Abstract
In vitro, bacteriophage N4 virion RNA polymerase (vRNAP) recognizes in vivo sites of transcription initiation on single-stranded templates. N4 vRNAP promoters are comprised of a hairpin structure and conserved sequences. Here, we show that vRNAP consists of a single 3500 amino acid polypeptide, and we define and characterize a transcriptionally active 1106 amino acid domain (mini-vRNAP). Biochemical and genetic characterization of this domain indicates that, despite its peculiar promoter specificity and lack of extensive sequence similarity to other DNA-dependent RNA polymerases, mini-vRNAP is related to the family of T7-like RNA polymerases.
Keywords: bacteriophage N4/polymerase domain/single-subunit polymerases/virion RNA polymerase
Introduction
DNA-dependent RNA polymerases (RNAPs) are divided into two classes based on structural composition and sequence similarity. One class consists of the large multisubunit enzymes of eubacteria and archaea, and the nuclear and chloroplast polymerases of eukaryotes (Severinov, 2000). The other class encompasses small, primarily single-subunit enzymes homologous to phage T7 RNAP (McAllister and Raskin, 1993).
The early region of the double-stranded linear DNA genome of lytic coliphage N4 is transcribed by a phage-coded, virion-encapsidated RNAP. vRNAP is a polypeptide with an estimated mol. wt of 320 kDa (Falco et al., 1980), which is injected into the host cell with the phage genome at the onset of infection (Rothman-Denes and Schito, 1974; Falco et al., 1977). vRNAP promoters are comprised of conserved sequences and a 5 bp stem, three base loop hairpin structure (Haynes and Rothman-Denes, 1985; Glucksmann et al., 1992). In contrast to other known RNAPs, vRNAP transcribes single-stranded, promoter-containing templates in vitro with in vivo specificity (Falco et al., 1978; Haynes and Rothman-Denes, 1985).
The vRNAP gene encodes a 3500 amino acid polypeptide lacking extensive sequence similarity to either of the two families of DNA-dependent RNAPs. Here, we show that a central 1106 amino acid domain of the vRNAP polypeptide (mini-vRNAP) possesses transcriptional activity. Mini-vRNAP displays all the transcriptional properties of full-length vRNAP. Mutational and biochemical analyses indicate that the N4 vRNAP catalytic site is related to that of the T7-like RNAPs, although vRNAP appears to be an evolutionarily highly diverged member of this class of enzymes.
Results
The vRNAP gene
Sequencing of the vRNAP gene revealed an open reading frame encoding a 3500 amino acid protein with a calculated size of 382.5 kDa that contains no cysteine residues. vRNAP is injected into the host cell at the onset of infection. We suggest that the lack of cysteine residues might reflect the requirement for passage through the host periplasm, which contains proteins that catalyze disulfide bond formation (Ritz and Beckwith, 2001).
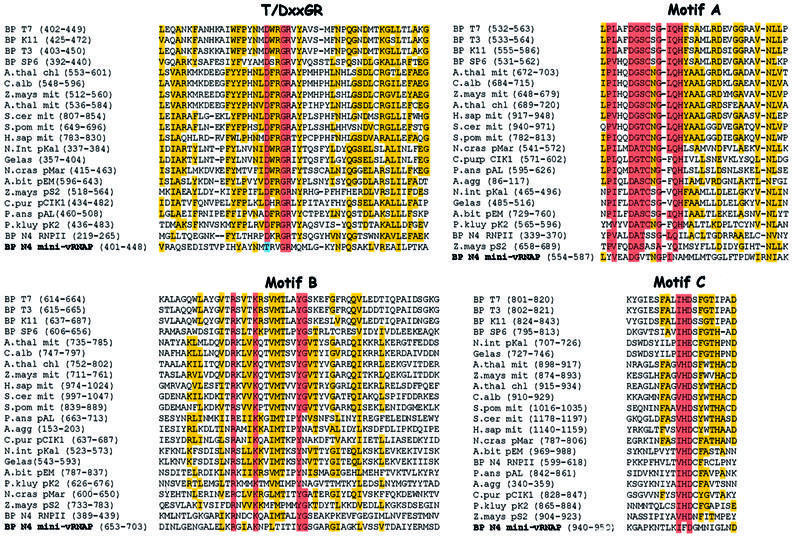
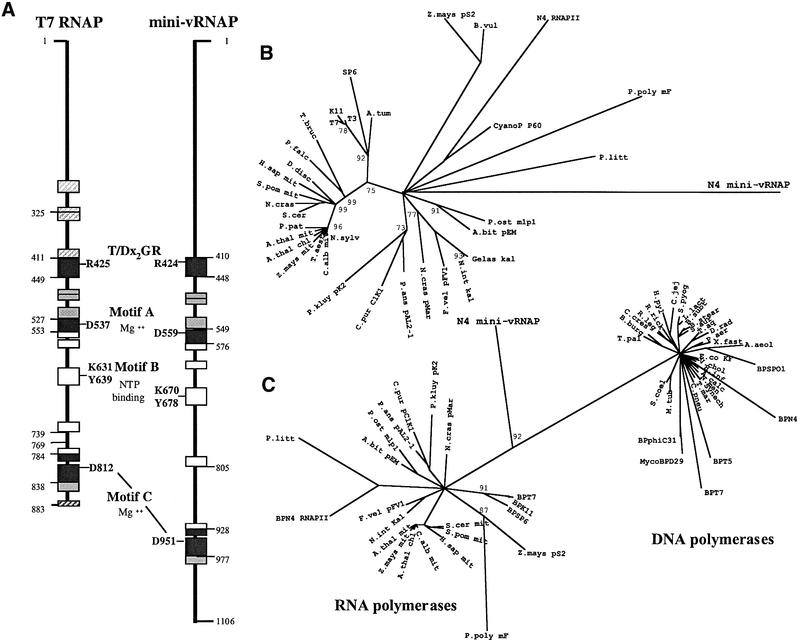
vRNAP’s size is comparable with the combined sizes of the β and β′ subunits of eubacterial RNAPs. Therefore, we expected that the vRNAP polypeptide might be homologous to these subunits, which comprise the catalytic center of multisubunit RNAPs (Mustaev et al., 1997; Zhang et al., 1999) and are synthesized as a fused polypeptide in some helicobacters and campylobacters (Zakharova et al., 1998). However, we found no extensive sequence similarity to members of either the multisubunit or single-subunit RNAP classes using standard comparison programs (BLAST, Altschul et al., 1997; FASTA, Pearson and Lipman, 1988). We next searched for regions of similarity using ‘profiles’, position-specific scoring matrices generated from conserved sequence domains characteristic of each RNAP class (Gribskov and Veretnik, 1996). Profile analysis revealed matches to two motifs common to the single-subunit (T7-like) DNA-dependent RNAPs and structurally related Pol I DNA polymerases (DNAPs), the T/DxxGR motif and motif B (Figure 1). Motif B lies between amino acids 1663 and 1676 in the vRNAP sequence. Matches to the T/DxxGR motif were found at amino acids 1417–1421 (TxxGR, present in DNAPs) and 2524–2528 (DxxGR, present in RNAPs). Alignments of the vRNAP sequence to these two motifs are shown in Figure 1. All catalytically important residues within these motifs are conserved in the vRNAP sequence; however, other highly conserved residues are not.
Fig. 1. Mini-vRNAP contains four motifs characteristic of T7-like RNA polymerases and Pol I DNA polymerases. Motifs TxxGR, A, B and C (Delarue et al., 1990; Mendez et al., 1994) are highlighted in red. Residues that are identical or conserved in 50% of the sequences are highlighted in yellow. Sequence abbreviations (in alphabetical order) and accession numbers are as follows: A.bit pEM, Agaricus bitorquis pEM plasmid-encoded RNAP (P33539); A.thal chl, Arabidopsis thaliana chloroplast RNAP (CAA69972); A.thal mit, Arabidopsis thaliana mitochondrial RNAP (CAA70210); A.agg, Ascosphaera aggregata plasmid-encoded RNAP (AAB64106); BP K11, bacteriophage K11 RNAP (P18147); BP SP6, bacteriophage SP6 RNAP (P06221); BP T3, bacteriophage T3 RNAP (CAA26719); BP T7, bacteriophage T7 RNAP (CAA24390); C.alb mit, Chenopodium album mitochondrial RNAP (CAA69305); C.pur pClK1, Claviceps purpurea pClK1 plasmid-encoded RNAP (P22372); Gelas pKal, Gelasinospora sp. kal plasmid-encoded RNAP (S62751); H.sap mit, Homo sapiens mitochondrial RNAP (AAB58255); BP N4 RNPII, bacteriophage N4 RNAP II (AAL71577, AAL71578); BP N4 mini-vRNAP (AY050713); N.cras pMar, Neurospora crassa Maranhar plasmid-encoded RNAP (P33540); N.int pKal, Neurospora intermedia Kalilo plasmid-encoded RNAP (P33541); P.kluy pK2, Pichia kluyveri plasmid-encoded RNAP (CAA72339); P.ans pAL, Podospora anserina pAL2-1 plasmid-encoded RNAP (S26945); S.pom mit, Schizosaccharomyces pombe mitochondrial RNAP (O13993); S.cer mit, Saccharomyces cerevisiae mitochondrial RNAP (P13433); Z.mays mit, Zea mays mitochondrial RNAP (CAA06488); Z.mays pS2, Zea mays pS2 plasmid-encoded RNAP (S22768). Mini-vRNAP amino acid 1 corresponds to amino acid 998 of vRNAP.
Identification of an active domain within N4 vRNAP
Enzymes belonging to the family of T7-like RNAPs are ∼100 kDa in size. In contrast, vRNAP is over three times larger. To determine whether a smaller transcriptionally active domain exists within the vRNAP polypeptide, we performed controlled trypsin proteolysis of vRNAP purified from virions, followed by a catalytic autolabeling assay (Grachev et al., 1987; Hartmann et al., 1988). In this assay, the proteolytic products are incubated with template DNA and a cross-linkable derivative of the initiating nucleotide, which becomes covalently linked to the ε-amino group of any lysine located within 12 Å of the nucleotide’s α-phosphate. Addition of the next template-directed [α-32P]NTP results in phosphodiester bond formation and radioactive labeling of the enzyme’s catalytic center. Three major proteolytic products were generated after 10 min of trypsinolysis (Figure 2A, lane 1); the two faster migrating products retain catalytic activity (lane 2, bands 2 and 3). Further incubation with trypsin yielded a single stable, active polypeptide with an estimated mol. wt of 122 kDa (lanes 3 and 4, band 3). N-terminal peptide sequencing of the products shown in lane 1 (Figure 2B) allowed identification of the shortest transcriptionally active peptide (Figure 2C). This peptide, designated mini-vRNAP, corresponds to vRNAP amino acids 998–2103 and contains the two previously identified polymerase motifs, TxxGR and B. The DxxGR motif is not functionally relevant since it is located in a region dispensable for RNAP activity.
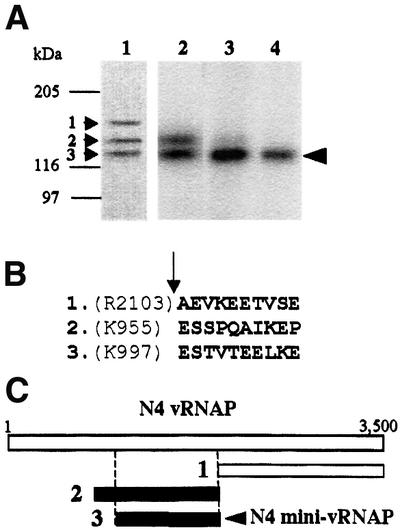
Fig. 2. Identification of the vRNAP transcriptional domain. (A) Proteolysis and catalytic autolabeling of vRNAP. Lane 1: trypsinolysis products (1, 2 and 3) visible by Coomassie R-250 staining after 10 min of digestion and transfer to PVDF. Lanes 2–4: catalytic autolabeling of proteolysis products, followed by SDS–PAGE and autoradiography. vRNAP was treated with trypsin for 10 min (lane 2), 1 h (lane 3) or 10 h (lane 4). Sizes and positions of protein standards are indicated. The arrowhead indicates the position of the shortest transcriptionally active proteolytic fragment. (B) N-terminal sequence obtained by Edman degradation of the proteolytic fragments shown in lane 1. The arrow indicates the position of the trypsin-cleaved peptide bond. (C) Location of the major proteolytic fragments (A) relative to the sequence of the vRNAP protein.
Expression and characterization of recombinant mini-vRNAP
The region encoding vRNAP amino acids 998–2103 was cloned with a His6 tag under the control of an arabinose-inducible promoter. After induction of protein expression, ∼98% pure recombinant mini-vRNAP was obtained after a single passage of the crude extract over a metal affinity column (Figure 3A). Recombinant mini-vRNAP is expressed at a 100-fold higher level than recombinant full-length vRNAP. At least 10 mg of nearly homogeneous mini-vRNAP are obtained from 1 l of induced cells. In addition, mini-vRNAP is stable after induction (Figure 3B).
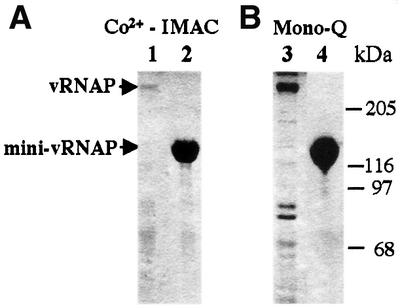
Fig. 3. Yield and relative stability of N-terminally His6-tagged recombinant vRNAP and mini-vRNAP. Enzymes were purified from 100 ml cultures and visualized by silver staining (Ausubel et al., 1999). Lanes 1 and 3, full-length vRNAP; lanes 2 and 4, mini-vRNAP. (A) Metal affinity chromatography of recombinant proteins; 1% of the total yield of each protein is shown. (B) Concentration of affinity-purified proteins. Ten percent of the total yield of vRNAP and 3% of the total yield of mini-vRNAP are shown. Sizes and positions of protein standards are indicated.
The transcription initiation properties of virion-purified vRNAP and recombinant mini-vRNAP were compared by catalytic autolabeling using similar molar concentrations of protein (Figure 4A, left). Reactions were carried out using two different combinations of template and derivatized NTP. A wild-type promoter-containing template (+1C) was used to assay incorporation of the GTP derivative as the initiating nucleotide, while one with a T at position +1 (+1T) was used to examine incorporation of ATP or AMP derivatives as the initiating nucleotide (Figure 4A, right). Mini-vRNAP is 5- to 10-fold more active than full-length vRNAP in phosphodiester bond formation by this assay (lanes 3 and 7). Both proteins preferentially initiate transcription with GTP (compare lanes 3, 5 and 6 and lanes 7, 9 and 10). In addition, both proteins discriminate against incorporation of deoxynucleotides to a similar extent (compare lanes 3 and 4 with lanes 7 and 8).
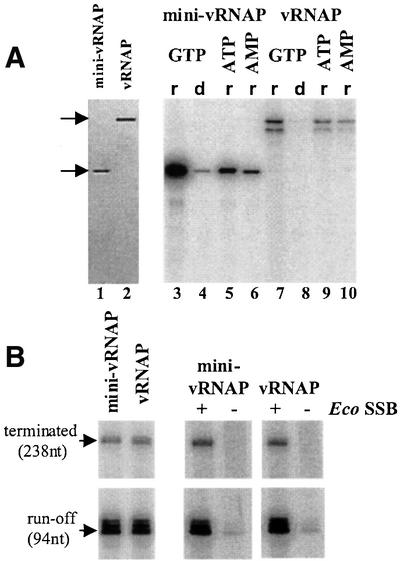
Fig. 4. Transcriptional properties of virion-purified vRNAP and affinity-purified recombinant mini-vRNAP. (A) Activity of vRNAP and mini-vRNAP enzymes in transcription initiation. Left: mini-vRNAP and vRNAP preparations used in catalytic autolabeling and in vitro transcription reactions (∼1 pmol visualized by silver staining). Right: transcription initiation properties of the mini-vRNAP and vRNAP enzymes determined by catalytic autolabeling. All reactions contained 1 µM each of enzyme and template and 1 mM 4-hydroxybenzaldehyde derivative of the initiating nucleotide. Reactions were carried out using derivatized GTP, the +1C template and [α-32P]rATP (r) or [α-32P]dATP (d), or derivatized ATP or AMP, the +1T template and [α-32P]rGTP. (B) Activity of vRNAP and mini-vRNAP enzymes in transcription elongation. Left: transcription at template excess conditions (10 nM of enzyme and 50 nM of pED7). Right: transcription at limiting template conditions (10 nM of enzyme and 1 nM of pED7). EcoSSB (Pharmacia) was added to a concentration of 1 µM. Lengths of the transcription products are indicated.
We next compared the ability of each enzyme to synthesize a 98 nucleotide run-off product and a 238 nucleotide product terminating at an N4 terminator (Figure 4B). Mini-vRNAP produces the same amount of run-off and terminated transcripts as vRNAP, indicating that both enzymes have similar elongation and termination properties (Figure 4B, left).
We previously showed that the product of vRNAP transcription on single-stranded templates is not displaced, resulting in formation of an extended RNA–DNA hybrid that limits synthesis to a single round (Falco et al., 1978). Addition of Escherichia coli single-stranded DNA- binding protein (EcoSSB) under these conditions activates vRNAP transcription by recycling of the template (Markiewicz et al., 1992; E.Davydova, unpublished data). EcoSSB activates transcription by vRNAP and mini-vRNAP to the same extent (Figure 4B, right panels).
These results indicate that mini-vRNAP and full-length vRNAP possess the same initiation, elongation and termination properties, as well as the need for EcoSSB-mediated product displacement.
Function of motifs TxxGR and B
T7-like RNAPs and Pol I DNAPs share a common overall structure often compared with a cupped right hand (Sousa et al., 1993; Joyce and Steitz, 1994). Two motifs present in these enzymes (T/DxxGR and motif B) were identified in mini-vRNAP.
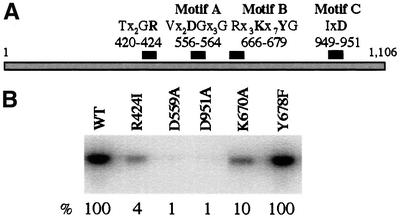
Escherichia coli Klenow fragment (KF) containing the R668A substitution in the TxxGR motif displayed a 300-fold reduction in Kcat value, presumably due to reduction in DNA binding affinity (Polesky et al., 1992). Structural studies of DNAPs from phage T7 (Doublie et al., 1998) and Bacillus stearothermophilus (Kiefer et al., 1998) indicate that the residue equivalent to KF R668 interacts with the terminal base on the primer strand. Substitutions of the corresponding arginine in T7 RNAP (R425) suggest that this motif plays a role in stabilizing the RNA–DNA hybrid during early stages of transcription (Imburgio et al., 2002). Substitution of arginine with isoleucine (R424I) within the mini-vRNAP motif resulted in severe reduction in transcription initiation as measured by catalytic autolabeling (Figure 5B). Substitution of the motif’s threonine with an aspartate characteristic of RNAPs resulted in a slight (10%) decrease in activity as measured by catalytic autolabeling and run-off transcription (T420D, not shown).
Fig. 5. Activity of N4 mini-vRNAP enzymes containing single amino acid substitutions in predicted motifs. (A) Location of the four polymerase motifs within mini-vRNAP. Mutant enzymes containing single amino acid substitutions of conserved residues (in bold) were constructed. (B) Phosphodiester bond formation by affinity-purified mutant mini-vRNAP enzymes as measured by catalytic autolabeling. All mutant enzymes were stable and synthesized at wild-type levels. Reactions contained 1 µM purified enzyme and +1C template, 1 mM derivatized GTP and [α-32P]ATP. The activity of each enzyme relative to wild-type is listed below the autoradiograph.
In DNA-dependent RNAPs and Pol I DNAPs, motif B has the sequence Rx3Kx7YG (Joyce and Steitz, 1994). Biochemical, genetic and structural evidence indicates that the lysine in T7 RNAP (K631) interacts with the initiating nucleotide (Maksimova et al., 1991; Osumi-Davis et al., 1992; Woody et al., 1998; Cheetham and Steitz, 1999). Mutation of the corresponding residue in the KF (K758A) resulted in reduced dNTP binding (Pandey et al., 1994; Astatke et al., 1995). The motif B tyrosine plays different roles in RNAPs and DNAPs. The Y639F mutation in T7 RNAP does not affect synthesis in the presence of rNTPs but reduces discrimination against dNTP incorporation 20-fold (Sousa and Padilla, 1995). In contrast, the corresponding residue in Pol I DNAPs is implicated in positioning of the template–primer and formation of a tight nucleotide-binding pocket, which consequently influences enzyme fidelity (Eom et al., 1996; Kiefer et al., 1998; Minnick et al., 1999).
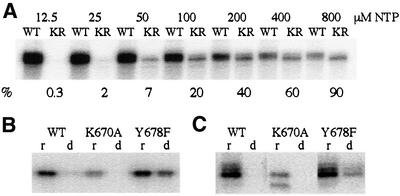
We constructed three mutant mini-vRNAP enzymes containing substitutions in the conserved motif B lysine (K670A and K670R) and tyrosine (Y678F) residues. All mutant enzymes retain wild-type promoter binding affinity (Kd = 0.9–1.4 nM, not shown). The Y678F enzyme is as active as wild-type polymerase in initiating transcription (Figure 5B). In contrast, the K670A (Figure 5B) and K670R (not shown) enzymes display a 10-fold reduction in activity. To explore further the role of residue K670 in enzyme function, transcription elongation reactions were performed in the presence of increasing concentrations of all four NTPs (Figure 6A). The K670R enzyme displays a considerable reduction in affinity for NTPs (Figure 6A). At high NTP concentration (800 µM of each NTP), the K670R and wild-type enzymes display comparable activities, indicating no change in Kcat. Therefore, the mini-vRNAP motif B lysine residue plays a role in NTP binding.
Fig. 6. Transcription properties of wild-type mini-vRNAP and enzymes containing mutations within motif B. (A) Transcription elongation by the wild-type and K670R mutant enzymes as a function of rNTP concentration. Reaction mixtures contained 10 nM purified mini-vRNAP (WT) or K670R mutant enzyme (KR), 2 µM of P2-51 deoxyoligonucleotide template and 0.2 µCi/µl of [α-32P]rATP. The concentration of each of the four rNTPs included in each reaction mix is listed above the autoradiograph. As the concentration of rNTPs increases, the specific activity of [α-32P]rATP decreases from 16 to 0.25 Ci/mmol. The activity of the K670R enzyme relative to wild-type at each rNTP concentration is listed below the autoradiograph. (B) Transcription initiation properties of wild-type, K670A and Y678F enzymes measured by catalytic autolabeling. Reactions contained 1 µM of purified enzyme and +1C template, 1 mM derivatized GTP and either [α-32P]rATP (r) or [α-32P]dATP (d). (C) Transcription elongation properties of the wild-type and mutant mini-vRNAP enzymes. Reactions contained 10 nM of purified enzyme, 1 µM of heat-denatured pED7 DNA template and either [α-32P]rATP (r) or [α-32P]dATP (d).
To explore the role of the Y678 residue in mini-vRNAP, catalytic autolabeling and run-off transcription reactions were carried out in the presence of dNTPs. The wild-type and K670A enzymes display <5% incorporation of dNTP relative to rNTP in phosphodiester bond formation. In contrast, the Y678F enzyme incorporates dNTP at a relative level of 60% (Figure 6B). However, incorporation of dNTPs into long products is not as efficient (Figure 6C), as has been observed for the T7 RNAP Y639F enzyme (Sousa and Padilla, 1995).
These results indicate that mini-vRNAP motifs B and TxxGR are functionally important, and suggest that residues K670 and Y678 play the same roles as the corresponding residues in T7 RNAP.
Identification and function of motifs A and C in mini-vRNAP
Both DNA- and RNA-dependent single-subunit polymerases contain two other short motifs, motifs A and C (Joyce and Steitz, 1994). The sequence of these motifs varies considerably between families; however, each motif contains 1–2 universally conserved carboxylate residues (Poch et al., 1989; Delarue et al., 1990). The positions of the carboxylate residues of motifs A and C are essentially identical in all single-subunit polymerases; the residues lie either within β-strands or in adjacent loop regions (Joyce and Steitz, 1994). Aspartate residues within these motifs bind catalytically essential Mg2+ ions, as suggested by mutagenesis (Polesky et al., 1990; Bonner et al., 1992; Osumi-Davis et al., 1992) and revealed by crystallography (Doublie et al., 1998; Kiefer et al., 1998).
The mini-vRNAP sequence does not contain exact matches to any of the known motif A or C consensus sequences. A 50% match to the T7-like motif A sequence is present at mini-vRNAP amino acids 554–587 (Figure 1). It includes the conserved metal-chelating aspartate and a conserved glycine located five residues downstream. To identify a candidate motif C, we used the PHD program to search the mini-vRNAP sequence for aspartate residues that are predicted to lie within or near β-strands (Rost and Sander, 1993; Rost et al., 1994). Four such residues (D929, D951, D1044 and D1051) lie downstream of motif B. An alignment of the region surrounding D951 to T7-like RNAP motif C sequences is shown in Figure 1.
A number of mini-vRNAP mutant enzymes were constructed to address the potential roles of residues D559 and D951. Substitution of either aspartate residue with alanine (D559A or D951A) resulted in severe deficiency in phosphodiester bond formation (Figure 5B). Both mutant enzymes bind promoter-containing templates with wild-type affinity (not shown), suggesting that these two residues play a role in catalysis. Mutant enzymes in which either aspartate was replaced with glutamate (D559E or D951E) displayed a comparable loss in activity both in vitro (not shown) and in vivo (I.Kaganman, E.K.Davyolova, K.M.Kazmierczak and L.B.Rothman-Denes, unpublished data).
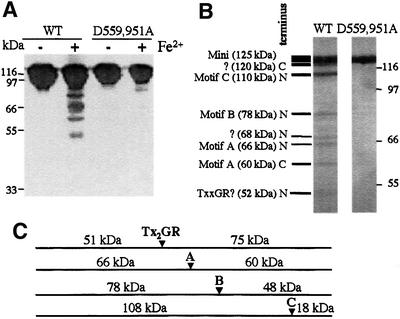
To determine whether residues D559 and D951 play a role in chelating the Mg2+ ions required for catalysis, wild-type mini-vRNAP and an enzyme in which both aspartate residues were replaced by alanine (D559,951A) were subjected to iron-induced localized peptide cleavage (Zaychikov et al., 1996). Upon addition of Fe2+, the wild-type enzyme was cleaved to yield a specific, reproducible pattern of fragments (Figure 7A). In contrast, the D559,951A enzyme was resistant to cleavage, strongly suggesting that these aspartate residues indeed play a role in metal ion binding. To map the cleavage sites, the reaction was carried out on wild-type mini-vRNAP enzymes containing N- or C-terminal His6 tags. Cleavage products were identified by metal affinity chromatography and SDS–PAGE (not shown). Products arising from cleavage in the vicinity of motifs A (Figure 7B, 66 and 60 kDa bands), B (78 kDa band), C (110 kDa band) and most probably TxxGR (52 kDa band) were identified. A fifth cleavage site was localized within the first 100 N-terminal amino acids of mini-vRNAP (120 kDa band).
Fig. 7. Aspartate residues in predicted mini-vRNAP motifs A and C chelate Fe2+. Iron-induced cleavage of wild-type and D559,951A mutant enzymes. Products were visualized by silver staining. (A) Cleavage was carried out for 1 h at room temperature. Products were electrophoresed on 10% SDS–polyacrylamide gels. (B) Cleavage was carried out for 2 h at 23°C. Products were electrophoresed on 6% SDS–polyacrylamide gels. (C) Expected products generated by cleavage of N-terminally His6-tagged mini-vRNAP in the vicinity of motifs A, B, C and TxxGR. Sizes of the expected products are indicated.
Discussion
Sequencing of the bacteriophage N4 vRNAP gene revealed that it encodes a 3500 amino acid polypeptide. We have defined a stable and transcriptionally active 1106 amino acid domain (mini-vRNAP) within the vRNAP polypeptide. It contains four short motifs (T/DxxGR, A, B and C) characteristic of single-subunit DNA-dependent polymerases. N- and C-terminal deletion analysis indicates that this is the shortest polypeptide possessing transcriptional activity (not shown). Mini-vRNAP displays the same transcription initiation, elongation and termination properties as full-length vRNAP. Mutational analysis of residues within the TxxGR and B motifs reveals that they are functionally relevant. Mutational and biochemical analyses indicate that the aspartate residues in motifs A and C are responsible for chelating the catalytic Mg2+ ions. These results strongly suggest that, in spite of a lack of extensive sequence similarity, N4 mini-vRNAP is a member of the single-subunit RNAP family.
Alignment of the mini-vRNAP sequence with 34 other members of the T7 RNAP family allowed us to identify a small number of highly conserved residues, in addition to those previously identified, common to all these enzymes. All enzymes contain aspartate and tyrosine at positions corresponding to T7 RNAP Asp569 and Tyr571; an aliphatic residue at amino acid 444; an arginine or lysine at amino acid 472; a small side chain (glycine or alanine) residue at amino acid 542; a glutamine or asparagine at amino acids 544 and 560; and an aliphatic residue at amino acid 561. A number of these residues lie in close proximity to each other within the T7 RNAP crystal structure (PDB 1QLN). The positions of the conserved side chains suggest that they may affect polymerase activity. However, the mini-vRNAP sequence lacks many conserved residues found in the T7-like RNAPs. All these RNAPs contain the motif sequence DxxGR, while mini-vRNAP possesses the DNA polymerase-like TxxGR motif. In addition, the central histidine residue of motif C (Figure 1) is replaced by a phenylalanine in mini-vRNAP.
Single-subunit RNAPs appear to have evolved from a common ancestor as they share blocks of conserved sequence in addition to the four catalytically important motifs. When analyzed by phylogenetic methods, the RNAP sequences cluster into three separate groups comprised of the mitochondrial and chloroplast enzymes, the linear plasmid-encoded enzymes and the bacteriophage enzymes. However, the order of evolutionary divergence of these enzymes cannot be determined (Cermakian et al., 1997). We generated phylogenetic trees using conserved sequence blocks identified within mini-vRNAP (Figure 8A). Though ours is a shorter alignment than used in similar analyses (Cermakian et al., 1997; Rousvoal et al., 1998), we too observe the three major groups of enzymes (Figure 8B). We see an RNAP presumed to be a prophage remnant found in the Agrobacterium tumefaciens chromosome clustering with the phage enzymes (Wood et al., 2001). Surprisingly, mini-vRNAP, a second N4-encoded RNAP (N4 RNAP II) (Willis et al., 2002) and a cyanophage enzyme fall within the cluster of linear plasmid-encoded enzymes. BLAST analysis suggests that N4 RNAP II is closely related to these enzymes (Willis et al., 2002). In contrast, mini-vRNAP displays 15–21% sequence identity to the various plasmid-encoded enzymes, 16–20% identity to the mitochondrial enzymes and 18–21% identity to the phage enzymes in the aligned regions analyzed. Therefore, N4 vRNAP’s exact evolutionary relationship to other RNAPs cannot be determined unambiguously, and its position in this phylogenetic tree is very probably due to a long branch attraction artefact (Felsenstein, 1978). We believe that N4 vRNAP is the most highly divergent member of this family of enzymes; confirmation of this evolutionary relationship rests on elucidation of the mini-vRNAP crystal structure. We have been unable to detect any additional degree of relatedness between mini-vRNAP and N4 RNAP II. These two enzymes were either acquired by the N4 phage at different times or were subjected to very different selective pressures.
Fig. 8. Phylogenetic analysis. (A) Comparison of mini-vRNAP and T7 RNAP. Locations and sizes of conserved sequence blocks are schematized. Numbers indicate the known boundaries of structural domains in T7 RNAP (Jeruzalmi and Steitz, 1998) and inferred boundaries in N4 mini-vRNAP. The locations of polymerase motifs and catalytically important residues within them are indicated. Striped blocks could not be identified unambiguously in the mini-vRNAP sequence. (B) Phylogenetic relationship of T7-like RNAP enzymes. The tree was generated using conserved sequence blocks common to all single-subunit RNAPs (A). Puzzling support values are shown at nodes when >75% (10 000 puzzling steps were performed). Abbreviations are as in Figure 1, with additions: A.tum, Agrobacterium tumefaciens; B.vul, Beta vulgaris; CyanoP P60, cyanophage P60; D.disc, Dictyostelium discoideum; F.vel, Flammulina velutipes; N.sylv, Nicotiana sylvestris; P.falc, Plasmodium falciparum; P.litt, Pylaiella littoralis; P.ost, Pleurotus ostreatus; P.pat, Physcomitrella patens; P.poly, Physarum polycephalum; T.aest, Triticum aestivum; T.bruc, Trypanosoma brucei. (C) Phylogenetic relationship of T7-like RNA polymerases and Pol I DNA polymerases. Puzzling support values are shown at nodes when >80% (25 000 puzzling steps were performed). Abbreviations as in (B), with additions: A.aeol, Aquifex aeolicus; A.calc, Acinetobacter calcoaceticus; B.subt, Bacillus subtilis; B.stear, Bacillus stearothermophilus; BPSPO1, bacteriophage SPO1; B.burg, Borrelia burgdorferi; C.cres, Caulobacter crescentus; C.jej, Campylobacter jejuni; C.pneu, Chlamydophila pneumoniae; D.rad, Deinococcus radiodurans; E.co KF, E.coli Klenow fragment; H.inf, Haemophilus influenzae; H.pyl, Helicobacter pylori; L.lact, Lactococcus lactis; M.tub, Mycobacterium tuberculosis; MycoBPD29, mycobacterium phage D29; N.men, Neisseria meningitidis; P.aer, Pseudomonas aeruginosa; R.leg, Rhizobium leguminosarum; R.rick, Rickettsia rickettsii; S.aur, Staphylococcus aureus; S.pyog, Streptococcus pyogenes; S.coel, Streptomyces coelicolor; Synech, Synechocystis sp PCC6803; T.aq, Thermus aquaticus; T.mar, Thermotoga maritima; T.pal, Treponema pallidum; V.chol, Vibrio cholerae; X.fast, Xylella fastidiosa.
RNAPs are believed to have evolved from either a DNAP or a reverse transcriptase based on these enzymes’ conserved structures and similar motif residues (Delarue et al., 1990; Joyce and Steitz, 1994; Cermakian et al., 1997). Because of its degree of divergence, we were interested in mini-vRNAP’s relationship to DNAPs. We therefore performed phylogenetic analysis of sequence regions encompassing the four polymerase motifs common to both T7-like RNAP and Pol I DNAP families. In our trees, RNAP and DNAP sequences cluster in two separate, distinct groups. The mini-vRNAP sequence falls on the branch connecting the two groups (Figure 8C). However, when we changed the mini-vRNAP TxxGR sequence to DxxGR and repeated the analysis, mini-vRNAP fell within the RNAP sequence cluster. Therefore, we do not believe that N4 vRNAP is an evolutionary intermediate between the two groups of enzymes.
How is the vRNAP sequence able to tolerate so many changes? Recent evidence suggests that some highly conserved polymerase motifs are in fact plastic. Random mutagenesis of motif A in Taq DNAP I and Klenow fragment revealed that all residues except for the catalytically essential aspartate could be changed to yield enzymes with near wild-type activity (Patel and Loeb, 2000; Shinkai et al., 2001). Patel and Loeb propose that such tolerance of mutation provides an evolutionary advantage and allows for the occurrence of ‘beneficial mutants’ under adverse conditions. They further suggest that the wild-type sequence could be restored by horizontal transfer. Analysis of bacteriophage genomes suggests that horizontal transfer played a significant role in phage evolution (Hendrix et al., 1999). Why then was the vRNAP domain’s sequence not restored to wild-type after divergence? It may be that any recombination event between the vRNAP and N4 RNAP II sequences or the sequence of a co-infecting phage could impair the function of N- and C-terminal vRNAP domains, thereby affecting phage viability. We have found recently that the vRNAP C-terminus comprises a stable domain that is required for polymerase encapsidation, while the N-terminal domain is required for genome injection (I.Kaganman, E.K.Davyolova, K.M.Kazmierczak and L.B.Rothman-Denes, unpublished data). It is also possible that the location of the mini-vRNAP domain within a multifunctional polypeptide allowed it to be maintained in the N4 genome while it acquired its unique properties (Murzin, 1998).
Materials and methods
Bacteria and phage
The E.coli K12 strain W3350 (F– cour r+m+ gal– lac–) was used to prepare wild-type N4 phage as described (Falco et al., 1980). The E.coli strain BL21 (F– ompT hsdSB rB–mB–) was used to produce recombinant proteins for purification.
Sequencing of the vRNAP gene and sequence analysis
N4 genomic fragments encompassing the vRNAP gene were subcloned into M13 replicative factor (RF) DNA. The complete manual sequence for both DNA strands was generated using Sequenase T7 DNAP (v.2.0, United States Biochemicals). The vRNAP gene was sequenced independently as part of the N4 genome (Pittsburgh Bacteriophage Institute, unpublished data).
Profiles corresponding to conserved domains from the T7-like RNAPs (McAllister and Raskin, 1993) and those found in the β and β′ subunits of multisubunit polymerases were constructed using the Wisconsin GCG software package, v.8. The β and β′ subunit sequence alignments were compiled manually from available sources (Patel and Pickup, 1989; Klenk and Zillig, 1994; Sonntag and Darai, 1996; unpublished β-subunit alignment was provided by Dr B.Hall, University of Washington).
T7-like RNAP alignments used for phylogenetic analysis were generated using ClustalX (Higgins et al., 1996) and refined manually. Domains were defined approximately as in Li et al. (2001). The alignments used are available upon request. Trees were generated using the quartet maximum-likelihood method (Strimmer and von Haeseler, 1996) as implemented by Tree-Puzzle 5.0 (Schmidt et al., 2002). Puzzling was performed using the BLOSUM62 matrix (Henikoff and Henikoff, 1992). Trees were drawn using DrawTree (PHYLIP package, Felsenstein, 1993). Sequences of conserved domains corresponding to T7 RNAP amino acids 413–449, 470–492, 506–527, 528–553, 554–564, 568–583, 621–654, 725–739, 740–743, 776–785, 786–799, 806–838 and 839–855 were joined to yield an alignment 246 positions long. For analysis of RNAP and DNAP sequences (58 sequences, 135 positions long), domains corresponding to T7 RNAP amino acids 413–449, 529–561, 624–651 and 806–837, and E.coli Klenow fragment amino acids 333–368, 374–407, 428–455 and 553–586 were used.
Purification of N4 vRNAP from virions
vRNAP was purified by the method of Falco et al. (1980) with the following modifications. The disrupted phage solution was loaded onto a Macro-Prep ceramic HAP resin (Bio-Rad) column equilibrated with 4 M guanidine-HCl, 10 mM potassium phosphate buffer pH 7; vRNAP was recovered in the flow-through fraction. Subsequent purification steps were as described (Falco et al., 1980) except that POROS-HE1 and POROS-50 DEAE resin (PerSeptive Biosystems) columns replaced the original heparin–Sepharose 4B and DEAE–Sephadex columns, respectively. Purified vRNAP was stored in aliquots at –80°C.
Polymerase activity assay
Transcription reaction mixtures (5–10 µl) contained 10 mM Tris–HCl pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol (DTT), 1 mM each ATP, CTP and GTP, 0.1 mM UTP, 0.5–1 µCi of [α-32P]UTP (3000 Ci/mmol, Amersham-Pharmacia) and 10–100 nM enzyme. Enzyme and template concentrations are specified in the figure legends. Mixtures were incubated at 37°C for 5 min, terminated by addition of 7 µl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF), and followed by electrophoresis on 6–10% polyacrylamide/8 M urea gels. Reaction products were quantitated by phosphoimaging.
Transcription template pED7 is a pET11d derivative containing N4 early promoter Pe2. An XbaI cleavage site and N4 transcription terminator t2 are located 94 and 238 bp downstream, respectively, of the transcription start site. Heat-denatured pED7 DNA was used to examine transcription termination. Heat-denatured, XbaI-linearized pED7 DNA or the P2-51 deoxyoligonucleotide (TCCCAGACAAAAGGTTAAGATTTCATACAGGATTGGATGCATTACTTCATCCAAAAGAAGCGGAGCTTCTTCTCTTTAAAG) were used as templates in run-off transcription reactions.
Proteolysis of vRNAP
Reaction mixtures (100 µl) containing 50 µg of vRNAP purified from virions, 0.5 µg of modified trypsin (Promega), 20 mM Tris–HCl pH 8, 200 mM NaCl were incubated at 37°C. Aliquots (10 µl) were removed at the indicated times and incubated on ice for 1 h after addition of Complete protease inhibitor (Roche) to 5× concentration. After trypsin inactivation, samples were autolabeled catalytically using the +1C deoxyoligonucleotide and GTP derivative.
Samples for N-terminal peptide sequencing were prepared by adding 25 µl of 5× loading buffer (250 mM Tris–HCl pH 6.8, 500 mM DTT, 10% SDS, 0.5% bromophenol blue, 50% glycerol) to the 100 µl reaction mixture after 10 min of incubation. After electrophoresis on an 8% polyacrylamide gel, peptides were electroblotted to Immobilon-PSQ membrane (PVDF, Millipore) using transfer conditions described in Matsudaira (1987). Peptide sequencing was performed by Dr Carol Beach (University of Kentucky).
Catalytic autolabeling assay
The promoter-containing deoxyoligonucleotides +1C (ATCCAAAAGAAGCGGAGCTTC) or +1T (ATCTAAAAGAAGCGGAGCTTC) were used as templates. The underlined bases indicate the transcription start site.
The assay was performed as described (Grachev et al., 1987; Hartmann et al., 1988) with modifications. The reaction (5 µl) contained 10 mM Tris–HCl pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 1 µM promoter oligonucleotide, 1 mM 4-hydroxybenzaldehyde derivative of the initiating nucleotide, and polymerase, and was incubated at 37°C for 15 min. After NaBH4 addition to 10 mM final concentration, the mixture was incubated at room temperature for 10 min. The next templated [α-32P]NTP or dNTP (5 µCi, 3000 Ci/mmol, Amersham-Pharmacia) was added to the reaction, which was incubated a further 15 min at 37°C. Reactions were terminated by adding 6 µl of 2× loading buffer; 5 µl aliquots were analyzed on 10% polyacrylamide gels. Reaction products were quantitated by phosphoimaging.
Cloning of the full-length vRNAP gene and mini-vRNAP
DNA fragments used for cloning were generated by PCR amplification using N4 genomic DNA as template. An amplicon encoding vRNAP was synthesized using TaKaRa Ex Taq polymerase (Oncor) according to the manufacturer’s recommendations. (primers: CGACGCAGATCTATGTCAGTATTTGATAGACTGGC; and GCGCGGAATTCTAGATCTTATTAGTGAGTAAGGTTG). An amplicon encoding mini-vRNAP was synthesized using Pfu DNA polymerase (Stratagene) according to the manufacturer’s recommendations (primers: CGACGCAGATCTGAAAGTACAGTTACAGAAG; and GCAGGAGATCTTACCTAGCCTTAGCGACTTTTTG).
The vRNAP amplicon was cloned into the BglII site of expression plasmid pBAD/His B (Invitrogen) to generate plasmid pKMK7. The mini-vRNAP amplicon was cloned into the BglII site of pKMK18 (pBAD/His B with PflMI, EcoRV, BstBI and HindIII sites removed) to generate pKMK22. The recombinant proteins contain a 37 amino acid vector-encoded, N-terminal leader sequence (MGGSHHHHHHGMASMTGGQQMGRDLYDDDDKDPSSRS). Numbering of vRNAP and mini-vRNAP residues in the text does not include the leader sequence.
To clone the mini-vRNAP gene with a C-terminal His6 tag, DNA corresponding to vRNAP amino acids 998–2102 was synthesized using Pfu DNA polymerase (primers: CGACGCAGATCTGAAAGTACAGTTACAGAAG; and GACTACGTAGGTACCAGCCTTAGCGACTTTTTG). This amplicon encodes mini-vRNAP minus the terminal arginine residue and was cloned into the BglII and KpnI sites of pBAD/Myc-HisC (Invitrogen) to generate plasmid pKMK25. The encoded polypeptide contains a seven amino acid N-terminal vector-encoded sequence (MDPSSRS) and a 31 amino acid C-terminal sequence (GTIWEFEAYVEQKLISEEDLNSAVDHHHHHH). The sequence of the vRNAP gene and N- and C-terminally His6-tagged mini-vRNAP constructs are found in DDBJ/EMBL/GenBank under accession numbers AY128691, AY050713 and AY128690, respectively.
Site-directed mutagenesis of mini-vRNAP
Site-directed mutagenesis was carried out on plasmid pKMK22 using the Transformer Site-Directed Mutagenesis Kit, 2nd version (Clontech). Mutant mini-vRNAP enzymes contain the following DNA sequence changes: T420D, ACC→GAT (pKMK41); R424I, CGT→ATT (pKMK42); D559A, GAT→GCG (pKMK44); D559E, GAT→GAA (pKMK70); K670A, AAG→GCG (pKMK29); K670R, AAG→CGC (pKMK36); Y678F, TAT→TTC (pKMK30); D951A, GAT→GCG (pKMK45); D951E, GAT→GAA (pKMK71). The D559A,951A enzyme is encoded by pKMK46. The sequence of all polymerase expression constructs was confirmed.
Expression and purification of His6-tagged recombinant proteins
Escherichia coli BL21 cells bearing pKMK7 were grown at 37°C to OD600 = 0.5 in 1 l of M9 minimal salts medium (Miller, 1972) supplemented with 0.2% casamino acids (Difco), 1 mM MgSO4, 0.1 mM CaCl2, 10 µg/ml thiamine, 4% Lenox L broth (LB), 100 µg/ml ampicillin and 0.4% glucose; cells bearing pKMK22 or mutant mini-vRNAP-expressing plasmids were grown in LB medium. After centrifugation, cell pellets were resuspended in supplemented M9 medium containing 0.2% arabinose and grown for 1 h at 37°C. After low speed centrifugation, pelleted cells were resuspended in sonication buffer [20 mM Tris–HCl pH 8.0, 20 mM NaCl, 1× Complete protease inhibitor, EDTA-free (Roche)] and sonicated in pulses on ice. After low speed centrifugation, the cleared lysate was applied to a Talon Co2+-IMAC resin column (Clontech) equilibrated in sonication buffer. The column was washed with 20 mM Tris–HCl pH 8.0, 1 M NaCl, followed by cold sonication buffer. Protein was eluted with 20 mM Tris–HCl pH 8.0, 20 mM NaCl, 60 mM imidazole, and concentrated on HiTrap Q (Amersham-Pharmacia). Eluted protein was diluted 1:1 (v/v) in glycerol and stored at –20°C.
Iron-induced protein cleavage
Reaction mixtures (5 µl) contained 10 µg of N- or C-His6-tagged wild-type or D559,951A mutant mini-vRNAP enzymes, 50 µM Fe(NH4)2(SO4)2, 20 mM HEPES pH 7.6, 50 mM NaCl and 5 mM DTT. Incubation and electrophoresis conditions are described in Figure 7. Metal affinity chromatography of cleavage products prior to SDS–PAGE allowed identification of N- or C-terminally derived fragments.
Acknowledgments
Acknowledgements
We thank Dr M.Long for advice on phylogenetic analysis, Dr W.McAllister for encouraging us to explore the evolutionary relationship between mini-vRNAP and DNA polymerases, and Drs R.Hendrix, G.Hatfull, J.Lawrence and members of the Pittsburgh Bacteriophage Institute for the sequence of the C-terminal 800 nucleotides of the vRNAP gene. This work was supported by NIH Grant AI 12575 to L.B.R.-D. K.M.K. was partially supported by United States Public Health Service Grant T32 GM07197. A.A.M. acknowledges the support of NIH grant GM49242.
References
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astatke M., Grindley,N.D.F. and Joyce,C.M. (1995) Deoxynucleoside triphosphate and pyrophosphate binding sites in the catalytically competent ternary complex for the polymerase reaction catalyzed by DNA polymerase I (Klenow fragment). J. Biol. Chem., 270, 1945–1954. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1999) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY.
- Bonner G., Patra,D., Lafer,E.M. and Sousa,R. (1992) Mutations in T7 RNA polymerase that support the proposal for a common polymerase active site structure. EMBO J., 11, 3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N., Ikeda,T.M., Miramontes,P., Lang,B.F., Gray,M.W. and Cedergren,R. (1997) On the evolution of the single-subunit RNA polymerases. J. Mol. Evol., 45, 671–681. [DOI] [PubMed] [Google Scholar]
- Cheetham G.M.T. and Steitz,T.A. (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science, 286, 2305–2309. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch,O., Tordo,N., Moras,D. and Argos,P. (1990) An attempt to unify the structure of polymerases. Protein Eng., 3, 461–467. [DOI] [PubMed] [Google Scholar]
- Doublie S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- Eom S.H., Wang,J. and Steitz,T.A. (1996) Structure of Taq polymerase with DNA at the polymerase active site. Nature, 382, 278–281. [DOI] [PubMed] [Google Scholar]
- Falco S.C., VanderLaan,K. and Rothman-Denes,L.B. (1977) Virion-associated RNA polymerase required for bacteriophage N4 development. Proc. Natl Acad. Sci. USA, 74, 520–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S.C., Zivin,R. and Rothman-Denes,L.B. (1978) Novel template requirements of N4 virion RNA polymerase. Proc. Natl Acad. Sci. USA, 75, 3220–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S.C., Zehring,W. and Rothman-Denes,L.B. (1980) DNA-dependent RNA polymerase from bacteriophage N4 virions. J. Biol. Chem., 255, 4339–4347. [PubMed] [Google Scholar]
- Felsenstein J. (1978) Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool., 27, 401–410. [Google Scholar]
- Felsenstein J. (1993) PHYLIP (Phylogeny Inference Package) Version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle.
- Glucksmann M.A., Markiewicz,P., Malone,C. and Rothman-Denes,L.B. (1992) Specific sequences and a hairpin structure in the template strand are required for N4 virion RNA polymerase promoter recognition. Cell, 70, 491–500. [DOI] [PubMed] [Google Scholar]
- Grachev M., Kolocheva,T., Lukhtanov,E. and Mustaev,A. (1987) Studies on the functional topography of E.coli RNA polymerase. Eur. J. Biochem., 163, 113–121. [DOI] [PubMed] [Google Scholar]
- Gribskov M. and Veretnik,S. (1996) Identification of sequence patterns with profile analysis. Methods Enzymol., 266, 198–212, [DOI] [PubMed] [Google Scholar]
- Hartmann G.R. et al. (1988) Initiation of transcription—a general tool for affinity labeling of RNA polymerases by autocatalysis. Biol. Chem. Hoppe-Seyler, 369, 775–788. [PubMed] [Google Scholar]
- Haynes L.L. and Rothman-Denes,L.B. (1985) N4 virion RNA polymerase sites of transcription initiation. Cell, 41, 597–605. [DOI] [PubMed] [Google Scholar]
- Hendrix R.W., Smith,M.C.M., Burns,R.N., Ford,M.E. and Hatfull,G.F. (1999) Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc. Natl Acad. Sci. USA, 96, 2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- Imburgio D., Anikin,M. and McAllister,W.T. (2002) A conserved DX2GR motif in T7 RNA polymerase stabilizes the RNA:DNA hybrid during transcription initiation. J. Mol. Biol., 319, 37–51. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D. and Steitz,T.A. (1998) Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J., 17, 4101–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C.M. and Steitz,T.A. (1994) Function and structure relationships in DNA polymerases. Annu. Rev. Biochem., 63, 777–822. [DOI] [PubMed] [Google Scholar]
- Kiefer J.R., Mao,C., Braman,J.C. and Beese,L.S. (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature, 391, 304–307. [DOI] [PubMed] [Google Scholar]
- Klenk H.-P. and Zillig,W. (1994) DNA-dependent RNA polymerase subunit B as a tool for phylogenetic reconstructions: branching topology of the Archaeal domain. J. Mol. Evol., 38, 420–432. [DOI] [PubMed] [Google Scholar]
- Li J., Maga,J.A., Cermakian,N., Cedergren,R. and Feagin,J.E. (2001) Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol. Biochem. Parasitol., 113, 261–269. [DOI] [PubMed] [Google Scholar]
- Maksimova T.G. et al. (1991) Lys631 residue in the active site of the bacteriophage T7 RNA polymerase. Eur. J. Biochem., 195, 841–847. [DOI] [PubMed] [Google Scholar]
- Markiewicz P., Malone,C., Chase,J.W. and Rothman-Denes,L.B. (1992) E.coli single-stranded DNA binding protein is a supercoiled-dependent transcriptional activator of N4 virion RNA polymerase. Genes Dev., 6, 2010–2019. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem., 262, 10035–10038. [PubMed] [Google Scholar]
- McAllister W.T. and Raskin,C.A. (1993) The phage RNA polymerases are related to DNA polymerases and reverse transcriptases. Mol. Microbiol., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Mendez J., Blanco,L., Lazaro,J.M. and Salas,M. (1994) Primer-terminus stabilization at the φ29 DNA polymerase active site. J. Biol. Chem., 269, 30030–30038. [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Minnick D.T., Bebenek,K., Osheroff,W.P., Turner,R.M.J., Astatke,M., Liu,L., Kunkel,T.A. and Joyce,C.M. (1999) Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment). J. Biol. Chem., 274, 3067–3075. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. (1998) How far divergent evolution goes in proteins. Curr. Opin. Struct. Biol., 8, 380–387. [DOI] [PubMed] [Google Scholar]
- Mustaev A., Kozlov,M., Markovtsov,V., Zaychikov,E., Denissova,L. and Goldfarb,A. (1997) Modular organization of the catalytic center of RNA polymerase. Proc. Natl Acad. Sci. USA, 94, 6641–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Davis P.A., de Aguilera,M.C., Woody,R.W. and Woody,A.-Y.M. (1992) Asp537, Asp812 are essential and Lys631, His811 are catalytically significant in bacteriophage T7 RNA polymerase activity. J. Mol. Biol., 226, 37–45. [DOI] [PubMed] [Google Scholar]
- Pandey V.N., Kaushik,N. and Modak,M.J. (1994) Role of lysine 758 of Escherichia coli DNA polymerase I as assessed by site-directed mutagenesis. J. Biol. Chem., 269, 13259–13265. [PubMed] [Google Scholar]
- Patel D.D. and Pickup,D.J. (1989) The second-largest subunit of the poxvirus RNA polymerase is similar to the corresponding subunits of procaryotic and eucaryotic RNA polymerases. J. Virol., 63, 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P.H. and Loeb,L.A. (2000) DNA polymerase active site is highly mutable: evolutionary consequences. Proc. Natl Acad. Sci. USA, 97, 5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Sauvaget,I., Delarue,M. and Tordo,N. (1989) Identification of four conserved motifs among RNA-dependent polymerase encoding elements. EMBO J., 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesky A.H., Steitz,T.A., Grindley,N.D.F. and Joyce,C.M. (1990) Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J. Biol. Chem., 265, 14579–14591. [PubMed] [Google Scholar]
- Polesky A.H., Dahlberg,M.E., Benkovic,S.J., Grindley,N.D.F. and Joyce,C.M. (1992) Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J. Biol. Chem., 267, 8417–8428. [PubMed] [Google Scholar]
- Ritz D. and Beckwith,J. (2001) Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol., 55, 21–48. [DOI] [PubMed] [Google Scholar]
- Rost B. and Sander,C. (1993) Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl Acad. Sci. USA, 90, 7558–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander,C. and Schneider,R. (1994) PHD—an automatic mail server for protein secondary structure. CABIOS, 10, 53–60. [DOI] [PubMed] [Google Scholar]
- Rothman-Denes L.B. and Schito,G.C. (1974) Novel transcribing activities in N4-infected Escherichia coli. Virology, 60, 65–72. [DOI] [PubMed] [Google Scholar]
- Rousvoal S., Oudot,M.-P., Fontaine,J.-M., Kloareg,B. and Loiseaux-de Goer,S. (1998) Witnessing the evolution of transcription in mitochondria: the mitochondrial genome of the primitive brown alga Pylaiella littoralis (L.) Kjellm. encodes a T7-like RNA polymerase. J. Mol. Biol., 277, 1047–1057. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer,K., Vingron,M. and von Haeseler,A. (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics, 18, 502–504. [DOI] [PubMed] [Google Scholar]
- Severinov K. (2000) RNA polymerase structure–function: insights into points of transcriptional regulation. Curr. Opin. Microbiol., 3, 118–125. [DOI] [PubMed] [Google Scholar]
- Shinkai A., Patel,P.H. and Loeb,L.A. (2001) The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J. Biol. Chem., 276, 18836–18842 [DOI] [PubMed] [Google Scholar]
- Sonntag K.-C. and Darai,G. (1996) Evolution of viral DNA-dependent RNA polymerases. Virus Genes, 11, 271–284. [DOI] [PubMed] [Google Scholar]
- Sousa R. and Padilla,R. (1995) A mutant T7 RNA polymerase as a DNA polymerase. EMBO J., 14, 4609–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R., Chung,Y.J., Rose,J.P. and Wang,B.-C. (1993) Crystal structure of bacteriophage T7 RNA polymerase at 3.3 Å resolution. Nature, 364, 593–599. [DOI] [PubMed] [Google Scholar]
- Strimmer K. and von Haeseler,A. (1996) Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol., 13, 964–969. [Google Scholar]
- Willis S.H., Kazmierczak,K.M., Carter,R.H. and Rothman-Denes,L.B. (2002) N4 RNA polymerase II, a heterodimeric RNA polymerase with homology to the single-subunit family of RNA polymerases. J. Bacteriol., 184, 4952–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.W. et al. (2001) The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science, 294, 2317–2323. [DOI] [PubMed] [Google Scholar]
- Woody A.-Y.M., Osumi-Davis,P.A., Hiremath,M.M. and Woody,R.W. (1998) Pre-steady-state and steady-state kinetic studies on transcription initiation catalyzed by T7 RNA polymerase and its active-site mutants K631R and Y639F. Biochemistry, 37, 15958–15964. [DOI] [PubMed] [Google Scholar]
- Zakharova N., Hoffman,P.S., Berg,D.E. and Severinov,K. (1998) The largest subunits of RNA polymerase from gastric Helicobacters are tethered. J. Biol. Chem., 273, 19371–19374. [DOI] [PubMed] [Google Scholar]
- Zaychikov E. et al. (1996) Mapping of catalytic residues in the RNA polymerase active center. Science, 273, 107–109. [DOI] [PubMed] [Google Scholar]
- Zhang G., Campbell,E.A., Minakhin,L., Richter,C., Severinov,K. and Darst,S.A. (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell, 98, 811–824. [DOI] [PubMed] [Google Scholar]