnicoat,A. and Horsthemke,B. (1999) A 5-kb imprinting center deletion in a family with Angelman syndrome reduces the shortest region of deletion overlap to 880 bp. Hum. Genet., 105, 665–666. [DOI] [PubMed] [Google Scholar]
- Buiting K., Barnicoat,A., Lich,C., Pembrey,M., Malcolm,S. and Horsthemke,B. (2001) Disruption of the bipartite imprinting center in a family with Angelman syndrome. Am. J. Hum. Genet., 68, 1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Cleary,M.A., Baker,C.C., Guan,X.-J. and Tilghman,S.M. (1998) Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol. Cell. Biol., 18, 3466–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.L., Trasler,J.M., Moss,S.B., Yang,G.J. and Bartolomei,M.S. (1999) Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics, 58, 18–28. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Olsson,T., Turner,B.M., Cranston,G. and Allshire,R.C. (1997) Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell, 91, 1021–1032. [DOI] [PubMed] [Google Scholar]
- El-Maarri O. et al. (2001) Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat. Genet., 27, 341–344. [DOI] [PubMed] [Google Scholar]
- Feil R. and Khosla,S. (1999) Genomic imprinting in mammals: an interplay between chromatin and DNA methylation? Trends Genet., 15, 431–435. [DOI] [PubMed] [Google Scholar]
- Fulmer-Smentek S.B. and Francke,U. (2001) Association of acetylated histones with paternally expressed genes in the Prader–Willi deletion region. Hum. Mol. Genet., 10, 645–652. [DOI] [PubMed] [Google Scholar]
- Glenn C.C., Saitoh,S., Jong,M.T., Filbrandt,M.M., Surti,U., Driscoll,D.J. and Nicholls,R.D. (1996) Gene structure, DNA methylation and imprinted expression of the human SNRPN gene. Am. J. Hum. Genet., 58, 335–346. [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg D., Selig,S., Brandeis,M., Simon,I., Keshet,I., Driscoll,D.J., Nicholls,R.D. and Cedar,H. (1993) Allele-specific replication timing of imprinted gene regions. Nature, 364, 459–463. [DOI] [PubMed] [Google Scholar]
- Knoll J.H.M., Cheng,S.-D. and Lalande,M. (1994) Allele specificity of DNA replication timing in the Angelman/Prader–Willi syndrome imprinted chromosomal region. Nat. Genet., 6, 41–46. [DOI] [PubMed] [Google Scholar]
- Li E., Beard,C. and Jaenisch,R. (1993) Role for DNA methylation in genomic imprinting. Nature, 366, 362–365. [DOI] [PubMed] [Google Scholar]
- Meguro M., Kashiwagi,A., Mitsuya,K., Nakao,M., Kondo,I., Saitoh,S. and Mitsuo,O. (2001) A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipd translocase associated with Angelman syndrome. Nat. Genet., 28, 19–20. [DOI] [PubMed] [Google Scholar]
- Nicholls R.D., Saitoh,S. and Horsthemke,B. (1998) Imprinting in Prader–Willi and Angelman syndromes. Trends Genet., 14, 194–199. [DOI] [PubMed] [Google Scholar]
- Nielsen S.J. et al. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature, 412, 561–565. [DOI] [PubMed] [Google Scholar]
- Ohta T., Buiting,K., Kokkonen,H., McCandless,S., Heeger,S., Driscoll,D.J., Cassidy,S.B., Horsthemke,B. and Nicholls,R.D. (1999a) Molecular mechanism of Angelman syndrome in two large families involves an imprinting mutation. Am. J. Hum. Genet., 64, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. et al. (1999b) Imprinting mutation mechanisms in Prader–Willi syndrome. Am. J. Hum. Genet., 64, 397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. and Cedar,H. (1994) DNA methylation and genomic imprinting. Cell, 77, 473–476. [DOI] [PubMed] [Google Scholar]
- Reis A., Dittrich,B., Greger,V., Buiting,K., Lalande,M., Gillessen-Kaesbach,G., Anvret,M. and Horsthemke,B. (1994) Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader–Willi syndromes. Am. J. Hum. Genet., 54, 741–747. [PMC free article] [PubMed] [Google Scholar]
- Rountree M.R., Bachman,K.E. and Baylin,S.B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet., 25, 269–277. [DOI] [PubMed] [Google Scholar]
- Saitoh S. and Wada,T. (2000) Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader–Willi syndrome. Am. J. Hum. Genet., 66, 1958–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A., Buiting,K., Zeschnigk,M., Doerfler,W. and Horsthemke,B. (1998) Methylation analysis of the PWS/AS region does not support an enhancer-competition model of genomic imprinting on human chromosome 15. Nat. Genet., 19, 324–325. [DOI] [PubMed] [Google Scholar]
- Schweizer J., Zynger,D. and Francke,U. (1999) In vivo nuclease hypersensitivity studies reveal multiple sites of parental origin-dependent differential chromatin conformation in the 150 kb SNRPN transcription unit. Hum. Mol. Genet., 8, 555–566. [DOI] [PubMed] [Google Scholar]
- Selig S., Okumura,K., Ward,D.C. and Cedar,H. (1992) Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J., 11, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer R., Eisenberg,S., Breslow,J.L. and Razin,A. (1991) Methylation patterns of the human apoAI–CIII–AIV gene cluster in adult and embryonic tissue suggest dynamic changes in methylation during development. J. Biol. Chem., 266, 23676–23681. [PubMed] [Google Scholar]
- Shemer R., Birger,Y., Dean,W.L., Reik,W., Riggs,A.D. and Razin,A. (1996) Dynamic methylation adjustment and counting as part of imprinting mechanisms. Proc. Natl Acad. Sci. USA, 93, 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Tenzen,T., Reubinoff,B.E., Hillman,D., McCarrey,J.R. and Cedar,H. (1999) Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature, 401, 929–932. [DOI] [PubMed] [Google Scholar]
- Szabo P.E. and Mann,J.R. (1996) Maternal and paternal genomes function independently in mouse ova in establishing expression of the imprinted genes Snrpn and Igf2r: no evidence for allelic trans-sensing and counting mechanisms. EMBO J., 15, 6018–6025. [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Puchyr,M., Gertsenstein,M., Harpal,K., Jaenisch,R., Rossant,J. and Nagy,A. (1999) Parental origin-specific expression of Mash2 is established at the time of implantation with its imprinting mechanism highly resistant to genome-wide demethylation. Mech. Dev., 87, 129–142. [DOI] [PubMed] [Google Scholar]
- Xin Z., Allis,C.D. and Wagstaff,J. (2001) Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader–Willi syndrome imprinting center. Am. J. Hum. Genet., 69, 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]