Abstract
To explore mechanisms for specificity of function within the family of E2F transcription factors, we have identified proteins that interact with individual E2F proteins. A two-hybrid screen identified RYBP (Ring1- and YY1-binding protein) as a protein that interacts specifically with the E2F2 and E2F3 family members, dependent on the marked box domain in these proteins. The Cdc6 promoter contains adjacent E2F- and YY1-binding sites, and both are required for promoter activity. In addition, YY1 and RYBP, in combination with either E2F2 or E2F3, can stimulate Cdc6 promoter activity synergistically, dependent on the marked box domain of E2F3. Using chromatin immunoprecipitation assays, we show that both E2F2 and E2F3, as well as YY1 and RYBP, associate with the Cdc6 promoter at G1/S of the cell cycle. In contrast, we detect no interaction of E2F1 with the Cdc6 promoter. We suggest that the ability of RYBP to mediate an interaction between E2F2 or E2F3 and YY1 is an important component of Cdc6 activation and provides a basis for specificity of E2F function.
Keywords: Cdc6 activation/E2F transcription factor/RYBP/YY1
Introduction
The E2F transcription factors play a critical role in regulating transcription in response to the stimulation of cell proliferation. The stimulation of cell growth leads to the activation of G1 cyclin-dependent kinase (CDK) activity that then phosphorylates retinoblastoma protein (Rb), resulting in the inactivation of its ability to control E2F accumulation (Dyson, 1998; Nevins, 1998). The mammalian E2F family consists of six distinct gene products (E2F1–E2F6) that function as heterodimers with members of the DP family of proteins. In contrast, the Drosophila genome encodes only two E2F genes, de2f1 and de2f2, suggesting that the increased diversity of the E2Fs in mammalian cells provides for a more complex transcriptional gene regulation in higher eukaryotes. At least in part, the diversity in the E2F family reflects distinct roles in transcriptional regulation. The E2F1, E2F2 and E2F3 proteins act as positive regulators of transcription, whereas the E2F4 and E2F5 proteins appear to function primarily as repressors of transcription, in concert with Rb family proteins (Dyson, 1998; Nevins, 1998).
Various experiments suggest that the individual E2F proteins might perform distinct functions in the control of cell growth and cell fate. For instance, E2F1 has the ability uniquely to promote apoptosis (DeGregori et al., 1997; Kowalik et al., 1998; Moroni et al., 2001). Moreover, recent work has shown that Myc requires distinct E2F activities to induce S phase and apoptosis, since the ability of Myc to induce S phase is impaired in the absence of either E2F2 or E2F3 but not E2F1 or E2F4 (Leone et al., 2001). In contrast, the ability of Myc to induce apoptosis is reduced markedly in cells deleted for E2F1 but not E2F2 or E2F3. In addition, previous work has shown that E2F1 is induced specifically in response to DNA damage (Lin et al., 2001), further highlighting a distinct functional role for E2F1 in signaling cell fate.
In contrast to the role of E2F1, the E2F3 protein appears to be critically important for cell proliferation. Previous work has shown that inhibition of E2F3 activity by antibody microinjection impaired S phase entry, whereas inhibition of E2F1 in a similar manner did not (Leone et al., 1998). Moreover, analysis of E2F3-null mouse embryo fibroblasts demonstrated a reduction in proliferative capacity and reduced expression of certain E2F-responsive genes (Humbert et al., 2000). In contrast, E2F1-null cells are not impaired for proliferation and do not exhibit a reduction in most of these RNAs.
These studies suggest that individual members of the E2F subclasses may have different biological properties and indicate that functional specificity occurs through the control of different sets of target genes. Although it is possible that the individual E2F family members might possess subtle but distinct DNA recognition properties, there is little evidence to support this and structural studies of an E2F–DNA complex would suggest that this is an unlikely mechanism for specificity (Zheng et al., 1999). An alternative mechanism for promoter specificity could involve the coordinated action of multiple transcription factors whereby specific combinations of factors are required for proper promoter function. This concept of combinatorial control has been put forward as a model for achieving the massive complexity of transcription control for the large number of protein-coding genes using a limited number of transcription factors (Yamamoto et al., 1998). Various studies have now provided evidence for such combinatorial specificity, involving both upstream binding transcription factors and components of the basal transcription machinery (Pilpel et al., 2001; Smale, 2001).
Using a yeast two-hybrid screen to select for proteins that specifically interact with E2F proteins, we identified RYBP (Ring1- and YY1-binding protein) as a protein that interacts specifically with E2F2 and E2F3, but not with E2F1 and E2F4, and dependent on the E2F marked box domain. Given the interaction of RYBP and YY1, together with the occurrence of YY1-binding sites in several E2F-regulated promoters, including Cdc6, proliferating cell nuclear antigen (PCNA), RR1 and cyclin A, the discovery of a factor that binds both YY1 and E2F suggests a possible mechanism for the formation of a transcription factor complex. We now provide evidence that the ability of RYBP to facilitate an interaction between E2F2/E2F3 and YY1 is a mechanism of combinatorial gene regulation of the Cdc6 promoter.
Results
Identification of E2F-interacting proteins
Fusion proteins containing the Gal4 DNA-binding domain (DBD) and amino acids 1–373 of E2F2, eliminating the transcription activation domain, were used to screen a human placental cDNA library. A total of 93 clones were identified by histidine nutritional selection and β-galactosidase (β-gal) activity. Inserts from each of the isolated clones were subjected to DNA sequence analysis to identify the interacting proteins, and a summary of these results is provided in Table I. To determine the specificity of the protein interactions, all clones were screened for interaction against E2F1, E2F2, E2F3 and E2F4, as well as mutants that eliminate the C-terminus of the E2F2 protein, which includes the marked box domain (Figure 1A). It is evident from these assays that a subset of the clones identified in the screen encode proteins that bind with some degree of specificity to E2F2. Of particular interest was the RYBP protein, which was seen to interact with E2F2 and E2F3, weakly with E2F4, and not at all with E2F1. The clone encoding RYBP, previously identified as a partner for the Ring1 and YY1 transcription factors (Kalenik et al., 1997), was isolated four independent times from the E2F2 screen. RYBP was of additional interest since several E2F-regulated promoters contain YY1-binding sites, and previous studies have provided evidence for a functional interaction between YY1 and E2Fs (van Ginkel et al., 1997).
Table I. Yeast two-hybrid screen for E2F-specific binding partners.
| Clone | E2F1 | E2F2 | E2F3 | E2F4 | E2F2ΔC | E2F3ΔC |
|---|---|---|---|---|---|---|
| BAC clone 174p21 | ++ | ++ | ++ | – | – | +/– |
| Ring1- and YY1-binding protein (RYBP) | – | ++ | ++ | +/– | – | – |
| PR/SET domain-containing protein 07 (SET 07) | – | ++ | – | – | – | – |
| Clone KIAA0161 | + | + | + | + | + | + |
| DRAL, LIM domain-containing protein | ++ | ++ | ++ | ++ | ++ | ++ |
| BAC clone 12p13 RPCI11-154121 | ++ | ++ | ++ | ++ | ++ | ++ |
| Clone DJ0614C10 | ++ | ++ | ++ | ++ | ++ | ++ |
| FHL2, LIM domain protein | ++ | ++ | ++ | ++ | ++ | ++ |
| Unknown (no BLAST match) | ++ | ++ | +/– | +/– | – | – |
| Unknown (no BLAST match) | ++ | + | + | + | + | + |
Interaction assays were performed by mating to transformants of the Matα strain bearing the various E2F constructs. Diploids were assayed by selecting for growth in the absence of histidine and for expression of the lacZ reporter by liquid β-gal activity as described in Materials and methods.
++ represents strong interaction measured by β-gal activity greater than the positive control; + represents β-gal activity equal to the positive control; +/– represents β-gal activity ≤50% of the positive control; – represents β-gal activity no greater than the negative control.
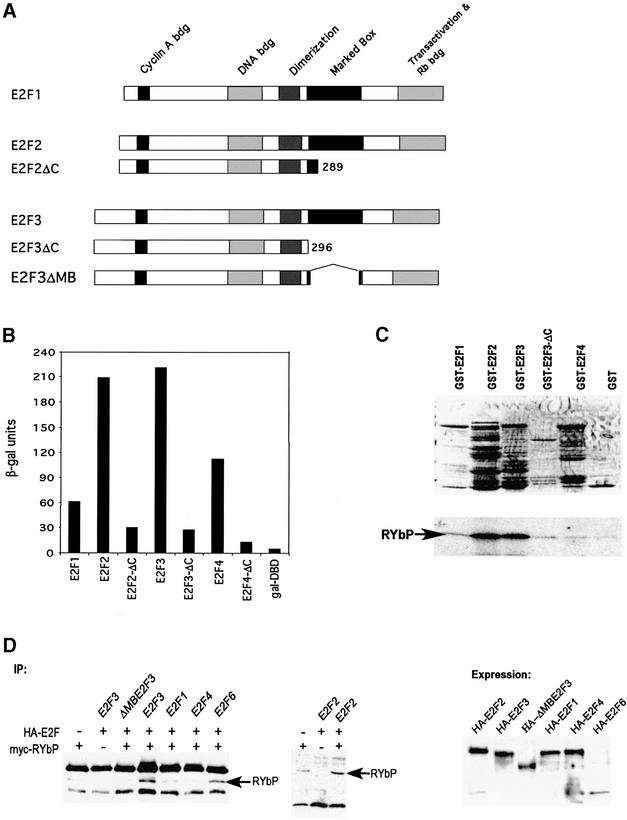
Fig. 1. Specificity in the interaction of RYBP with E2F proteins. (A) Schematic depiction of the structure of the E2F family of transcription factors. Functional domains previously defined are indicated. (B) Analysis of human full-length RYBP protein–protein interactions with various E2F proteins (E2F2ΔC and E2F3ΔC are C-terminal truncations including the marked box domain). Matα yeast (PJ69-a) harboring RYBP fused to the Gal4 activation domain was mated to transformants of PJ69-α bearing the various E2F constructs fused to the Gal4 DBD. Diploids were assayed for expression of β-gal as measured in a quantitative liquid culture assay. (C) RYBP was synthesized (35S-labeled) and incubated with GST–hE2F1, GST–hE2F2, GST–hE2F3, GST–hE2F3ΔC and GST–hE2F4 fusion proteins and GST alone coupled to glutathione–Sepharose beads. Proteins were resolved by 10% SDS–PAGE and stained with Coomassie Blue followed by autoradiography. (D) Asynchronously growing NIH-3T3 mouse fibroblast cells were transfected with either Myc-tagged RYBP or HA-E2F3 alone, and co-transfected with HA-E2F3, HA-E2F3ΔMB, HA-E2F1, HA-E2F2, HA-E2F4 and HA-E2F6. Following immunoprecipitation (IP) from transfected NIH-3T3 whole-cell extracts with α-HA antibodies as indicated, precipitated proteins were resolved by SDS–PAGE, transferred to a nitrocellulose membrane and probed with α-myc antibodies. Expression of the various co-transfected E2Fs was verified by western analysis.
Identification of RYBP as an E2F2- and E2F3-specific interacting protein, dependent on the marked box domain
As seen in Figure 1B, RYBP interacts strongly with E2F2 and E2F3, as measured by activation of the lacZ gene and production of β-gal activity. In contrast, both E2F1 and E2F4 were less efficient in interaction with RYBP. These assays also demonstrated that the interaction of RYBP with the E2F proteins was dependent on the C-terminal region, which includes the marked box domain.
As further evidence for the interaction of RYBP with E2F proteins, we carried out in vitro interaction assays using GST fusions of the various E2F proteins and in vitro translated RYBP. Consistent with the yeast assays, 35S-labeled full-length RYBP associated with both E2F2 and E2F3 proteins but not with E2F1 nor E2F4 (Figure 1C). Moreover, the interaction was dependent on the marked box domain, as indicated by the failure of the E2F3 C-terminal mutant to interact with RYBP.
To determine whether the RYBP protein could interact with E2F2 or E2F3 when co-expressed in a mammalian cell, we carried out co-immunoprecipitation assays with transfected cells. As shown in Figure 1D, both hemagglutinin (HA)-E2F2 and HA-E2F3 co-immunoprecipitated Myc-RYBP protein. In contrast, there was little or no evidence for an interaction of RYBP with any of the other E2F proteins, with the exception of E2F6, consistent with a recent report describing the binding of RYPB to this E2F family member (Trimarchi et al., 2001). In addition, these assays also demonstrated the role of the marked box domain, since an E2F3 mutant in which a portion of the domain was deleted did not interact with RYPB.
A YY1-binding element is essential for Cdc6 promoter activity
Several E2F-regulated promoters, including PCNA, RR1, cyclin A and Cdc6, contain potential YY1 elements. We have chosen to focus on the Cdc6 promoter in light of past work that has detailed the characteristics of Cdc6 promoter function and the role of E2F activities in the control of Cdc6 activity (Hateboer et al., 1998; Ohtani et al., 1998; Yan et al., 1998). As shown in Figure 2A, the Cdc6 promoter contains a pair of E2F elements that flank a YY1 consensus sequence. The identity of the E2F sequences as authentic E2F-binding sites has been verified in past work (Yan et al., 1998). To confirm that the putative YY1 site in the Cdc6 promoter was capable of binding YY1, we measured the binding of GST–YY1 protein to a Cdc6 promoter fragment by gel mobility shift assays. Binding was readily detected and could be competed with the wild-type promoter sequence but not a mutant sequence (Figure 2B).
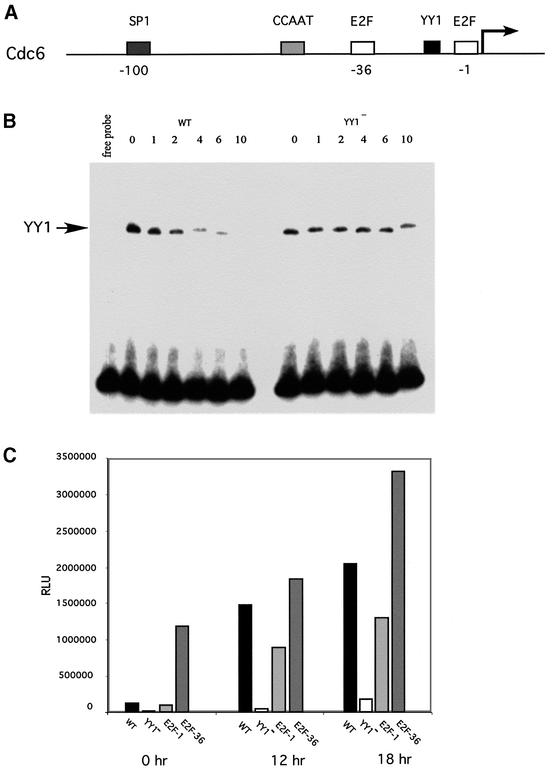
Fig. 2. Role of E2F and YY1 in the growth-dependent activation of the Cdc6 promoter. (A) Schematic of the Cdc6 promoter. Representation of transcription factor-binding sites in the –130 to +21 bp human Cdc6 promoter. The transcription start site is depicted with an arrow. A putative YY1 element (YY1) is located near the transcription start site between two E2F elements (E2F) in addition to an upstream CCAAT box and a putative SP1 element (SP1). (B) Specific binding of YY1 to the Cdc6 promoter. Specific binding of the YY1 protein to the Cdc6 promoter fragment (–130 to +21) was measured by competition with wild-type oligo (WT) and mutant oligo (YY1–) in a gel mobility shift assay. WT oligo (60 bp) contains the wild-type sequence of the Cdc6 promoter including the putative YY1 element mutant. Mutant oligo (YY1–) contains the WT sequence with a two base mutation in the YY1 core element. A 50 ng aliquot of purified YY1–GST fusion protein, cleaved from the GST by protease, was used in the binding reaction with increasing concentrations of competitor oligo DNA: 0 = 0 ng, 1 = 50 ng, 2 = 100 ng, 4 = 200 ng, 6 = 300 ng, 10 = 500 ng. (C) Role of E2F- and YY1-binding sites in Cdc6 promoter function. Transient transfection assays were performed in subconfluent REF52 cells. Cells were harvested under conditions of growth arrest induced by serum deprivation (t = 0 h), and at subsequent time points after addition of serum (12 and 18 h). Reporter gene activity (mean of triplicate determinations) was calculated and normalized by β-gal activity. WT = wild-type Cdc6 promoter (–1700 to +7 bp); YY1– = mutated YY1 site; E2F-1 = E2F site mutated at position –1 upstream of the start site; E2F-36 = E2F site mutated at position –36 upstream of the start site.
To investigate the role of these sequences in the control of Cdc6 promoter activity, we assayed a series of mutant constructs, in comparison with the wild-type promoter (–1.7 to +7 kb), for activity following stimulation of cell growth. REF52 cells were transfected with reporter constructs and the cells were then brought to quiescence by serum deprivation for 48 h. Cells were stimulated to grow by serum addition and then harvested at either 0, 12 or 18 h after stimulation. As shown in Figure 2C, the stimulation of cell growth led to a substantial activation of the wild-type Cdc6 promoter (an ∼20-fold increase), consistent with previous work (Hateboer et al., 1998; Ohtani et al., 1998; Yan et al., 1998). Mutation of the –36 E2F element resulted in a derepression of the promoter in quiescent cells, consistent with many studies that have identified a role for E2F in negative control of transcription, particularly in quiescent cells. In contrast, mutation of the –1 E2F element reduced the activation of the promoter following growth stimulation, suggesting distinct roles for the two E2F elements and a role for the –1 element in positive control of the promoter. Mutation of the YY1 element abolished Cdc6 promoter activity, thus demonstrating a critical role for this sequence in the activity of this promoter. Based on these results, we conclude that E2F activity, probably an E2F–Rb family complex acting through the –36 E2F element, functions to repress transcription in a quiescent cell; subsequently, both E2F and YY1 contribute to positive activation of the promoter following growth stimulation, probably utilizing the –1 E2F site and the YY1 site. Indeed, expression of either YY1 or E2F2 can activate the Cdc6 promoter (Figure 3D; Supplementary data available at The EMBO Journal Online).
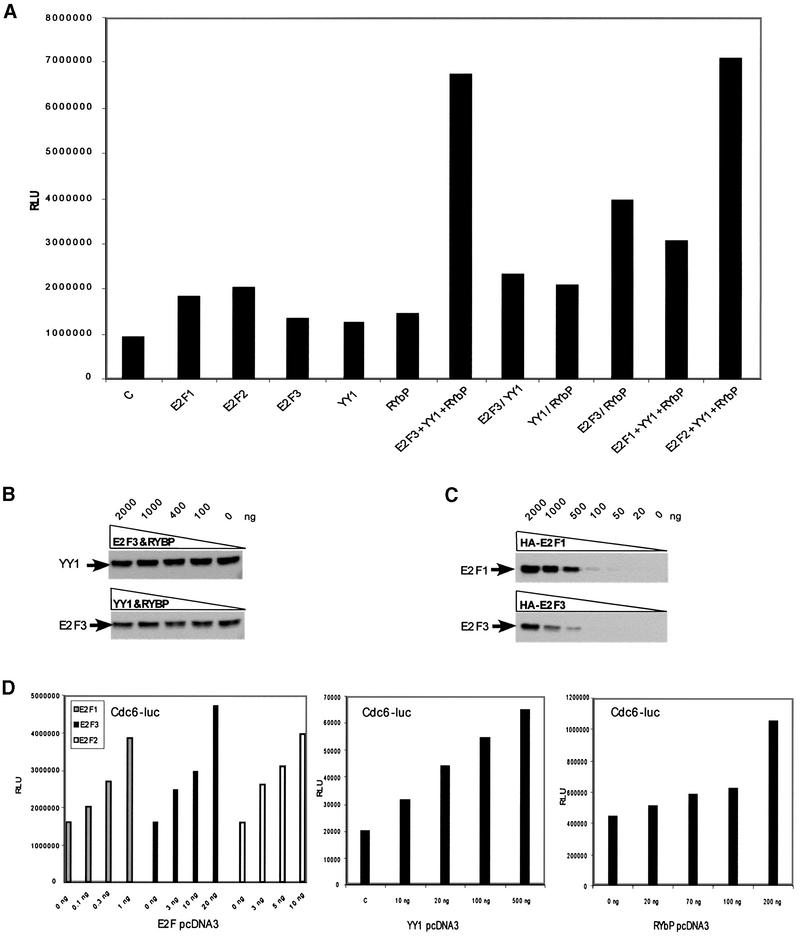
Fig. 3. Activation of the Cdc6 promoter by YY1 and E2F2 or E2F3. (A) Synergistic activation of the Cdc6 promoter by YY1, RYBP, and E2F2 or E2F3. Cells were transfected with Cdc6 reporter plasmid and low amounts of either YY1 alone, E2F1, E2F2 or E2F3 alone, RYBP alone or various combinations of the plasmid. The amounts of plasmid required for activation of the Cdc6 promoter were: E2F1, 0.3 ng; E2F3, 3 ng; E2F3, 3 ng; YY1, 10 ng; and RYBP, 70 ng. (B) E2F3 and YY1 protein level in the presence of overexpressed YY1/RYBP and E2F3/RYBP. NIH-3T3 cells were transfected with increasing amounts of YY1/RYBP plasmid or E2F3/RYBP plasmid as indicated. To achieve efficient transfection (∼50%), Superfect transfection reagent (Qiagen) was used. Cells were harvested 24 h post-transfection and nuclear extracts were resolved by SDS–PAGE, western blotted and analyzed with antibody against endogenous E2F3 and YY1 protein. (C) Western blot analysis of increasing amounts of transfected HA-E2F1 and HA-E2F3. NIH-3T3 cells were transfected with the indicated concentration of plasmid encoding HA-E2F1 and HA-E2F3. Protein level was detected by western blotting using α-HA antibody. (D) Titration of E2F1, E2F2, E2F3, YY1 and RYBP. Increasing concentrations as indicated of either E2F1, E2F2, E2F3, YY1 or RYBP pcDNA3 were transfected into NIH-3T3 cells together with Cdc6 reporter plasmid. The amount of plasmid required for activation of the Cdc6 promoter was determined: E2F1, 0.3 ng; E2F2, 3 ng, E2F3; 3 ng; YY1, 10 ng; and RYBP, 70 ng.
A role for YY1 together with E2F2 or E2F3 in synergistic activation of the human Cdc6 promoter
Although it is clear that either E2F2 or YY1 can stimulate Cdc6 promoter activity, it is likely that the ability of the single proteins to activate is a function of their overexpression. Previous work has demonstrated that the interaction of the adenovirus E4 ORF6/7 protein with E2F, mediated through the marked box domain, leads to a stimulation of E2F-dependent transcription (Neill et al., 1990; Raychaudhuri et al., 1990; Neill and Nevins, 1991; Jost et al., 1996). Given the previous data linking RYBP with YY1, together with the fact that the activity of the Cdc6 promoter is dependent on both YY1 and E2F, we have investigated the potential synergistic effect of E2F with YY1 in the activation of Cdc6 promoter activity. Cells were transfected with the Cdc6 reporter plasmid and either YY1 alone, E2F2 or E2F3 alone, RYBP alone, or various combinations of the plasmids. In each case, the concentration employed was below that required for activation of the promoter, as defined by preliminary titrations of the plasmids (see Figure 3D). As seen from the data in Figure 3A, it is apparent that while neither of these proteins could activate the Cdc6 promoter on their own, there was a synergistic activation of transcription when E2F2 or E2F3 was co-expressed with YY1 and RYBP. Omission of RYBP, YY1 or E2F from these assays greatly reduced promoter activity, indicating that all three proteins are required for full promoter activity.
To rule out the possibility that the synergistic increase in activity by E2F3, YY1 and RYBP was due to altered E2F protein levels, we transfected NIH-3T3 cells with increasing amounts of YY1 pcDNA3 and Myc-RYBP plasmid. Cells were harvested 24 h post-transfection and processed for nuclear extracts. Extracts were resolved by SDS–PAGE and analyzed with antibody to endogenous E2F3 and YY1. As shown in Figure 3B, overexpression of E2F3 and RYBP did not alter YY1 protein levels, nor did overexpression of YY1 and RYBP alter E2F3 protein levels.
The in vivo and in vitro binding data demonstrate that the interaction of E2F proteins with RYPB was restricted to E2F2, E2F3 and E2F6. To determine whether the synergistic activation of transcription by YY1, RYBP and E2F reflected the specificity of this interaction, we assayed the ability of E2F1 to participate in the synergistic activation of transcription with YY1 and RYBP. As shown in Figure 3A, E2F1 was not able to activate along with YY1 and RYBP despite the fact that E2F1, when expressed at high levels, could activate the Cdc6 promoter on its own (Figure 3D). The comparison of the capacity of E2F1 and E2F3 to synergize with YY1 and RYBP used plasmid concentrations that yielded equal levels of the two E2F proteins as determined by titration of the plasmids and western blotting (Figure 3C).
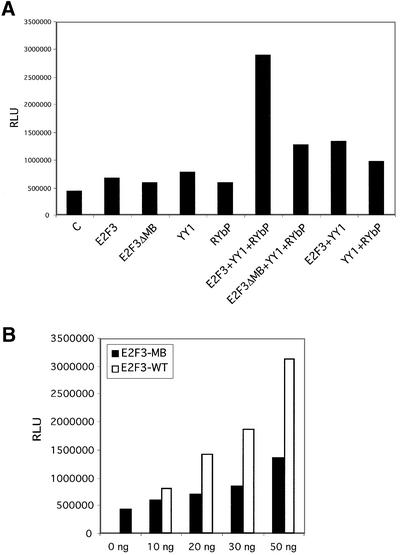
Finally, we have also assayed the contribution of the marked box domain of E2F3 to cooperation with YY1 in the activation of the Cdc6 promoter. As shown by the analysis in Figure 4A, the combination of E2F3, YY1 and RYBP again led to synergistic activation of Cdc6 promoter activity. In contrast, the E2F3ΔMB mutant failed to cooperate with YY1 and RYBP. This mutant nevertheless was functional and capable of activating the Cdc6 promoter when overexpressed (Figure 4B), demonstrating that the mutant has the capacity for transcription activation.
Fig. 4. Synergistic activation of the Cdc6 promoter by YY1, RYBP and E2F3 requires the E2F3 marked box domain. (A) The E2F3Δ marked box mutant fails to cooperate with YY1 and RYBP. NIH-3T3 cells were transfected with Cdc6 reporter plasmid and low amounts of either YY1 alone, E2F3 alone, E2F3Δ marked box mutant (E2F3ΔMB) alone, RYBP alone or combinations of the plasmid. The plasmid concentrations were: E2F3, 3 ng; E2F3ΔMB, 10 ng; YY1, 10 ng; and RYBP, 70 ng. Reporter gene activity (mean of triplicate determinations) was calculated and normalized to β-gal activity. (B) A high level of expression of the E2F3ΔMB mutant activates the Cdc6 promoter. NIH-3T3 cells were transfected with Cdc6 reporter plasmid together with the indicated amount of pcDNA3-E2F3ΔMB. Reporter gene activity (mean of triplicate determinations) was calculated and normalized to β-gal activity.
Taken together, these results thus demonstrate a specific role for E2F2 and E2F3 in activating the Cdc6 promoter in combination with YY1 and RYBP, dependent on the marked box domain of the E2F proteins. These observations suggest that formation of an E2F–RYBP–YY1 complex facilitates the activation of this promoter during G1/S phase.
Chromatin immunoprecipitation assays reveal an interaction of E2F2 and E2F3, together with YY1 and RYBP, on the Cdc6 promoter
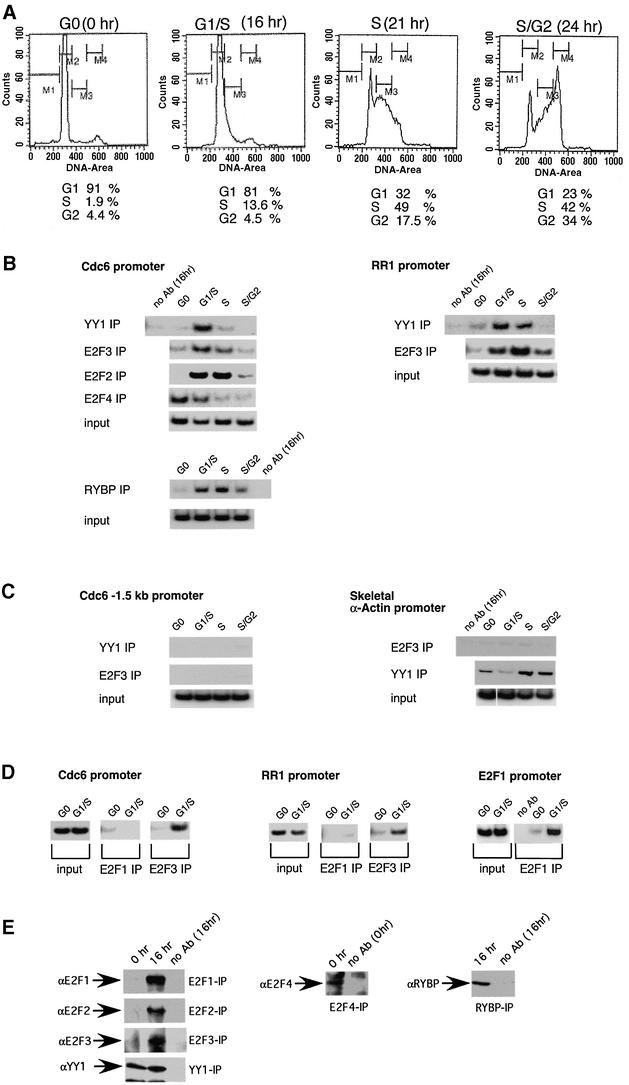
We have made use of chromatin immunoprecipitation (ChIP) assays to examine the interaction of the E2F2 or E2F3 proteins with the Cdc6 promoter as a direct measure of the in vivo specificity of these interactions. Human T98G cultures were brought to quiescence and then stimulated to grow by the addition of fresh media with serum. Cells were harvested either at quiescence or 16, 21 or 24 h following growth stimulation. As shown by the fluorescence-activated cell sorting (FACS) analysis in Figure 5A, these time points represent cells at G1/S, S and S/G2 of the cell cycle. The cells were cross-linked with formaldehyde, extracts were prepared, DNA was fragmented and then chromatin material was immunoprecipitated with antibodies specific for YY1, RYBP, E2F2, E2F3 and E2F4. DNA was released from the immunoprecipitates and then used for PCR assays to measure the presence of the Cdc6 promoter.
Fig. 5. ChIP assays reveal an interaction of E2F2 and E2F3, as well as YY1, with the Cdc6 promoter. (A) Cell cycle synchronization. FACS analysis of T98G cells that were rendered quiescent by serum starvation (0 h) and subsequently re-stimulated with serum for the indicated times and allowed to enter the cell cycle. DNA content is plotted versus cell number (counts). (B) The binding of YY1, E2F and RYBP proteins to the Cdc6 and RR1 promoter changes during the cell cycle. ChIPs using extracts from synchronized T98G cells (as in A) were performed with antibodies as indicated. The resulting precipitated DNA was amplified with primer pairs as indicated. Input represents 2% of the total amount of chromatin added to each immunoprecipitation. No antibody serves as a control for the immunoprecipitation. PCR products were detected by autoradiography. PCR products in the input, no antibody control and specific immunoprecipitation lanes were obtained in the same experiment. Each experiment was performed at least three independent times, and a representative example is shown. (C) Cdc6 1.5 kb and skeletal α-actin promoter sequences. The Cdc6 1.5 kb promoter sequence was detected with primers corresponding to regions –1.5 kb upstream of the E2F- and YY1-binding sites. The skeletal α-actin promoter sequences served as a control for an E2F3-independent promoter. ChIPs and analysis of DNA were as described in (B). (D) E2F3 but not E2F1 binds to the Cdc6 and RR1 promoter at G1/S. ChIPs were performed as described with antibodies and primer pairs as indicated. T98G cells were serum starved (G0) and released into the cell cycle for 16 h (G1/S). (E) IP-western. One-third of the immunoprecipitated material in (B), (C) and (D) was analyzed by western blotting. Immunoprecipitations were performed as described. The resulting precipitates were probed with the same antibodies as indicated.
As shown in Figure 5B, Cdc6 promoter sequences were detected in the E2F2 and E2F3 immunoprecipitates from the 16 and 21 h samples (G1/S and S phase of the cell cycle) but not in the quiescent sample, and the amount recovered in the 24 h sample (S/G2 transition) was reduced compared with that of the 21 h sample. Likewise, the YY1 immunoprecipitate revealed the Cdc6 promoter sequence with kinetics that coincided with those of E2F2 and E2F3 at G1/S transition and S phase of the cell cycle. The interactions of E2F2, E2F3 and YY1 are thus coincident with the time of activation of the Cdc6 promoter. In contrast, the E2F4 immunoprecipitate revealed an interaction with the Cdc6 promoter in the quiescent sample that then diminished as the cells entered the cell cycle. These results are thus consistent with a role for E2F4, together with an Rb family member, functioning to repress transcription in quiescent cells and then released from the promoter as cells are stimulated to grow. Coincident with this release of E2F4 is the binding of either E2F2 or E2F3 together with YY1 to the promoter at G1/S. Although the functional analysis of the Cdc6 promoter (Figure 2) would suggest a role for the –36 E2F site in the interaction with E2F4, and the –1 site interacting with E2F2 or E2F3, the precise nature of the actual sites of interaction has not been established from these assays.
The same samples were also assayed for interactions involving the RYBP protein. As shown in Figure 5B, RYBP immunoprecipitation also revealed an interaction of this protein with the Cdc6 promoter and with kinetics that mirrored those of E2F2, E2F3 and YY1, and thus the activation of Cdc6 promoter activity.
As further evidence for the joint interaction of the E2F proteins with YY1, we also assayed the RR1 promoter in which E2F sites are also found together with YY1 sites (Johansson et al., 1998). As shown by the data in Figure 5B (right panel), both E2F3 and YY1 could be seen to interact with the RR1 promoter and with kinetics similar to those for the interaction with the Cdc6 promoter.
Additionally, we performed controls to ensure that the ChIP results were produced by specific enrichment of the Cdc6 and RR1 promoter sequence that flanked the E2F and YY1 sites. As shown in Figure 5C, PCR amplification of Cdc6 promoter regions –1.5 kb upstream from the E2F- and YY1-binding sites revealed no evidence for interaction with either YY1 or E2F3. A further example of the specificity can be seen by the interaction of YY1 with the skeletal α-actin promoter, previously shown to be a target for negative control by YY1 (Lee et al., 1992). In this instance, YY1 was found to interact with the skeletal α-actin promoter in quiescent cells and at late time points at S phase and S/G2 transition (Figure 5C). In contrast, there is no evidence for an E2F3 interaction. Thus, in this context, the interaction of YY1 is E2F3 independent.
Finally, as further evidence for the specificity in the chromatin associations, we also assayed for interaction of the E2F1 protein with both the Cdc6 promoter and the RR1 promoter. As shown in Figure 5D, we found no evidence for an E2F1 interaction with either the Cdc6 promoter or the RR1 promoter at a time when E2F3 was clearly seen to interact. The absence of an association of E2F1 with the Cdc6 and RR1 promoters is not due to an inability of anti-E2F1 antibody to detect this protein when bound to chromatin since the same samples did show an association of E2F1 with the E2F1 promoter (Figure 5D). In addition, immunoprecipitation of chromatin isolated from cross-linked cells resulted in the detection of each E2F family member in the appropriate samples (Figure 5E). These experiments also attest to the specificity of each antibody used in the ChIPs. Importantly, the anti-E2F1 antibody (KH95; Santa Cruz) used in the immunoprecipitations is specific to E2F1 protein and not cross-reactive with E2F3 (see Supplementary data).
We conclude that E2F2 and E2F3 do indeed interact with the Cdc6 promoter in vivo, coincident with the interaction of YY1 and RYBP. We also note that whereas the kinetics of these chromatin interactions do coincide with the accumulation of the E2F proteins, the YY1 protein is present in both quiescent and G1/S samples (see Figure 5E), suggesting a role for the E2F proteins in the recruitment of YY1. Moreover, the absence of an interaction of E2F1 with the Cdc6 promoter demonstrates a specificity that reflects the specificity of interaction with RYBP and activation of Cdc6 promoter activity.
Discussion
The structural complexity of the E2F family reflects a complexity in function in which individual family members have been shown to perform distinct functions. For instance, E2F1-null mice have defects in thymocyte development and are tumor prone (Field et al., 1996; Yamasaki et al., 1996), phenotypes not seen upon deletion of other E2F genes. Other work has shown a unique ability of E2F1 to signal apoptosis and to induce p53 accumulation (DeGregori et al., 1997; Kowalik et al., 1998; Moroni et al., 2001). E2F3-null fibroblasts exhibit a reduced rate of proliferation and deregulation in the expression of E2F-responsive genes (Humbert et al., 2000), classifying E2F3 clearly as a critical regulator for normal cellular proliferation and consistent with previous work in which the inhibition of E2F3 activity blocked cell cycle progression (Leone et al., 1998). Although E2F2-null mice do not exhibit an overt phenotype, and cells prepared from E2F2-null embryos proliferate normally, the combined loss of E2F2 and E2F3 does result in early embryonic lethality, and cells that are null for both E2F2 and E2F3 are more impaired for proliferation than those only null for E2F3 (Wu et al., 2001). Moreover, both E2F2 and E2F3 are required for Myc to induce S phase, whereas E2F1 activity is not required for Myc-induced S phase but is required for Myc-induced apoptosis (Leone et al., 2001). Taken together, these observations provide evidence to support the distinct function of E2F family members and also suggest the possibility that E2F2 and E2F3 function might overlap. The work presented here provides biochemical evidence for promoter-specific interactions that could help to explain these functional differences.
Promoter-specific complexes involving E2F2 and E2F3
Based on the work presented here, and in combination with other recent findings, we propose a model whereby E2F-dependent gene regulation would result from interaction of specific E2F proteins with other transcriptional regulatory proteins in a promoter-specific fashion. In particular, our observation that E2F2 and E2F3 form a specific interaction with the YY1-binding protein RYBP, together with the role of YY1 in the control of Cdc6 promoter activity, provides a mechanism for selective gene control within the E2F family. We suggest that YY1 and any of the E2F proteins have the potential to interact with the respective binding sites in the Cdc6 promoter independently of the partner, but that such interactions are non-productive. This could reflect an inherently weak binding of the protein on its own or the inability to create a platform for recruitment of other proteins. Regardless, it would only be when the proper pair of proteins interact jointly, bridged by the RYBP factor, that a functional complex is formed. In this model, it is only when E2F2 or E2F3, together with YY1 and RYBP, interact jointly and form a promoter complex that a functional transcription complex is formed and the gene is activated (Figure 6). Based on other observations, including the paradigm of the E2F–E4 ORF6/7 interaction, as well as recent findings that demonstrate a specific interaction of E2F3 with the TFE3 transcription factor (P.Giangrande and J.Nevins, unpublished data), we favor the notion that the synergy reflects an enhanced DNA binding of the factors. Indeed, this would be consistent with the observation that YY1 is present in both quiescent and growing cells but is only recruited to the Cdc6 promoter at G1/S, coincident with the accumulation of the E2F proteins. A mechanism involving enhanced binding, for instance an increase in the stability of the co-complex, would then mean that only when all of the proteins are present is a functional complex generated, and thus the specificity of the interaction is essentially the combination of the two binding sites.
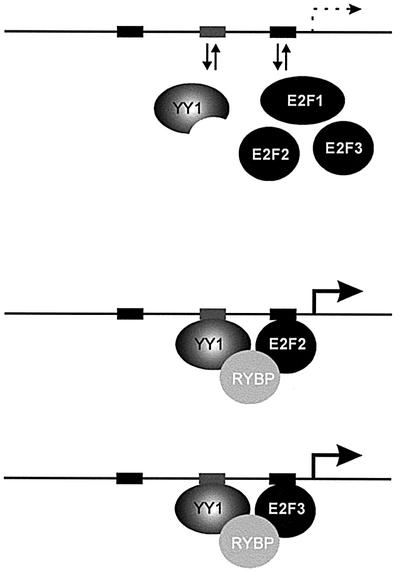
Fig. 6. Cooperative activation of Cdc6 by YY1, RYBP and E2F2/E2F3. A model for protein interactions involving YY1, RYBP and E2Fs based on stabilization of otherwise weak interactions.
The ChIP assays also provide evidence for specificity in the recruitment of a promoter complex. While both E2F2 and E2F3 could be found in association with YY1 and RYBP on the Cdc6 promoter, we could detect no evidence of an E2F1 interaction with this promoter. Although this is in contrast to previous ChIP results that did suggest an E2F1 interaction with the Cdc6 promoter at G1/S (Takahashi et al., 2000), more recent data also suggest a lack of E2F1 binding to Cdc6 (Ren et al., 2002). One possible explanation for the discrepancy of these results could lie in the use of antibodies since, in our experience, the E2F1-C20 antibody (Santa Cruz) employed in certain past ChIP assays can cross-react with the E2F3 protein (data not shown). Thus, to ensure specificity in the detection of E2F1, we have used the E2F1-KH95 antibody (Santa Cruz) that does not cross-react with E2F3.
The binding specificity of RYBP with E2F2 and E2F3, but not E2F1 and E2F4, provides a molecular basis for gene-specific transcriptional activation. Restricting individual E2Fs to only a few promoter-specific factors represents a basis for how a particular E2F protein might activate specific sets of target genes. In this instance, promoters that contain YY1 sites adjacent to E2F sites would be potential E2F2/E2F3 targets. As such, it would be the combination of a YY1 site and an E2F site that would dictate the specificity of transcription control. In addition, other E2F-controlled promoters, such as DHFR, E2F2 and Cdk2, are not responsive to YY1 (for E2F2 data, see Supplementary data), indicating that YY1 would not be the only partner for these E2F proteins; indeed, our recent data have identified the TFE3 transcription factor as a partner for E2F3. We would extend the model to other E2F proteins, such as E2F1, by suggesting that these factors interact with yet other transcriptional control proteins to impart specificity. The summation of this series of interactions thus describes a model of combinatorial gene regulation that creates specificity with a limited number of proteins and specific protein interactions.
Interestingly, recent results have also described an interaction between RYBP and the E2F6 family member, although these authors did not detect an interaction with either E2F2 or E2F3 (Trimarchi et al., 2001). The basis for this difference is unclear, but could reflect the nature of the constructs used as well as the assays employed. The structure of the E2F6 gene product, when compared with the other E2F family members, has suggested a role for E2F6 as an Rb-independent transcriptional repressor since it is lacking the C-terminal domain responsible for transcription activation and interaction with Rb family members. Indeed, functional assays have provided evidence for an ability of E2F6 to repress transcription independently of Rb (Morkel et al., 1997; Cartwright et al., 1998; Gaubatz et al., 1998; Trimarchi et al., 1998). Given the common ability to interact with RYBP, and thus the potential to form a complex with the polycomb protein Ring1, it seems possible that E2F6 could have a specific overlapping function with that of E2F2 and E2F3. Thus, rather than activating genes such as Cdc6, E2F6 might represent a specific repressor of Cdc6 transcription.
Materials and methods
Yeast two-hybrid assay
The yeast two-hybrid screen was performed as recommended by the manufacturer (Clontech, Palo Alto, CA) using the yeast strain PJ69-4a expressing DBD-E2F2 as the bait. After transformation with the human placenta cDNA library (MatchMaker; Clontech), 2 × 106 transformants were screened initially by histidine nutritional selection and β-gal activity. Plasmid DNA from 90 His+ clones was rescued from yeast by transformation into the Escherichia coli strain MH1066. The library plasmid DNA was sequenced and compared with the National Center for Biotechnology Information (NCBI) database using the basic local alignment search tool (BLAST) algorithm. To test whether the identified clones were specific interactors with the other DBD-E2F proteins and DBD-marked box mutants, the library plasmid DNA was transformed into the mating strain PJ69-4α (James et al., 1996). Mating was performed with the PJ69-4a culture harboring the individual DBD-E2F1, 2, 3 and 4, and marked box mutant proteins DBD-E2F2ΔC and DBD-E2F3ΔC. Interactions were assayed by replica plating, growth selection and β-gal activity. Plasmids constructed for interaction assays are described in the Supplementary data.
Expression vectors and reporter plasmids
Myc-tagged RYBP pcDNA3, YY1pcDNA3, HA-E2F1/2/3/4/6 pcDNA3, E2F1/2/3/4-GSTpGEX and E2F3ΔCpGEX were cloned as described in the Supplementary data. Wild-type and E2F site mutant versions of the human Cdc6 promoter–luciferase constructs (–1.700 to +7 kb) were described previously (Yan et al., 1998). Nucleotide substitutions were introduced into the putative YY1 site at position –15 (GGCCATTC converted to GGCGGTTC) using a site-directed mutagenesis kit (GeneEditor; Promega). The minimal Cdc6 promoter region (–130 to +21 kb) was generated by PCR and ligated upstream of the luciferase gene in pGLBasic (Promega). The DNA sequence of all PCR-generated constructs was verified by sequencing.
Yeast two-hybrid liquid β-galactosidase assays
Quantitative two-hybrid liquid culture β-gal assays (Reynolds et al., 1997) were performed in the yeast strain PJ69-4. Expression levels of the GAD and GBD fusion proteins were evaluated by western blot analysis. Experiments were performed a minimum of three times. Values shown are representative results from individual experiments.
GST pull-down
GST fusion protein expression vectors (E2F1, 2, 3 and 4-GSTpGEX and E2F3ΔMB-pGEX-6P1) were transformed into E.coli strain BL21 (DE3), and purification of the GST proteins was performed as described previously (Burke et al., 2001). Myc-tagged RYBP was in vitro transcribed and translated (TNT reticulocyte lysate system; Promega) in the presence of [35S]methionine. Equal amounts of GST, GST–E2F1, GST–E2F2, GST–E2F3, GST–E2F4 and GST–E2F3ΔMB coupled to glutathione–Sepharose beads were incubated with [35S]RYBP overnight at 4°C in 20 mM Tris pH 8, 70 mM NaCl, 1 mM EDTA pH 8, 0.5% NP-40, 1 mM dithiothreitol (DTT; NETN-Buffer) and protease inhibitors. Beads were washed extensively six times with NETN-Buffer, boiled in SDS sample buffer and proteins were analyzed by 10% SDS–PAGE followed by autoradiography.
Cell culture
NIH-3T3 cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% bovine calf serum. REF52 cells were grown in DMEM containing 10% fetal bovine serum (FBS). To bring REF52 cells to quiescence, cultures at 40% confluence were washed once with DMEM and then incubated in DMEM plus 0.1% FBS for 24 h. Cells were serum stimulated in DMEM containing 20% FBS.
FACS analysis
T98G human glioblastoma cells (ATCC) were grown in DMEM containing 10% FBS. Cells were rendered quiescent by serum deprivation in DMEM containing 0.1% serum for 48 h and stimulated with 20% serum to allow cell cycle re-entry. Propidium iodide staining and FACS analysis were performed as previously described (Smith et al., 1996).
Transient transfection assays
Cells were transfected by SuperFect Transfection Reagent (Qiagen) according to the manufacturer’s instructions. The transfected cells were split into nine 60 mm dishes in DMEM containing 0.1% FBS for 24 h. Serum was added (20% final) and harvested at the indicated time points. For transactivation assays, NIH-3T3 cells were plated into 60 mm dishes, transfected by SuperFect Transfection Reagent and harvested 22 h after transfection. β-galactosidase assays were performed as described previously (Johnson et al., 1993). Luciferase assays were performed using a luciferase reporter assay system (Promega). All transfection assays were repeated at least three times, and representative results are shown.
Antibodies and immunoprecipitations
HA-E2F1, 2, 3, 4 and 6 proteins containing an N-terminal influenza HA epitope tag were detected using polyclonal antibody α-HA (Y-11; Santa Cruz). Human RYBP protein containing an N-terminal Myc epitope tag was detected using mouse monoclonal antibody α-c-myc (9E10; Santa Cruz). Immunoprecipitations were performed by transfecting NIH-3T3 cells with the expression vectors pcDNA3-HAE2F1, 2, 3, 4 and 6, and myc-RYBPpcDNA3 as indicated. Equal expression levels of the HA-E2F proteins were confirmed by western blot analysis using α-HA antibodies. Whole-cell extracts were prepared by sonication (2 × 20 s) in CoIP buffer [0.5% NP-40, 50 mM Tris pH 8, 25% glycerol, 0.1 mM EDTA, 70 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 1 µg/ml apoprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin]. Extracts were in cubated with α-HA-coupled affinity matrix (Convance) for 2 h at 4°C. After extensive washes with CoIP buffer, immunoprecipitated proteins were resolved by SDS–PAGE and detected by immunoblotting and fluorography.
E2F DNA binding assays
Gel retardation assays were as described previously (Chellappan et al., 1991; Ikeda and Nevins, 1993), with the following modifications. The gel contained 4% acrylamide (75:1 acrylamide/bisacrylamide) and 5% glycerol. The purified GST fusion proteins used in the gel shift assays were prepared as described and cleaved from GST by protease (Factor Xa) at room temperature for 1 h. The DNA probes contain the Cdc6 promoter sequences from positions –130 to +21.
Chromatin immunoprecipitations
Two subconfluent 150 cm2 dishes of T98G cells were used for each ChIP, and cells were cross-linked by the addition of formaldehyde to 1% final concentration. The procedure was performed as described previously (Takahashi et al., 2000) with the following modifications. Prior to immunoprecipitation, chromatin material was sonicated on ice to an average length of 700 bp and adjusted in 1× RIPA buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitors). Chromatin was pre-cleared with protein A– and protein G–Sepharose [blocked with 0.1 µg/ml poly(dI–dC) and 1 mg/ml bovine serum albumin (BSA)] at 4°C for 4 h. Pre-cleared chromatin was incubated with 2–5 µg of each antibody (E2F1, SC-251x; E2F2, SC-633; E2F3, SC-878x; E2F4, SC-1082x; YY1, SC-7341x; RYBP, as described in Garcia et al., 1999) at 4°C overnight. A 15 µl aliquot of 50% slurry of pre-blocked protein A/G–Sepharose was added and immune complexes were recovered after 3 h incubation at 4°C. Immunoprecipitates were washed seven times with RIPA buffer. Pellets were resuspended in TE and cross-links reversed at 65°C with 10 µg of RNase A and proteinase K overnight. Following phenol:chloroform extraction and ethanol precipitation, pellets were resuspended in 50 µl of H2O and analyzed by PCR. PCRs were performed with 2 µl of immunoprecipitated material, 50 pmol of each primer set, 0.5 U of Taq Gold DNA polymerase (Applied Biosystems) and 0.01 µCi of [32P]dGTP using primer sets as described in the Supplementary data. PCR products were electrophoresed on 6% polyacrylamide gels. Proteins in chromatin immunoprecipitates were detected by western blotting. Immuno precipitation reactions were performed as described above. After washing with RIPA buffer, the samples were resuspended in 100 µl of Tris/EDTA containing 1% SDS and incubated at room temperature for 15 min. Samples were boiled for 30 min in 1× sample buffer, and one-third of the samples were electrophoresed on an SDS–polyacrylamide gel and transferred to a membrane. To detect YY1, RYBP and the E2F proteins, the following antibodies were used: anti-E2F1 (sc-251x; Santa Cruz); anti-E2F2 [Ab2 (TFE25) and Ab1 (TFE22); NeoMarkers]; anti-E2F3 [Ab1 (TFE31) and Ab2 (TFE36); NeoMarkers]; anti-E2F4 (sc-511; Santa Cruz), anti-YY1 (sc-7341x; Santa Cruz) and anti-RYBP (Garcia et al., 1999).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank J.Cook, T.Burke, P.Giangrande and P.Black for invaluable advice, discussion and technical assistance, Drs D.Picard, S.Gaubatz, R.Martell, T.Shenk and W.Krek for reagents, and Kaye Culler for help with the preparation of the manuscript. J.R.N. is an Investigator of the Howard Hughes Medical Institute. M.V. was supported by the Ministry of Science and Technology (grants SAF2001-2211-C02-01 and CAM 08.1/0050/2000). S.S. was supported by NIH grant CA79983.
References
- Burke T.W., Cook,J.G., Asano,M. and Nevins,J.R. (2001) Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem., 276, 15397–15408. [DOI] [PubMed] [Google Scholar]
- Cartwright P., Muller,H., Wagener,C., Holm,K. and Helin,K. (1998) E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene, 17, 611–623. [DOI] [PubMed] [Google Scholar]
- Chellappan S.P., Hiebert,S., Mudryj,M., Horowitz,J.M. and Nevins,J.R. (1991) The E2F transcription factor is a cellular target for the RB protein. Cell, 65, 1053–1061. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Leone,G., Miron,A., Jakoi,L. and Nevins,J.R. (1997) Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl Acad. Sci. USA, 94, 7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Field S.J., Tsai,F.-Y., Kuo,F., Zubiaga,A.M., Kaelin,W.G.,Jr, Livingston, D.M., Orkin,S.H. and Greenberg,M.E. (1996) E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell, 85, 549–561. [DOI] [PubMed] [Google Scholar]
- Garcia E., Marcos-Gutierrez,C., del Mar Lorente,M., Moreno,J.C. and Vidal,M. (1999) RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex and with the transcription factor YY1. EMBO J., 18, 3404–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz S., Wood,J.G. and Livingston,D.M. (1998) Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl Acad. Sci. USA, 95, 9190–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G., Wobst,A., Petersen,B.O., Cam,L.L., Vigo,E., Sardet,C. and Helin,K. (1998) Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol., 18, 6679–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert P.O., Verona,R., Trimarchi,J.M., Rogers,C., Dandapani,S. and Lees,J.A. (2000) E2f3 is critical for normal cellular proliferation. Genes Dev., 14, 690–703. [PMC free article] [PubMed] [Google Scholar]
- Ikeda M.-A. and Nevins,J.R. (1993) Identification of distinct roles for separate E1A domains in the disruption of E2F complexes. Mol. Cell. Biol., 13, 7029–7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E., Hjortsberg,K. and Thelander,L. (1998) Two YY-1-binding proximal elements regulate the promoter strength of the TATA-less mouse ribonucleotide reductase R1 gene. J. Biol. Chem., 273, 29816–29821. [DOI] [PubMed] [Google Scholar]
- Johnson D.G., Schwarz,J.K., Cress,W.D. and Nevins,J.R. (1993) Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature, 365, 349–352. [DOI] [PubMed] [Google Scholar]
- Jost C.A., Ginsbreg,D. and Kaelin,W.G.,Jr (1996) A conserved region of unknown function participates in the recognition of E2F family members by the adenovirus E4 ORF 6/7 protein. Virology, 220, 78–90. [DOI] [PubMed] [Google Scholar]
- Kalenik J.L., Chen,D., Bradley,M.E., Chen,S.-J. and Lee,T.-C. (1997) Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res., 25, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik T.F., DeGregori,J., Leone,G. and Nevins,J.R. (1998) E2F1-specific induction of apoptosis and p53 accumulation is modulated by mdm2. Cell Growth Differ., 9, 113–118. [PubMed] [Google Scholar]
- Lee T.-C., Shi,Y. and Schwartz,R.J. (1992) Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal α actin transcription in embryonic myoblast. Proc. Natl Acad. Sci. USA, 89, 9814–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G., DeGregori,J., Yan,Z., Jakoi,L., Ishida,S., Williams,R.S. and Nevins,J.R. (1998) E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev., 12, 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G. et al. (2001) Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell, 8, 105–113. [DOI] [PubMed] [Google Scholar]
- Lin W.-C., Lin,F.-T. and Nevins,J.R. (2001) Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev., 15, 1833–1845. [PMC free article] [PubMed] [Google Scholar]
- Morkel M., Wenkel,J., Bannister,A.J., Kouzarides,T. and Hagemeier,C. (1997) An E2F-like repressor of transcription. Nature, 390, 567–568. [DOI] [PubMed] [Google Scholar]
- Moroni M.C., Hickman,E.S., Denchi,E.L., Caprara,G., Colli,E., Cecconi,F., Muller,H. and Helin,K. (2001) Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol., 3, 552–558. [DOI] [PubMed] [Google Scholar]
- Neill S.D. and Nevins,J.R. (1991) Genetic analysis of the adenovirus E4 6/7 trans activator: interaction with E2F and induction of a stable DNA–protein complex are critical for activity. J. Virol., 65, 5364–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S.D., Hemstrom,C., Virtanen,A. and Nevins,J.R. (1990) An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc. Natl Acad. Sci. USA, 87, 2008–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J.R. (1998) Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ., 9, 585–593. [PubMed] [Google Scholar]
- Ohtani K., Tsujimoto,A., Ikeda,M. and Nakamura,M. (1998) Regulation of cell growth-dependent expression of mammalian CDC6 gene by the cell cycle transcription factor E2F. Oncogene, 17, 1777–1785. [DOI] [PubMed] [Google Scholar]
- Pilpel Y., Sudarsanam,P. and Church,G.M. (2001) Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet., 29, 153–159. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri P., Bagchi,S., Neill,S.D. and Nevins,J.R. (1990) Activation of the E2F transcription factor in adenovirus-infected cells involves E1A-dependent stimulation of DNA-binding activity and induction of cooperative binding mediated by an E4 gene product. J. Virol., 64, 2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Cam,H., Takahashi,Y., Volkert,T., Terragni,J., Young,R.A. and Dynlacht,B.D. (2002) E2F integrates cell cycle progression with DNA repair, replication and G2/M checkpoints. Genes Dev., 16, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A., Lundblad,V., Dorris,D. and Keaveney,M. (1997) Yeast vectors and assays for expression of cloned genes. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A., Struhl,K. and Benson Chanda,V. (eds), Current Protocols in Molecular Biology. John Wiley and Sons, Inc., Boston, MA, pp. 13.6.2–13.6.3. [DOI] [PubMed]
- Smale S.T. (2001) Core promoters: active contributors to combinatorial gene regulation. Genes Dev., 15, 2503–2508. [DOI] [PubMed] [Google Scholar]
- Smith E.J., Leone,G., DeGregori,J., Jakoi,L. and Nevins,J.R. (1996) The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 from a G1 cell state. Mol. Cell. Biol., 16, 6965–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Rayman,J.B. and Dynlacht,B.D. (2000) Analysis of promoter binding by the E2F and Rb families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev., 14, 804–816. [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild,B., Verona,R., Moberg,K., Andon,N. and Lees,J.A. (1998) E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc. Natl Acad. Sci. USA, 95, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild,B., Wen,J. and Lees,J.A. (2001) The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl Acad. Sci. USA, 98, 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ginkel P.R., Hsiao,K.M., Schjerven,H. and Farnham,P.J. (1997) E2F-mediated growth regulation requires transcription factor cooperation. J. Biol. Chem., 272, 18367–18374. [DOI] [PubMed] [Google Scholar]
- Wu L. et al. (2001) The E2F1–3 transcription factors are essential for cellular proliferation. Nature, 414, 457–462. [DOI] [PubMed] [Google Scholar]
- Yamamoto K.R., Darimont,B.D., Wagner,R.L. and Iniguez-Lluhi,J.A. (1998) Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harb. Symp. Quant. Biol., 63, 587–598. [DOI] [PubMed] [Google Scholar]
- Yamasaki L., Jacks,T., Bronson,R., Goillot,E., Harlow,E. and Dyson,N.J. (1996) Tumor induction and tissue atrophy in mice lacking E2F-1. Cell, 85, 537–548. [DOI] [PubMed] [Google Scholar]
- Yan Z., DeGregori,J., Shohet,R.V., Leone,G., Stillman,B., Nevins,J.R. and Williams,R.S. (1998) Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Fraenkel,E., Pabo,C.O. and Pavletich,N.P. (1999) Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev., 13, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]